Abstract
Vesicular stomatitis virus (VSV) is a prototypic negative sense single-stranded RNA virus. The bullet-shape appearance of the virion results from tightly wound helical turns of the nucleoprotein encapsidated RNA template (N-RNA) around a central cavity. Transcription and replication require polymerase complexes, which include a catalytic subunit L and a template-binding subunit P. L and P are inferred to be in the cavity, however lacking direct observation, their exact position has remained unclear. Using super-resolution fluorescence imaging and atomic force microscopy (AFM) on single VSV virions, we show that L and P are packaged asymmetrically towards the blunt end of the virus. The number of L and P proteins varies between individual virions and they occupy 57 ± 12 nm of the 150 nm central cavity of the virus. Our finding positions the polymerases at the opposite end of the genome with respect to the only transcriptional promoter.
Introduction
Vesicular Stomatitis Virus (VSV) possesses a single-stranded negative sense RNA genome and is the prototypic model for understanding transcription and replication of potent human pathogens including Ebola and rabies. VSV is also an effective oncolytic agent since some attenuated VSV strains replicate preferentially in malignant cells [1]. Taken together, VSV is emerging as a useful model and potent tool in the arsenal of modern medicine.
VSV virions are 180 nm long and 80 nm wide and resemble a bullet with one tapered and one blunt end. The VSV genome is tightly encapsidated by nucleoprotein (N), forming the N-RNA genome template. Recent CryoEM studies showed N-RNA forms a left-handed helix that winds around a cavity, with the 3′ end at the tapered end and the 5′ end at the blunt end [2,3,4] (please see corrections to ref 5 for the correct RNA orientation). Like all other non-segmented negative sense (NNS) RNA viruses, VSV packages multiple copies of a polymerase that consists of the L protein catalytic subunit (241 kD) and a template-binding P protein subunit (30 kD). The polymerases transcribe and replicate the N-RNA genome by engaging at promoter sites at or near the 3′ end of the genome template [5].
On average ~50 L and ~400 P proteins are packaged within each virion [6]. The position of the L and P proteins within the bullet shaped virion has not been established previously. L organizes itself into a ring structure with an approximate diameter of 10 nm and does not bind the template by itself [7,8,9]. P proteins form dimers through their central domains and also bind L and the N-RNA template [8,10,11]. In recent CryoEM studies of VSV [3], either symmetry mismatches between the N-RNA helix, L and P, or random positioning of L and P, resulted in averaging out of the L and P density within the central cavity of VSV. Therefore to resolve the position of L and P, single virion imaging techniques with sufficiently high resolution are required.
While the resolution of simultaneously imaged multiple fluorophores is limited by diffraction, the position of a single fluorescent molecule can be localized with nanometer precision. The precision of localization is inversely proportional to the square root of the number of photons collected from the single molecule [12]. It is therefore possible to reconstruct an image by successively photoactivating subsets of molecules that are further apart from each other than the diffraction limit and localizing their positions with nanometer precision. This principle was developed in photoactivatable localization microscopy (PALM, [13]) fluorescence photoactivatable localization microscopy (fPALM, [14]) and stochastic optical reconstruction microscopy (STORM, [15]) to achieve in plane resolutions of ~20nm. The axial resolution of these techniques can be extended either through introduction of astigmatism associated with the out of plane images [16] or using Biplane imaging [17] both of which report an axial resolution of ~50nm.
Although fluorescence is specific, it does not report the density of unlabeled protein molecules. Atomic force microscopy (AFM) is sensitive to the overall protein density, via changes in the mechanical properties of the sample: the protein density within an object can be probed by measuring the deformation of a sample in response to a force applied by the AFM cantilever. AFM has been used in this way to measure the stiffness of single virions [18,19,20]. In general, the non-enveloped viruses are very stiff with a Young’s modulus in the range of GPa [20] while the enveloped viruses can be an order of magnitude softer in the range of 100 MPa [18].
Since the N-RNA is packaged with specific orientation of its 5′ end at the blunt end of the virion [3] and since the polymerases can only engage a promoter at the 3′ end [21,22], positioning of the polymerases within the bullet will inform our understanding of the early transcription of the N-RNA genome immediately upon delivery to the host cytoplasm and to some extent the final moments of packaging the genome in budding virions.
Materials and Methods
Super-resolution imaging and Fluorescence Localization
Images were recorded with a SR 200 microscope (Vutara, Inc.) based on the Biplane approach [17]. The envelope of VSV virions was mapped by labeling ~75% of the VSV-G protein on the exterior of the virion with Alexa-Fluor®647 (Invitrogen A21245). Alexa fluorophores can be efficiently initialized to their dark state in imaging buffer [SI] and can then be photo-activated through application of 405 nm UV light. This method was used to generate the viral envelopes shown in Figs. 1, 2.
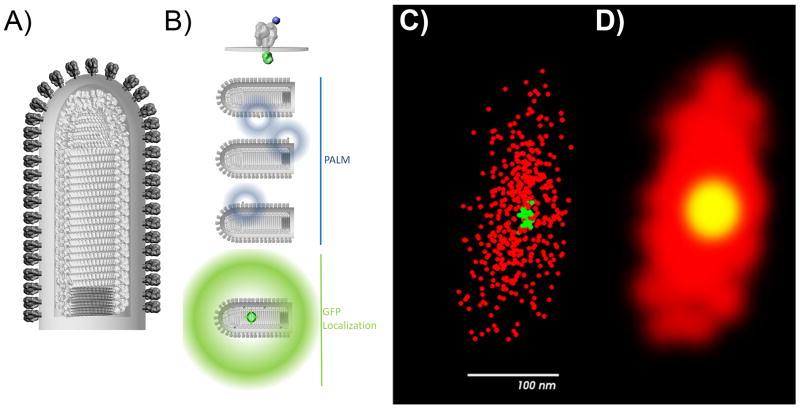
Fig. 1.
Locating eGFP center of fluorescence within reconstruction of VSV envelope. A) A model of VSV virion constructed partly from CryoEM data [3]. B) VSV-G is tagged from the exterior with Alexa 647 and internally with eGFP, ensuring that the eGFP center of fluorescence coincides with the center of the virion. C) Super-resolution fluorescence microscopy results in localization of VSV-G proteins (Red) and location of the center of fluorescence of eGFP (green). D) Volumetric rendering of the VSV envelope from super-resolution fluorescence imaging data along with the location of the center of fluorescence of eGFP.
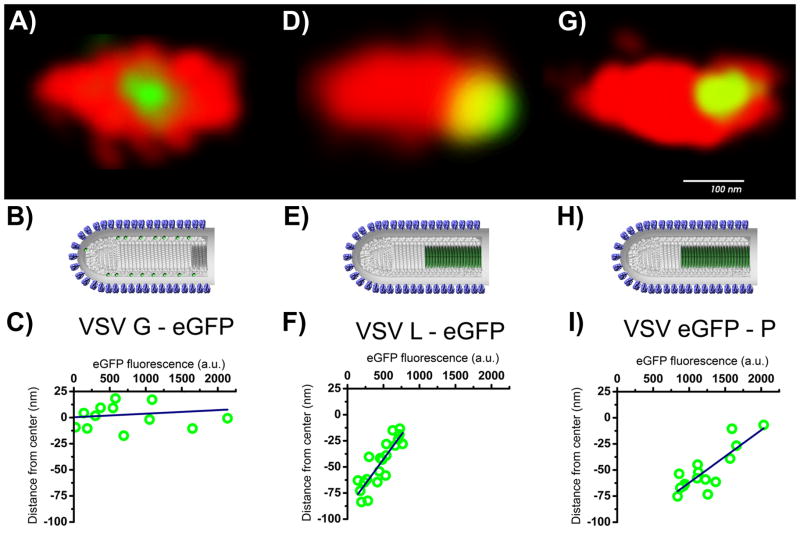
Fig. 2.
Center of fluorescence of the eGFP-linked proteins (G, P, and L, respectively A, D and G). Images in the center (B,E, and H) show the model of the VSV constructed partly from CryoEM data [3] in comparison to representative volumetric rendering of virions (Red) and G-eGFP, eGFP-P, and L-eGFP (Green). The center of fluorescence of eGFP is shown in green. At the bottom (C, F, and I), the distributions of the center of fluorescence of the virions with respect to the amount of eGFP fluorescence detected from each virion shows packaging with a constant density.
To locate the internal proteins within individual virions, we used recombinant viruses encoding enhanced Green Fluorescent Protein (eGFP) linked to the viral proteins P or L. In these recombinant viruses, all of the P or L proteins are respectively replaced by P linked to eGFP (eGFP-P) or eGFP linked to L (L-eGFP) and these viruses have been previously characterized [23,24]. The center of mass of the eGFP labeled proteins was determined in each virion and its relative distance to the center of the high resolution envelope was measured. The eGFP proteins associated with these internal proteins were not photoswitched, rather their total fluorescence intensity and the position of their center of fluorescence with respect to the center of the envelope was determined.
Excitation and activation of single fluorescent molecules was achieved via a speckle-free illumination with an even intensity distribution, which was realized by a specialized beam homogenizer. Two color channels were detected sequentially at 50 frames/sec. eGFP data was collected first over 500 frames at 30% power (1.2kW/cm2), then Alexa 647 was collected over 15,000 frames at 100% power (4kW/cm2). Data analysis was performed using the Vutara SRX software (Version 4.01).
Validation of super-resolution imaging method
First we have validated the resolution of the Biplane microscope by imaging beads on the microscope for 1000 imaging frames with an average fluorescence signal of 500 photons (comparable to our sample fluorescence). The analysis of this data as shown in detail in (SI) shows that we can localize these beads with less than 10 nm precision in XY and 25 nm precision along the optical axis. Resolution of the microscope is defined by the full width half maximum analysis and is 20 nm in plane and 50 nm along the optical axis.
Since the VSV G proteins are uniformly distributed on the surface of the envelope, we used them for our control experiments. VSV virions were pseudotyped with eGFP linked to the C terminus of VSV G protein (SI). Since virions are assembled on different cells within the population with varying levels of expression of VSV G-eGFP expression, the collected virions have a distribution of G-eGFP incorporation between virions. Each virion incorporates 1200 copies of G independent of whether the G has a GFP tag or not and G proteins are uniformly distributed on the surface of each virion [6]. We expect that the center of fluorescence of G-eGFP to co-localize with the center of the viral envelope independent of the number of G-eGFP proteins packaged on the surface of the virion. Because the virus is pseudotyped on a population of cells with varying expression levels of G-eGFP (SI), we also expect that the amount of G-eGFP in each virion will vary. As Figs. 1 and 2(A–C) show, the G-eGFP center of fluorescence overlapped with the center of the virion to within ±11 nm, measured for 13 VSV:G-eGFP virions.
We also verified this method on HIV virions. HIV forms spherical virions with a diameter of 120–150 nm [25]. The membrane of these virions is coated internally with a lattice of Gag proteins as observed through CyoEM. The Gag lattice is uniformly distributed inside the virions with the exception of a few gaps [26], therefore we expect that the center of mass of Gag be very close to the center of the virion, if not exactly on the center of virions due to the possible empty patches. We verified our super-resolution fluorescence imaging method on HIV Virus Like Particles (VLPs) pseudotyped with VSV G which incorporated Gag-eGFP proteins (SI). G proteins were labeled with Alexa 647 tagged antibodies to mark the envelope of the VLP and 12 HIV virions pseudotyped with G incorporating Gag-eGFP showed that the center of the virion co localized with the center of the Gag signal to within 12 nm precision as shown in (SI).
AFM Materials and Methods
We used atomic force microscopy (AFM) for iso-force scans across the surface of several virions. The AFM (Asylum Research MFP-3D in a dark environmental chamber) consists of a pyramidal gold-coated tip (radius of curvature 20 nm; height 10 μm) attached to the underside of a pliable cantilever (force constant 3N/m; length = 200 μm), a piezoelectrically actuated sample scanner (<1 nm precision), and associated digital control and data acquisition electronics. On stiff samples, the AFM cantilever bends easily to accommodate changes in the sample height, so the path of the tip faithfully reports sample topography. When the sample is more pliant than the cantilever, the sample will deform under the force of the tip, so the height signal becomes more indicative of the sample’s stiffness rather than its topography, therefore, the height signal provides an isoforce map of the sample surface rather than a true topographical map.
Results
Super-resolution fluorescence imaging shows P and L are localized at the blunt end of VSV
To measure the center of mass of P and L proteins within the virus, we used recombinant viruses encoding enhanced Green Fluorescent Protein (eGFP) linked to the viral proteins P or L. In these recombinant viruses, all of the P or L proteins are respectively replaced by P linked to eGFP (eGFP-P) or eGFP linked to L (L-eGFP) and these viruses have been previously characterized [23,24]. VSV: L-eGFP is temperature sensitive, replicates in 32C and packages 1/3 of the wild type levels of the polymerase L. VSV: GFP-P grows at 37C to a lower titer than WT and packages half the L and P proteins when compared to WT virus. These fluorescent viruses although imperfect, allow localization of their L and P. Later, we will examine the effect of protein density on localization by comparing these viruses to WT using AFM.
The total fluorescence intensity detected from the virions is directly proportional to the copy number of packaged eGFP linked proteins; however, it cannot be used as an absolute measure of the copy number due to fluorescence quenching at high concentrations. More importantly for this work, the center of fluorescence of the eGFP pinpoints the center of mass of the L or P protein distributions, which can be determined with ~10 nm precision [12].
For all 15 VSV:eGFP-P virions measured, the eGFP-P center of fluorescence was located toward the blunt end of the virion. We observed a 30 percent variation in the total fluorescence between individual virions of VSV:eGFP-P, which represents variations in the number of P-eGFP packaged. The average fluorescence signal for 15 virions was 1233 ± 351 photons. As shown in Fig. 2D, virions with fewer eGFP-P copies showed the largest displacement of the eGFP localization with respect to the center of the virion, while virions with the maximum number of eGFP-P copies had almost no displacement. The linear relationship between the displacement and the number of packaged eGFP-P molecules shown in Fig. 2D indicates a constant packing density of P proteins as they fill the cavity starting from the blunt end. In average the center of mass of the P-eGFP was displaced from the center of the virion by a distance of 50 ± 21 nm.
Because P proteins associate with L, we next measured the distribution of L. We also found variation in the fluorescence signal between individual VSV:L-eGFP virions of 461 ± 202 photons. Furthermore, as with the eGFP-P, we observed a linear relationship between the L-eGFP fluorescence intensity and the displacement of the fluorescence center from the virus center as shown in Fig. 2F. Again, this indicates uniform packaging of L-eGFP proteins at the blunt end of the virus. The average center of fluorescence for 20 VSV:L-eGFP virions was located 46 ± 22 nm from the geometric center of the virus toward the blunt end. We conclude that P and L package asymmetrically the same volume at the blunt end of VSV.
Note that neither VSV:eGFP-P nor VSV:L-eGFP package wild type levels of L and P (see below), so the fluorescent measurements cannot be used to estimate either the density or the exact length of the volume in which L and P package in wild type virions.
AFM reveals protein density at the blunt end of VSV
We next used a fluorescence-free assay both to corroborate the above results and to estimate the size of the volume occupied by L and P proteins within wild type virions. Any variation in the protein density along the length of a virus particle should lead to a change in its mechanical properties, specifically its stiffness. Stiffness of the envelopes viruses [18] is almost an order of magnitude less than non-enveloped viruses like bacteriophage [20] or adenovirus [27]. The proteins associated with the envelope of VSV (G, M and N) have a symmetric helical organization [3], therefore, the AFM tip can deform the envelope and probe the stiffness of the underlying structure which should have no variance along the central axis of the virus. Internal proteins and genomic structure of the viruses have direct effects on their mechanical properties, as shown previously for DNA binding within the capsid of parvovirus of mice [28] and maturation of HIV virions [18,29]. We hypothesized that the virus should be stiffer at the blunt end due to the presence of an over density of P and L proteins there. Using a relatively stiff cantilever with tapping-mode AFM under ambient conditions, we performed an iso-force scan of the virions tethered on glass surfaces as explained in the methods. The stiffness measurements rely on using the AFM probe as a force sensor. In particular for soft samples, the probe depresses the surface, and the degree of depression is related to the stiffness of the sample relative to the spring constant of the AFM cantilever (SI). We were able to press considerably into the virus and detected an increase in the height of the virus at the blunt end. Since the scans are iso-force, the increase in height indicates an increase in stiffness from extra protein density at the blunt end. AFM was able to resolve the asymmetric bullet shape of the virions, thus placing the region of elevated stiffness at the blunt end (Fig. 3 and SI).
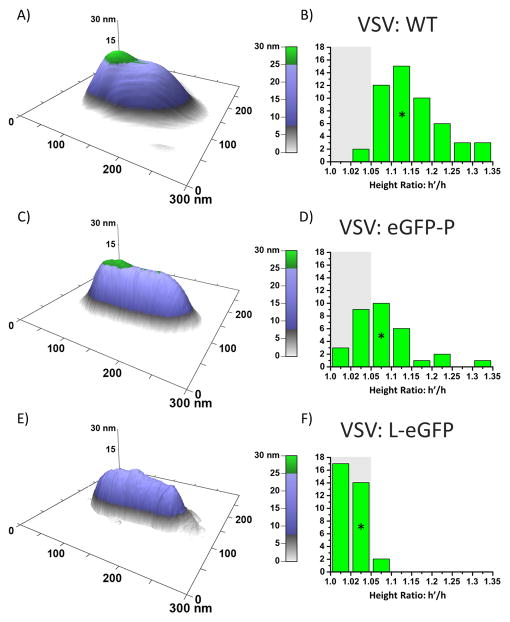
Fig. 3.
AFM surfaces of VSV show two distinct regions with respect to structural composition, specifically a variation in elastic response to the force exerted by the tip. A) Wild type VSV virions have the stiffest bump region, indicated by the tallest bump. B) The ratio of the two regions’ respective heights (h′ for the bump, h for the main body) is 1.14 ± 0.07 for 50 WT virions. C) A representative VSV:eGFP-P virion with a similarly distinctive bump. D) VSV:eGFP-P h′/h is 1.07 ± 0.06 for 30 virions. E) VSV:L-eGFP virions have little or no distinguishable regions towards the rear, and F) have a measured h′/h of 1.02 ± 0.01 for 30 virions.*Indicates the representative virions.
As shown in Fig. 3, we observed virions with 180×80nm bullet shapes, each with recognizable tapered and blunt ends distinguished by analyzing the AFM cross sections (SI). During AFM scans, the tip locally compresses the virions to a height of~30 nm due to deformation of the soft viral envelope, but the measured widths (~100 nm) accurately reflect the true diameter of the virions with a 20 nm convolution. This demonstrates that the virions were not “pancaked” onto the substrate due to surface interactions or dehydration. This was confirmed by increasing the force and pushing the virus into the non-elastic region which resulted in fully deformed virions (data not shown).
For all wild type virions measured, there was a region at the blunt end that exhibited less depression (higher stiffness) compared to the rest of the virus, as shown in green. This region extended 57 ± 12 nm in length from the blunt end, obtained from a sample of 50 wild type viruses.
Detected density at the blunt end of the virus correlates with the amount of P and L packaged
Recombinant VSV:eGFP-P viruses exhibit slower than wild type growth kinetics and package about half the number of P and L proteins compared to the wild type. Their bullet-shaped morphology is, however, indistinguishable from the wild type when viewed in EM [23,30]. For 30 VSV:eGFP-P virions, the region of higher stiffness extended 68 ± 21 nm, but the magnitude of elevated stiffness was much lower compared to wild type, as shown in Fig. 3. These results suggest that VSV:eGFP-P forms a protein complex of lower density compared to the wild type. A GFP-labeled P cannot package as densely as wild type because eGFP likely sterically hinders packaging. This may explain why the eGFP-P virions have a volume with lower stiffness than wild type that nonetheless extends significantly within the cavity.
VSV:L-eGFP viruses are only partially functional, package only about a third as much L as wild type(but normal amounts of P), and do not transcribe their RNA in vitro. They do, however, replicate well at 32°C and form VSV virions with the correct morphology[24]. Increased stiffness at the blunt end of VSV:L-eGFP was almost undetectable via AFM, consistent with a much lower density volume of polymerase complexes compared to wild type.
Discussion
Using high-resolution fluorescence imaging and center of mass calculations on recombinant viruses in which all wild type L or P proteins were genetically replaced with GFP linked fusion proteins of L and P, we show asymmetric packaging of the polymerase complex of VSV at its blunt end. L and P packaged within the blunt end of the virus create a region of higher stiffness within the blunt end of the virus. We measured this region of higher stiffness using AFM and in WT virions report its average length at 57 nm with a variation of ± 12 nm among virions. This variation reflects the variable number of L and P packaged in each virion as also measured with fluorescence and shown in Figure 2.
The present study has focused on identifying the position of the polymerases within the cavity of VSV which resulted in identifying the asymmetric distribution of polymerases at the blunt end of the virus. Transcription of the virion N-RNA is the first step required for virus multiplication after entry into the host cytosol, and this can only initiate at or near the 3′ end [21,22]. Recent CryoEM studies showed the N-RNA winds around a cavity in a left-handed helix, starting with its 3′ end at the virus′s tapered end and culminating with the 5′ end at the blunt end of the virus [2,3]. Based on our observations, the polymerases are therefore stacked at the 5′ end of the genome rather than at the 3′ end where the transcriptional promoter is located. We envision two ways in which the polymerase units can reach the 3′ transcriptional promoter. In the first model, we hypothesize it would be possible to start transcription through formation of an N-RNA template loop that brings the 3′ end of the genome close to the 5′ end of the N-RNA where the polymerases are located. Such N-RNA loops have been identified in the cytoplasm of cells infected with VSV before [31]. The observed asymmetry of the polymerase localization along the genome however cannot rule out the traditional model of transcription in which L-Px complexes dissociate from the N-RNA and re-engage the promoter at the 3′ end.
Supplementary Material
Highlights.
The VSV polymerases L are localized to the blunt end of the virus.
The VSV phosphoproteins P are localized to the blunt end of the virus.
There are different amounts of L and P proteins in each virus.
Acknowledgments
We thank: Prof. Stephen Harrison for fruitful discussions and comments in preparation of this manuscript, Prof. Sean Whelan for VSV:PeGFP virus and Jeff Ballew for providing support and technical assistant. This work was supported by NSF grants 1121972 (SS) and DBI-0845193 (JMG).
References
- 1.Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nature Biotechnology. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the Vesicular Stomatitis Virus Nucleoprotein-RNA Complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 3.Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. Cryo-EM Model of the Bullet-Shaped Vesicular Stomatitis Virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desfosses A, Ribeiro EA, Schoehn G, Blondel D, Guilligay D, Jamin M, Ruigrok RWH, Gutsche I. Self-organization of the vesicular stomatitis virus nucleocapsid into a bullet shape. Nat Commun. 2013;4:1429. doi: 10.1038/ncomms2435. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov I, Yabukarski F, Ruigrok RWH, Jamin M. Structural insights into the rhabdovirus transcription/replication complex. Virus Research. 2011;162:126–137. doi: 10.1016/j.virusres.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Jayakar HR, Jeetendra E, Whitt MA. Rhabdovirus assembly and budding. Virus Research. 2004;106:117–132. doi: 10.1016/j.virusres.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Morin B, Kranzusch PJ, Rahmeh AA, Whelan SPJ. The polymerase of negative-stranded RNA viruses. Current Opinion in Virology. 2013;3:103–110. doi: 10.1016/j.coviro.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahmeh AA, Morin B, Schenk AD, Liang B, Heinrich BS, Brusic V, Walz T, Whelan SPJ. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc Natl Acad Sci USA. 2012;109:14628–14633. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SPJ. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding H, Green TJ, Lu S, Luo M. Crystal Structure of the Oligomerization Domain of the Phosphoprotein of Vesicular Stomatitis Virus. Journal of Virology. 2006;80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov I, Crépin T, Jamin M, Ruigrok RWH. Structure of the Dimerization Domain of the Rabies Virus Phosphoprotein. Journal of Virology. 2010;84:3707–3710. doi: 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson RE, Larson DR, Webb WW. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaksonen M, Drubin DG. PALM reading: Seeing the future of cell biology at higher resolution. Developmental Cell. 2006;11:438–439. doi: 10.1016/j.devcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Hess ST, Girirajan TPK, Mason MD. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang B, Wang W, Bates M, Zhuang X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Meth. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 18.Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I. A Stiffness Switch in Human Immunodeficiency Virus. Biophysical Journal. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snijder J, Reddy VS, May ER, Roos WH, Nemerow GR, Wuite GJL. Integrin and Defensin Modulate the Mechanical Properties of Adenovirus. Journal of Virology. 2013;87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernando-Pérez M, Miranda R, Aznar M, Carrascosa JL, Schaap IAT, Reguera D, de Pablo PJ. Physical Virology: Direct Measurement of Phage phi29 Stiffness Provides Evidence of Internal Pressure (Small 15/2012) Small. 2012;8:2365–2365. doi: 10.1002/smll.201200664. [DOI] [PubMed] [Google Scholar]
- 21.Iverson LE, Rose JK. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 22.Whelan SPJ, Wertz GW. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc Natl Acad Sci USA. 2002;99:9178–9183. doi: 10.1073/pnas.152155599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruedas JB, Perrault J. Insertion of Enhanced Green Fluorescent Protein in a Hinge Region of Vesicular Stomatitis Virus L Polymerase Protein Creates a Temperature-Sensitive Virus That Displays No Virion-Associated Polymerase Activity In Vitro. J Virol. 2009;83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundquist WI, Kräusslich H-G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harbor Perspectives in Medicine. 2012;2 doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggs JAG, Kräusslich H-G. The Molecular Architecture of HIV. Journal of Molecular Biology. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Ortega-Esteban A, Perez-Berna AJ, Menendez-Conejero R, Flint SJ, Martin CS, de Pablo PJ. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci Rep. 2013;3 doi: 10.1038/srep01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrasco C, Carreira A, Schaap IAT, Serena PA, Gómez-Herrero J, Mateu MG, de Pablo PJ. DNA-mediated anisotropic mechanical reinforcement of a virus. Proceedings of the National Academy of Sciences. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang H-B, Hevroni L, Kol N, Eckert D, Tsvitov M, Kay M, Rousso I. Virion stiffness regulates immature HIV-1 entry. Retrovirology. 2013;10:4. doi: 10.1186/1742-4690-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das SC, Pattnaik AK. Role of the Hypervariable Hinge Region of Phosphoprotein P of Vesicular Stomatitis Virus in Viral RNA Synthesis and Assembly of Infectious Virus Particles. J Virol. 2005;79:8101–8112. doi: 10.1128/JVI.79.13.8101-8112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naeve CW, Summers DF. Electron Microscopy of Vesicular Stomatitis Virus Replicative Ribonucleoproteins. Journal of Virology. 1980;34:764–771. doi: 10.1128/jvi.34.3.764-771.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.