Abstract
Background
Autism Spectrum Conditions (ASC) are neurodevelopmental conditions characterized by difficulties in communication and social interaction, alongside unusually repetitive behaviours and narrow interests. Asperger Syndrome (AS) is one subgroup of ASC and differs from classic autism in that in AS there is no language or general cognitive delay. Genetic, epigenetic and environmental factors are implicated in ASC and genes involved in neural connectivity and neurodevelopment are good candidates for studying the susceptibility to ASC. The aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2) gene encodes a transcription factor involved in neurodevelopmental processes, neuronal connectivity and cellular responses to hypoxia. A mutation in this gene has been identified in individuals with ASC and single nucleotide polymorphisms (SNPs) have been nominally associated with AS and autistic traits in previous studies.
Methods
In this study, we tested 34 SNPs in ARNT2 for association with AS in 118 cases and 412 controls of Caucasian origin. P values were adjusted for multiple comparisons, and linkage disequilibrium (LD) among the SNPs analysed was calculated in our sample. Finally, SNP annotation allowed functional and structural analyses of the genetic variants in ARNT2. We tested the replicability of our result using the genome-wide association studies (GWAS) database of the Psychiatric Genomics Consortium (PGC).
Results
We report statistically significant association of rs17225178 with AS. This SNP modifies transcription factor binding sites and regions that regulate the chromatin state in neural cell lines. It is also included in a LD block in our sample, alongside other genetic variants that alter chromatin regulatory regions in neural cells.
Conclusions
These findings demonstrate that rs17225178 in the ARNT2 gene is associated with AS and support previous studies that pointed out an involvement of this gene in the predisposition to ASC.
Keywords: Autism spectrum conditions (ASC), Asperger Syndrome (AS), Aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2), Single nucleotide polymorphisms (SNPs)
Background
Autism Spectrum Conditions (ASC) are a group of neurodevelopmental conditions defined by difficulties in communication and social interaction, alongside unusually repetitive behaviours and narrow interests. Asperger Syndrome (AS) is a subtype of these conditions, where there is no delay in language or cognitive development [1]. The incidence of ASC in the general population is approximately 1 in 100 [2]. Male:female sex ratios of 9:1 and 4:1 are observed in individuals with AS [3] and classic autism [4], respectively, although the true sex ratio in AS may be closer to the sex ratio in classic autism because of under-detection of AS in females [5]. ASC are heritable [6,7] and multifactorial conditions, where genetic, epigenetic and environmental factors are involved [8,9].
ASC are neurodevelopmental in origin, and accordingly, genes involved in neural development and connectivity are good candidate genes for studying the genetics of these conditions [10,11]. The aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2) gene encodes a transcription factor of the basic helix-loop-helix (bHLH)-Per-ARNT-Sim (PAS) family, which is mainly expressed in the kidney and central nervous system [12,13]. ARNT2 dimerizes with the hypoxia-inducible factor-1α (HIF-1α) and regulates cellular responses to hypoxia [14]. It also dimerizes with single-minded homolog 1 (SIM1) and participates in the development of paraventricular and supraoptic nuclei in mice [15]. Variants in the SIM1 gene have been associated with obesity in humans and have been shown to reduce transcriptional activity of SIM1 combined with ARNT2. Interestingly, 11 individuals carrying these variants have reported atypical behaviours, such as ASC [16]. SIM1 controls the expression of methyl CpG binding protein 2 (MECP2) gene, which is disrupted in individuals with Rett Syndrome who display autistic features [17]. Mecp2 mouse models show typical features of Rett Syndrome [18,19], and knockout mice with a deletion of Mecp2 in Sim1-expressing neurons display altered social behaviour [20]. Heterozygous Arnt2 mutant mice lack secretory neurons in paraventricular and supraoptic nuclei, implicating the involvement of ARNT2 in neurodevelopmental processes [21]. Arnt2 and Sim1 are also involved in long-range axonal guidance and hence lay down the connections between the hypothalamus and the spinal cord in zebrafish [22]. ARNT2 is a transcription factor in mouse hippocampus [23] and regulates the brain-derived neurotrophic factor (Bdnf) gene that controls neural activity and is related to several human neurodevelopmental conditions [24]. A recent study by Lin and colleagues proposed a role of ARNT2 in the predisposition to ASC. They tested the levels of expression of candidate genes for ASC in neural aggregates in human stem cell models (comparable to a telencephalon in the early stages of development) after heat shock and they observed an alteration of ARNT2 expression [25]. One study reported a missense mutation in ARNT2 in individuals with ASC [26] and a previous study from our laboratory identified nominal associations between single nucleotide polymorphisms (SNPs) rs3901896 and rs4778599 with AS and autistic traits, respectively [27]. A recent study has reported nominally significant association for SNPs in ARNT2 and SIM1 with autistic traits in a general population sample [28].
In the current study, we tested 34 SNPs in ARNT2 for association with AS in an independent sample, composed of 118 cases and 412 controls. We found a significant association of rs17225178 with AS after correcting for multiple comparisons and a nominal association of rs3848173 with AS. To test the replicability of this observation, we analysed genome-wide association studies (GWAS) data from the Psychiatric Genomics Consortium (PGC), which tested SNPs for association with ASC in case–control and family-based samples [29]. Our study provides further support for a role of ARNT2 in ASC and in particular it points out an involvement of a SNP (that is rs17225178) in the predisposition to AS.
Methods
Participants
Participants were recruited by advertisement from the volunteer database of the Autism Research Centre in Cambridge (United Kingdom). They included both individuals with an AS diagnosis and individuals who were not diagnosed with AS and did not have any close family member with a diagnosis of ASC. All individuals completed the Autism Spectrum Quotient (AQ) online [30] to separate cases and controls from the two extreme ends of the AQ continuum. Eighty percent of individuals with an ASC diagnosis score above 32 on the AQ [31]. Cases were previously diagnosed by independent clinicians in the United Kingdom for having AS using the International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10) [32] or the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) [1] criteria, at recognized clinics in the United Kingdom, by trained psychiatrists or clinical psychologists. A proportion of these were diagnosed at our clinic in Cambridge (the CLASS Clinic). Individuals without an ASC diagnosis and with an AQ score below or equal to 24 were selected as controls. This ensured that individuals with high autistic traits but without a diagnosis of ASC (the so-called “Broader Autism Phenotype” (BAP) [33]) were not included in the study, as this can confound the analysis. The AQ measures autistic traits, has high heritability [34] and has a normal distribution in the general population [30]. Individuals in the general population show a mean AQ score of 16.4 ± 6.3, while individuals with AS have a mean AQ score of 35.8 ± 6.5 [31]. All participants resided in the United Kingdom when the study was conducted and reported Caucasian ancestry for at least three generations. Overall, we tested 530 individuals, among them 118 cases (74 males and 44 females) with a mean AQ score of 35.6 ± 8.9 (males mean: 35.1 ± 8.7, females mean: 36.6 ± 8.8) and 412 controls (185 males and 227 females) with a mean AQ score of 14.9 ± 5.0 (males mean: 16.0 ± 4.4, females mean: 13.9 ± 5.1) (Table 1).
Table 1.
Schematic description of the participants enrolled in the current study
| Status of the participants | Total number | Number of males | Number of females | Total AQ score | AQ score in males | AQ score in females |
|---|---|---|---|---|---|---|
| Cases | 118 | 74 | 44 | 35.6 ± 8.9 | 35.1 ± 8.7 | 36.6 ± 8.8 |
| Controls | 412 | 185 | 227 | 14.9 ± 5.0 | 16.0 ± 4.4 | 13.9 ± 5.1 |
Total numbers of participants and total AQ scores for cases and controls are reported. These values stratified by sex are also reported. AQ, Autism Spectrum Quotient.
SNP selection and genotyping
Thirty-four SNPs in the ARNT2 gene were selected from 80704652 bp to 80889940 bp on chromosome 15 with a mean inter-SNP distance of 7 Kb (GRCH37.p10 Primary Assembly, National Center for Biotechnology Information [NCBI]). We did not have a specific hypothesis about any SNP in particular and tested multiple SNPs within the same gene to identify a possible association of the gene with a given phenotype [35]. All chosen SNPs had a minor allele frequency (MAF) > 0.05 in the HapMap CEU (Utah residents with northern and western European ancestry) population (HapMap genome browser, release 27) and were available in the TaqMan SNP Genotyping Assays (Applied Biosystems Inc., Foster City, CA, USA), that was used for genotyping in the current study. rs1446336, rs8034535 and rs7175825 are tag SNPs, as indicated by the HapMap genome browser (release 27). The most upstream SNP is rs1446336 (Chr 15: 80704652) and the most downstream is rs1139650 (Chr 15: 80889940) (see Table 2). Buccal samples were collected by mail from the participants and DNA was extracted and anonymized [36]. SNP genotyping was performed following the protocol previously reported [27]. Hardy-Weinberg Equilibrium (HWE) was tested in controls using Plink v1.07 [37] at α = 0.05, and all selected SNPs were in HWE.
Table 2.
SNPs in ARNT2 analysed in the current study
| SNP ID | Chromosomal position | Major/minor allele | MAF (CEU) |
|---|---|---|---|
| rs1446336 | 80704652 | T/A | 0.407 |
| rs16972073 | 80709303 | G/C | 0.075 |
| rs12594558 | 80709766 | C/T | 0.259 |
| rs1026016 | 80710955 | C/T | 0.153 |
| rs3910982 | 80712122 | C/A | 0.333 |
| rs1020397 | 80718738 | G/C | 0.283 |
| rs4778790 | 80721271 | A/G | 0.389 |
| rs1912 | 80723361 | C/T | 0.138 |
| rs8034535 | 80725974 | A/G | 0.208 |
| rs8036233 | 80729687 | T/A | 0.474 |
| rs3901896 | 80734097 | C/T | 0.394 |
| rs3848173 | 80734588 | C/T | 0.129 |
| rs17788120 | 80742881 | T/C | 0.411 |
| rs17225178 | 80743866 | T/A | 0.081 |
| rs3848175 | 80747103 | G/A | 0.281 |
| rs895444 | 80749761 | A/G | 0.111 |
| rs12591546 | 80754740 | G/T | 0.275 |
| rs4778795 | 80760069 | G/T | 0.226 |
| rs12905523 | 80760688 | T/C | 0.240 |
| rs4778798 | 80770223 | T/C | 0.042 |
| rs7181179 | 80778586 | T/C | 0.009 |
| rs4778599 | 80781763 | G/A | 0.296 |
| rs7175825 | 80795419 | G/C | 0.442 |
| rs11858186 | 80801472 | C/T | 0.491 |
| rs12439920 | 80804741 | G/A | 0.103 |
| rs11072922 | 80806183 | C/T | 0.159 |
| rs4778604 | 80837819 | C/T | 0.492 |
| rs4423382 | 80854992 | A/C | 0.155 |
| rs10851935 | 80859935 | T/C | 0.341 |
| rs7403706 | 80861365 | T/C | 0.168 |
| rs7403013 | 80874261 | G/A | 0.127 |
| rs4072568 | 80884025 | G/A | 0.137 |
| rs6495511 | 80888783 | A/G | 0.243 |
| rs1139650 | 80889940 | A/G | 0.093 |
The table shows SNP ID and chromosomal position of genetic variants analysed in this study. Major/minor allele and MAF values in the CEU population for each SNP are also reported. CEU, Utah residents with northern and western European ancestry; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Statistical analysis
The Chi-Square statistics was applied to test association between 34 SNPs in ARNT2 and AS, using Plink v1.07. We used Bonferroni correction to adjust P values at the α threshold of significance (α = 0.05). Since Bonferroni correction is conservative when the tests are not independent, we used the SNPSpD web interface [38], to account for linkage disequilibrium (LD) patterns between all SNPs in the current sample. The effective number of independent loci was 21 and the subsequent threshold of significance after Bonferroni correction was α = 0.0024. We analysed LD blocks in our sample using Haploview [39].
Functional annotation of significant SNPs was conducted using the following softwares: HaploReg v2 [40] allows analysis of non-coding genetic variants in terms of their effect on the organization of chromatin regions and regulatory motifs; F-SNP [41] is a tool which combines multiple databases for providing the functional roles of genetic variants; the Genetic Association Database [42] gives information about genetic studies which previously identified association between the SNPs we analysed and ASC; SNPnexus [43] is an online database which was used to define regulatory elements and conserved regions; the University of California Santa Cruz (UCSC) genome browser [44] permits to validate functions of the SNPs analysed.
Analysis of replication data
We had access to the summary data of the PGC database and we checked for effect sizes and P values for rs17225178 in the ASC cohort. The PGC analysed genome-wide SNPs for association with ASC in 161 cases, 526 controls, 4,788 trio cases and 4,788 trio pseudocontrols all of Caucasian ancestry [29]. Details of methods, ancestry of the participants and analyses for this cohort are provided in reference [29].
Consent and ethics
Participants gave their informed consent before participating in this study. The current study had ethics approval from the Cambridge Psychology Research Ethics Committee and the NHS Research Ethics Committee (UK) and was conducted in accordance with the principles of the Declaration of Helsinki.
Results
rs17225178 was significantly associated with AS in our sample with P value = 0.001, and rs3848173 was nominally associated with AS in our sample. Minor alleles of these two SNPs are more prevalent in cases than in controls with odds ratio (OR) > 1 and are associated with an increased risk of AS (Table 3). To replicate our association, we searched for the P value and effect direction of rs17225178 in the PGC dataset. rs17225178 was significant (P value = 0.0121), and the effect direction was same between the two datasets (OR > 1) (Table 3).
Table 3.
Results of SNP association analyses in our sample and in the PGC database
| SNP ID | MAF (cases) | MAF (controls) | SNP association analysis in our sample | SNP association analysis in the PGC database | ||||
|---|---|---|---|---|---|---|---|---|
| OR | X 2 | P value | OR | SE | P value | |||
| rs3848173 | 0.2288 | 0.1667 | 1.484 | 4.768 | 0.029 | |||
| rs17225178 | 0.1966 | 0.1173 | 1.842 | 9.773 | 0.001771 | 1.115 | 0.0433 | 0.0121 |
Nominally significant P values after genetic association analysis between 34 SNPs in ARNT2 and AS in our sample are reported (α = 0.05). Significant P value after Bonferroni correction for total number of SNPs is written in bold and italicized (α = 0.0024). Major allele is the reference allele for the estimated OR. Results of genetic association analysis between rs17225178 and ASC in the PGC database are also reported. AS, Asperger Syndrome; ASC, Autism Spectrum Conditions; MAF, minor allele frequency; OR, odds ratio; PGC, Psychiatric Genomics Consortium; SE, standard error for the odds ratio; SNP, single nucleotide polymorphism.
rs17225178 is included in a LD block (25 Kb) in our sample, alongside seven other SNPs (rs3848173, rs8036233, rs3901896, rs17788120, rs3848175, rs895444 and rs12591546) (Figure 1). rs17225178 is located in intron 2 of ARNT2, alongside rs3848175 and rs895444. Intron 1 includes rs8036233, rs3901896, rs3848173 and rs17788120; while rs12591546 is located in intron 3 (Figure 2).
Figure 1.
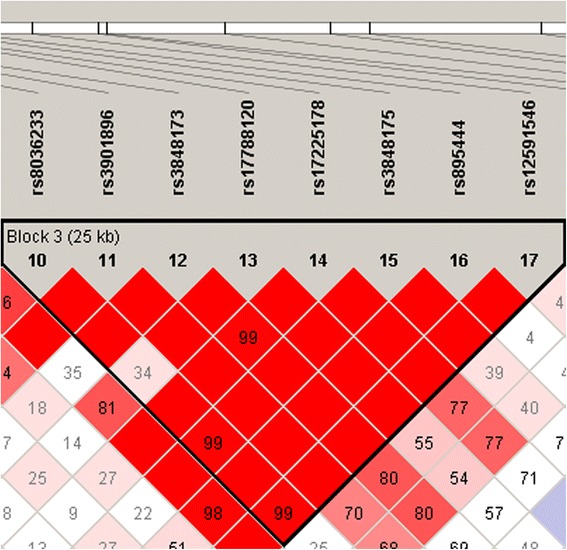
Linkage disequilibrium blocks in the ARNT2 gene calculated in our sample (detail). Block 3 is shown. It includes eight SNPs analysed in the current study. Numbers into the squares indicate D’ values.
Figure 2.
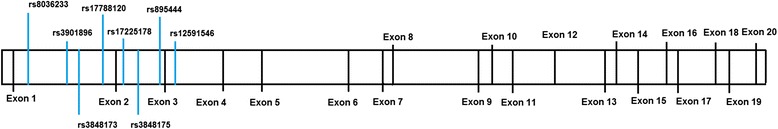
Schematic diagram of the ARNT2 gene. White boxes indicate introns. SNP significantly associated with AS in the current study and those in linkage disequilibrium with this genetic variant in our sample are indicated by blue lines.
Discussion
The aim of this study was to investigate the role of common genetic variants in the predisposition to AS, which is a high-functioning form of ASC. The complex genetic nature of ASC has led previous studies to examine multiple genes, mainly involved in neural development and connectivity, sex-hormones signaling and social-emotional pathways [10,11,27]. In the current study, we tested for a genetic association between 34 SNPs in ARNT2 with AS, and then replicated our significant SNP in a large ASC cohort. Our results show nominal association of rs3848173 with AS and significant association between rs17225178 and AS. We replicated the association of rs17225178 in the PGC ASC cohort, corroborating previous reports that showed an involvement of ARNT2 in ASC. This is consistent with a previous study from our laboratory which indicated nominal association of rs3901896 with AS, and of rs4778599 with autistic traits, measured using the AQ [27]. Recent studies have reported a mutation in the ARNT2 gene in ASC [26] and common variants associated with autistic traits [28].
ARNT2 is located on chromosome 15q24 and includes 20 exons and 19 introns (Figure 2). It spans 193.587 Kb and has eight splice variants. Four transcript variants are protein-coding: one is composed of the first 20 exons and 19 introns; two transcripts include the first 19 exons and 18 introns; one transcript includes the first 6 exons and 5 introns (UCSC genome browser). rs17225178 is located in intron 2 (Figure 2) and modifies transcription factor binding sites (TATA, GATA and TAL1 motifs) and genomic regions that regulate the chromatin state in neural cell lines (hippocampus and brain angular gyrus) (HaploReg v2). SNP annotation did not indicate the presence of miRNA binding sites or other structural variants at the level of this SNP and did not provide information about the effect of rs17225178 on protein structure and function (F-SNP, SNPnexus). We hypothesize that the presence of the risk allele of rs17225178 could influence the binding of different transcription factors or could allow the modification of chromatin states, regulating chromatin accessibility and dynamics. This genetic variant is included in a LD block in our sample, where seven other SNPs (rs8036233, rs3901896, rs3848173, rs17788120, rs3848175, rs895444 and rs12591546) that alter chromatin regulatory regions in neural cell lines are located (HaploReg v2). Two previous studies have reported nominal association of rs3901896 with AS [27] and autistic traits (for example, language impairment) [28]. This genetic variant is in LD with rs17225178 in our sample (Figure 1). An interesting next step will be to perform molecular studies to analyse possible chromatin modifications and binding of specific transcription factors at the level of regulatory regions of ARNT2, carrying the risk allele or LD region.
This is the first study that shows a statistically significant association of rs17225178 with AS and supports previous findings that indicate an involvement of ARNT2 in ASC. This is consistent with the role of ARNT2 in neural development, as is known from animal models. In particular, ARNT2 is involved in the development of oxytocinergic neurons [15]. The oxytocinergic system has been implicated in autism through multiple lines of evidence [45-51]. This result provides further support for the atypical oxytocinergic system in ASC by implicating a gene involved in the development of oxytocinergic neurons. In our study, we find a significant association with rs17225178, which is included in a LD block, alongside rs3901896 that has been nominally associated with AS [27] and autistic traits [28]. In the previous genetic study carried out in our laboratory [27], we did not find association between rs17225178 and AS, as we performed SNP association analysis at the gene level and not for each SNP independently.
The limitation of this study is the small sample size, so further research is required to corroborate these results. Animal models of Rett Syndrome have established a role for Mecp2 [18-20], and it will be important to further test if Arnt2 play an important role in underlying the autistic phenotype. Future work in animals should employ well-designed behavioural assays relevant to autism in Arnt2 knockout mice, such as social-preference paradigms [52]. In humans, larger association studies of ARNT2 SNPs with endophenotypes linked to autism will be particularly relevant.
Conclusions
In the current study, we report a statistically significant association between the genetic variant rs17225178 in ARNT2 and AS. Using the PGC ASC dataset, we were able to replicate our results in a larger cohort. The direction of effect was the same in both the cohorts. Our findings provide evidence for the role of ARNT2 in ASC and show the involvement of rs17225178 in this gene in AS, a subgroup of ASC.
Acknowledgments
We are grateful to Target Autism Genome, the Nancy Lurie Marks (NLM) Family Foundation, the Autism Research Trust, the Medical Research Council UK, the Max Planck Institute for Psycholinguistics and the Wellcome Trust Sanger Centre. BC was funded by the Medical Research Council UK and VW was funded by the Nehru Trust for Cambridge University and the Cambridge Commonwealth Trust. We thank Robert Plomin, Frank Dudbridge, Lindsey Kent and Ian Craig for relevant discussions, and Jon Breidbord, Allen Chan, Sylvia Lakatosova, Sally Wheelwright, Carrie Allison, Uma Mallya, Alex Politt and Leena Peltonen for support at different points of the project. The study was carried out in collaboration with the NIHR CLAHRC EoE.
Abbreviations
- AQ
Autism Spectrum Quotient
- ARNT2
aryl-hydrocarbon receptor nuclear translocator 2
- AS
Asperger Syndrome
- ASC
Autism Spectrum Conditions
- BAP
broader autism phenotype
- BDNF
brain-derived neurotrophic factor
- bHLH-PAS
basic helix-loop-helix-Per-Arnt-Sim
- CEU
Utah residents with northern and western European ancestry
- DSM-IV-TR
Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision
- GWAS
genome-wide association studies
- HIF-1α
hypoxia-inducible factor-1α
- HWE
Hardy-Weinberg Equilibrium
- ICD-10
International Statistical Classification of Diseases and Related Health Problems, Tenth Revision
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MECP2
methyl CpG binding protein 2
- NCBI
National Center for Biotechnology Information
- OR
odds ratio
- PGC
Psychiatric Genomics Consortium
- SIM1
single-minded homolog 1
- SNP
single nucleotide polymorphism
- UCSC
University of California Santa Cruz
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BC and SBC designed the study. ADN analysed the data. VW and BC co-supervised data analysis, and SBC received funding for the study. ADN, BC, SBC and VW wrote the paper. All authors read and approved the final manuscript.
Authors’ information
ADN and VW are joint first authors; SBC and BC are joint senior authors.
Contributor Information
Agnese Di Napoli, Email: agnesedinapoli@outlook.com.
Varun Warrier, Email: vw260@medschl.cam.ac.uk.
Simon Baron-Cohen, Email: sb205@cam.ac.uk.
Bhismadev Chakrabarti, Email: b.chakrabarti@reading.ac.uk.
References
- 1.American Psychiatric Association . DSM-IV Diagnostic and Statistical Manual of Mental Disorders, Text revision. 4. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 2.Baron-Cohen S, Scott FJ, Allison C, Williams JG, Bolton P, Matthews FE, et al. Prevalence of autism-spectrum conditions: UK school-based population study. Br J Psychiatry. 2009;194(6):500–9. doi: 10.1192/bjp.bp.108.059345. [DOI] [PubMed] [Google Scholar]
- 3.Gillberg C, Cederlund M, Lamberg K, Zeijlon L. Brief report: “the autism epidemic”. The registered prevalence of autism in a Swedish urban area. J Autism Dev Disord. 2006;36:429–35. doi: 10.1007/s10803-006-0081-6. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children. JAMA-J Am Med Assoc. 2001;285:3093–9. doi: 10.1001/jama.285.24.3093. [DOI] [PubMed] [Google Scholar]
- 5.Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9(6):e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:255–74. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 8.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14(12):1499–506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tordjman S, Somogyi E, Coulon N, Kermarrec S, Cohen D, Bronsard G, et al. Gene x environment interactions in autism spectrum disorders: role of epigenetic mechanisms. Frontiers in Psychiatry. 2014;5(53):1–17. doi: 10.3389/fpsyt.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24(42):9228–31. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–55. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, et al. cDNA cloning and tissue-specific expression of a novel basic Helix-Loop-Helix/PAS Factor (Arnt2) with close sequence similarity to the Aryl Hydrocarbon Receptor Nuclear Translocator (Arnt) Mol Cell Biol. 1996;16(4):1706–13. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb EA, AlMutair A, Kelberman D, Bacchelli C, Chanudet E, Lescai F, et al. ARNT2 mutation causes hypopituitarism, post-natal microcephaly, visual and renal anomalies. Brain. 2013;136:3096–105. doi: 10.1093/brain/awt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maltepe E, Keith B, Arsham AM, Brorson JR, Simon MC. The role of ARNT2 in tumor angiogenesis and the neural response to hypoxia. Biochem Biophys Res Commun. 2000;273:231–8. doi: 10.1006/bbrc.2000.2928. [DOI] [PubMed] [Google Scholar]
- 15.Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech Dev. 2000;90(2):253–61. doi: 10.1016/S0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 16.Ramachandrappa S, Raimondo A, Cali AMG, Keogh JM, Henning E, Saeed S, et al. Rare variants in single-minded 1 (SIM1) are associated with severe obesity. J Clin Invest. 2013;123(7):3042–50. doi: 10.1172/JCI68016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoghbi HY. MeCP2 dysfunction in humans and mice. J Child Neurol. 2005;20(9):736–40. doi: 10.1177/08830738050200090701. [DOI] [PubMed] [Google Scholar]
- 18.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–6. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 19.Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35(2):243–54. doi: 10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 20.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59(6):947–58. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoya T, Oda Y, Takahashi S, Morita M, Kawauchi S, Ema M, et al. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells. 2001;6:361–74. doi: 10.1046/j.1365-2443.2001.00421.x. [DOI] [PubMed] [Google Scholar]
- 22.Schweitzer J, LöhR H, Bonkowsky JL, Hübscher K, Driever W. Sim1a and Arnt2 contribute to hypothalamo-spinal axon guidance by regulating Robo2 activity via a Robo3- dependent mechanism. Development. 2013;140(1):93–106. doi: 10.1242/dev.087825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19(2):255–65. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci. 2011;31(9):3295–308. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin M, Zhao D, Hrabovsky A, Pedrosa E, Zheng D, Lachman HM. Heat shock alters the expression of schizophrenia and autism candidate genes in an induced pluripotent stem cell model of the human telencephalon. PLoS One. 2014;9(4):e94968. doi: 10.1371/journal.pone.0094968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90(1):133–41. doi: 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, et al. Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger Syndrome. Autism Res. 2009;2:157–77. doi: 10.1002/aur.80. [DOI] [PubMed] [Google Scholar]
- 28.Hovey D, Zettergren A, Jonsson L, Melke J, Anckarsäter H, Lichtenstein P, et al. Associations between oxytocin-related genes and autistic-like traits. Soc Neurosci. 2014;9(4):378–86. doi: 10.1080/17470919.2014.897995. [DOI] [PubMed] [Google Scholar]
- 29.Cross-disorder group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson E, Munir K, Munafò MR, Hughes M, McCormick M, Koenen KC. Stability of autistic traits in the general population: further evidence for a continuum of impairment. J Am Acad Child Adolesc Psychiatry. 2011;50(4):376–84. doi: 10.1016/j.jaac.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E. The Autism-Spectrum Quotient (AQ): evidence from Asperger Syndrome/high-functioning autism, males and females, scientists and mathematicians. J Autism Dev Disord. 2001;31(1):5–17. doi: 10.1023/A:1005653411471. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization . The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. Geneva: World Health Organization; 1992. [Google Scholar]
- 33.Wheelwright S, Auyeung B, Allison C, Baron-Cohen S. Defining the broader, medium and narrow autism phenotype among parents using the Autism Spectrum Quotient (AQ) Molecular Autism. 2010;1:10. doi: 10.1186/2040-2392-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoekstra RA, Bartels M, Verweij CJH, Boomsma DI. Heritability of autistic traits in the general population. Arch Pediatr Adolesc Med. 2007;161:372–7. doi: 10.1001/archpedi.161.4.372. [DOI] [PubMed] [Google Scholar]
- 35.Collins FS, Guyer MS, Chakravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278(5343):1580–1. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 36.Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet. 2003;33(1):67–72. doi: 10.1023/A:1021055617738. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyholt DR. A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(D1):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25(8):1048–55. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 42.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36(5):431–2. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 43.Chelala C, Khan A, Lemoine NR. SNPnexus: A web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25(5):655–61. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102.ArticlepublishedonlinebeforeprintinMay2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green JJ, Hollander E. Autism and oxytocin: new developments in translational approaches to therapeutics. Neurotherapeutics. 2010;7(3):250–7. doi: 10.1016/j.nurt.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guastella AJ, Einfeld SL, Gray KM, Rinehart NJ, Tonge BJ, Lambert TJ, et al. Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry. 2010;67(7):692–4. doi: 10.1016/j.biopsych.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 47.Hollander E, Novotny S, Hanratty M, Yaffe R, DeCaria CM, Aronowitz BR, et al. Oxytocin infusion reduces repetitive behaviors in adults with autistic and Asperger’s disorders. Neuropsychopharmacology. 2003;28:193–8. doi: 10.1038/sj.npp.1300021. [DOI] [PubMed] [Google Scholar]
- 48.Insel TR, O’Brien DJ, Leckman JF. Oxytocin, vasopressin and autism: is there a connection? Biol Psychiatry. 1999;45:145–57. doi: 10.1016/S0006-3223(98)00142-5. [DOI] [PubMed] [Google Scholar]
- 49.Modahl C, Green L, Fein D, Morris M, Waterhouse L, Feinstein C, et al. Plasma oxytocin levels in autistic children. Biol Psychiatry. 1998;43(4):270–7. doi: 10.1016/S0006-3223(97)00439-3. [DOI] [PubMed] [Google Scholar]
- 50.Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han Population. Biol Psychiatry. 2005;58:74–7. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Yrigollen C, Han SS, Kochetkova A, Babitz T, Chang JT, Volkmar FR, et al. Genes controlling affiliative behavior as candidate genes for autism. Biol Psychiatry. 2008;63:911–6. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverman JL, Yang M, Turner SM, Katz AM, Bell DB, Koenig JI, et al. Low stress reactivity and neuroendocrine factors in the BTBR T + tf/J mouse model of autism. Neuroscience. 2010;171(4):1197–208. doi: 10.1016/j.neuroscience.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]


