Abstract
Background
Few studies in epidemiology have evaluated the effects of gene-environment interaction on oxidative stress, even though this interaction is an important etiologic factor in lung carcinogenesis. We investigated the effects of the genetic polymorphisms of paraoxonase 1 (PON1), smoking, and the interaction between the two on lung cancer risk and oxidative stress.
Methods
This study’s subjects consisted of 416 newly diagnosed lung cancer patients and an equal number of matched controls. The GoldenGate assay was used for genotypic analyses of the PON1 gene. Urinary 8-hydroxydeoxyguanosine (8-OHdG) and thiobarbituric acid reactive substances levels were measured as indicators of oxidative stress.
Results
The PON1 rs662 AA genotype showed a significantly lower risk of lung cancer than the GG genotype (OR = 0.60, 95% CI: 0.36–0.99). The protective effect of the PON1 rs662 AA genotype on lung cancer risk was limited to non-smokers. Lung cancer patients who had the rs662 A allele showed a dose-dependent association between smoking status and oxidative stress markers. Among non-smoking lung cancer patients, urinary 8-OHdG levels were significantly lower in individuals with the rs662 GA and AA genotypes than in those with the GG genotype. Furthermore, we found a significant interaction effect between PON1 rs662 and smoking status on urinary 8-OHdG levels in lung cancer patients.
Conclusions
Our results suggest that the protective effect of PON1 rs662 SNP against lung carcinogenesis and the induction of oxidative stress might be modulated by the interaction between PON1 genetic polymorphisms and tobacco smoking.
Introduction
Of all types of cancer, lung cancer has the highest incidence and mortality worldwide [1]. In Korea, lung cancer is the leading cause of cancer-related death, accounting for approximately one-fourth of all cancer-associated deaths [2]. Tobacco smoking is the most important risk factor for lung cancer, as it is the most likely cause of approximately 90% of lung cancer cases [3,4]. However, only a small proportion of smokers (less than 15%) are diagnosed with lung cancer in their lifetime [3], and approximately 30% of lung cancer patients in Korea are lifelong non-smokers [5]. Thus, although tobacco smoking is a major determinant of lung cancer, it is not sufficient to cause cancer in the absence of additional factors, such as genetic susceptibility and exposure to other carcinogens (i.e., asbestos, nickel, chromium, arsenic, or radon). The carcinogenic effect of smoking is a result of an interaction with additional factors, especially genetic factors [3,4].
Tobacco smoking induces oxidative stress, which can cause severe damage to cellular macromolecules (i.e., DNA, proteins, and lipids) and is a pivotal carcinogenic mechanism linked to various diseases, including lung cancer [6,7]. Oxidative stress is mitigated by exogenous and endogenous antioxidants and antioxidant-defense enzymes (i.e., superoxide dismutase, catalase, glutathione peroxidase, etc.) [8]. Genetic polymorphisms of antioxidant enzymes have an impact on the inter-individual variability in antioxidant defense, which is also associated with susceptibility to various cancers [9].
Paraoxonase 1 (PON1), one of the antioxidant enzymes acting in the blood, plays a key role in preventing the effects of systemic oxidative stress [10–13]. In addition, PON1 is well known for detoxifying the activity of oxons, which are toxic metabolites of organophosphate pesticides [11]. PON1 is predominantly synthesized in the liver and secreted into the bloodstream after binding to high-density lipoproteins (HDL) [11]. The PON1 protein in humans is found in various types of tissue, including lung tissue [14], and PON1 in lung tissue is mainly localized in Clara cells, endothelial cells, and type I cells of the alveolar epithelium [15]. These cells, located in the respiratory portion of the lung, can be exposed to tobacco smoke and reactive oxygen substances released by environmental toxicants [15]. PON1 activity is modulated by various factors, such as genetic polymorphisms, environmental chemicals, pharmaceutical compounds, smoking, alcohol consumption, and dietary factors [12]. It is known that the major determinant of serum PON1 activity is the PON1 polymorphism [16]. Our previous study showed that approximately 54% of the variance of serum PON1 activity was regulated by the PON1 Arg192Glu (R192Q, rs662) polymorphism in a Korean population [17].
Recently, a meta-analysis suggested that the PON1 R192Q polymorphism is a significant risk factor for all cancers, including breast, brain and prostate cancers, especially in Asian populations [18]. At present, only three studies have been conducted to examine the effect of the PON1 genetic polymorphism on lung cancer risk [19–21]. Although gene-environment interaction could be an important etiologic factor in lung carcinogenesis, there are no epidemiological studies assessing this interaction on oxidative stress.
In this study, we investigated the effects of genetic polymorphisms of PON1, smoking, and the interaction between the two on lung carcinogenesis and oxidative stress.
Materials and Methods
Study Subjects
The study subjects consisted of 416 newly diagnosed lung cancer patients and an equal number of age- (within 5 years) and sex-matched controls. These patients were histologically confirmed to have lung cancer between January 2001 and October 2008 at the Chungbuk National University Hospital or at the Dankook University Hospital in the Republic of Korea. Control subjects that did not have a previous diagnosis of any type of cancer were selected from individuals receiving routine medical examinations at the two hospitals. After written informed consent was obtained from all subjects, trained interviewers collected information on demographic characteristics, lifestyle factors, and medical and occupational history. Items concerning smoking on the questionnaire included current smoking status, average number of cigarettes smoked daily, total duration of smoking, age of initiating smoking, and age of quitting smoking. Cumulative smoking amount was measured in pack-years (average number of cigarettes smoked daily / 20 × total duration of smoking in years). Non-smokers were defined as individuals who had never smoked cigarettes or who had not smoked more than 100 cigarettes in their lifetime [22]. Peripheral blood and urine were collected from all subjects and then stored at-80°C until the experiment.
Ethics Statement
This study was approved by the Institutional Review Board of Chungbuk National University Hospital, Republic of Korea (IRB No. 2011–09–072).
Single nucleotide polymorphisms selection and genotyping analysis
The candidate SNPs (single nucleotide polymorphisms) were selected from 3 public databases: the International HapMap Project database (http://hapmap.ncbi.nlm.nih.gov/), the Functional Element SNPs database (http://sysbio.kribb.re.kr:8080/fesd/index.jsp) [23], and the SNPinfo Web Server (http://snpinfo.niehs.nih.gov/). The selection criteria were as follows: (i) haplotype-tagging SNPs with an R-square cutoff of 0.9 and minimum minor allele frequency in CHB and JPT population of 0.05; (ii) SNPs located in functional regions, such as the promoter, start codon, splice site, coding exon, and stop codon; and (iii) non-synonymous SNPs. Finally, we selected seven SNPs (rs662, rs13306698, rs854572, rs854573, rs854552, rs854565, and rs854568) in the PON1 gene for genotyping.
Genomic DNA was isolated from peripheral blood using the QuickGene-810 Nucleic Acid Isolation System (Fujifilm, Tokyo, Japan) and the QuickGene DNA Whole Blood Kit in accordance with the manufacturer’s protocol; the DNA samples were stored at-70°C until analysis. SNP genotyping was performed using the VeraCode GoldenGate assay (Illumina, San Diego, CA, USA). All SNPs were in Hardy-Weinberg equilibrium in cases and controls, and the call rate for the seven SNPs was 100%. S1 Table presents detailed information on the seven SNPs and allele frequencies.
Analysis of biomarkers for oxidative stress
8-Hydroxydeoxyguanosine. The level of 8-hydroxydeoxyguanosine (8-OHdG) was measured using an 8-OHdG enzyme-linked immunosorbent assay (ELISA) kit (8-OHdG Check; Japan Institute for the Control of Aging, Fukuroi, Japan). Briefly, urine samples were centrifuged, and 50 μl of the supernatant and 50 μl of an aliquot of the primary antibody were added to an 8-OHdG precoated microplate and incubated at 37°C for 1 hour. The plate was washed 3 times with phosphate-buffered saline. Horseradish peroxidase-conjugated secondary antibody was added to each well, incubated at 37°C for 1 hour, and subsequently washed three times. A 100 μl enzyme substrate containing 3,3′,5,5′-tetra-methyl-benzidine was added, and the plates were incubated at room temperature for 15 minutes under dark conditions. The reaction was terminated by adding of 1 M phosphoric acid, and the absorbance at 450 nm was measured using a microplate reader (GENios; TECAN, Grödig/Salzburg, Austria). The concentration of 8-OHdG was calculated using a standard curve.
Thiobarbituric acid reactive substances. Urinary thiobarbituric acid reactive substances (TBARS) were determined using a high-performance liquid chromatographic (HPLC) system with a fluorescence detector [24].21 Briefly, 50 μl of 0.05% butylatedhydroxytoluene, 150 μl of 0.1125 N nitric acid (HNO3), and 150 μl of 42 mM thiobarbituric acid were added to a 50μl aliquot of the urine sample or 50 μl of 1,1,3,3-tetramethoxypropane standard and mixed using a vortex. The samples were then heated on a heat block (100°C) for 1 hour and then placed in ice water for 5 minutes to cool; 300 μl of n-butanol was subsequently added for the extraction of TBARS, and the samples were then centrifuged at 10,000 × g for 5 minutes. Ten microliters of the supernatant was injected into the HPLC system, which consisted of a pump (Lsp 930; Younglin, Seoul, Korea), an automatic injector (SIL 10Avp; Shimadzu, Kyoto, Japan), a fluorescence detector (RF-10AxL; Shimadzu), and a data acquisition module (Autochro-200; Younglin). The columns used were a 150-mm reverse-phase column (TSK-GEL ODS-80TM; Tosoh), and the mobile phases were potassium dihydrogen phosphate:methanol:acetonitrile (60:25:15, v/v/v) at a flow-rate of 1 ml/minute. The excitation/emission wavelengths were 515/553 nm.
Statistical analysis
Statistical power was calculated using Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) [25]. The parameters were set as follows: risk allele frequency of less than 0.15, alpha error of less than 0.05, and a disease prevalence of less than 0.1%. The power of a dominant model was 72.4% when the odds ratio for a genotype with one or two risk allele(s) was taken as 1.5.
Student’s t-test was used to compare continuous variables between the patients and control subjects. Associations between lung cancer and putative risk factors were estimated by odds ratios (ORs) and their corresponding 95% confidence intervals (95% CI) derived from multivariate conditional logistic regression models after adjusting for potential confounding factors, such as age, sex, smoking history, and occupational history. A stratified analysis was used to estimate the combined effects of genotypes and smoking status. The P-values for the interactions between the genotypes and smoking status were assessed using the Wald test for the cross-product term in a model containing the main effects of genotype and exposure variable. Multiple testing corrections were carried out using the Benjamini-Hochberg procedure for controlling the false discovery rate (FDR) [26]. All of the statistical analyses were performed using SAS Version 9.2 (SAS Institute, Cary, NC, USA). Linkage disequilibrium statistics and haplotype blocks were obtained using the Haploview program (http://www.broad.mit.edu/mpg/haploview) [27]. Haplotype frequency estimation and the analysis of the association of haplotypes with lung cancer risk were conducted with SNPStats (http://bioinfo.iconcologia.net/SNPStats_web) [28].
Results
The general characteristics of the 416 lung cancer cases and the 416 age- (within 5 years) and sex-matched controls are presented in Table 1. The lung cancer cases were on average almost 1.8 years older than the controls. The proportions of ex-smokers and current smokers were significantly higher in the lung cancer cases than in the controls, and smoking status was significantly associated with an increased risk of lung cancer. The mean of the cumulative smoking amount in lung cancer cases was about twice as high than that of controls, and cumulative smoking amount significantly increase the risk of lung cancer in a dose-dependent manner. The geometric means for urinary TBARS and 8-OHdG were not significantly different between lung cancer cases and controls.
Table 1. General characteristics of lung cancer cases and controls.
| Variables | Cases | Controls | P-value or OR (95% CI) |
|---|---|---|---|
| Age, years, mean ± SD | 66.2 ± 9.8 | 64.4 ± 9.9 | 0.011 |
| Gender, N (%) | 1.000 | ||
| Male | 324 (77.9) | 324 (77.9) | |
| Female | 92 (22.1) | 92 (22.1) | |
| Body mass index, kg/m2, mean ± SD | 22.3 ± 3.4 | 23.7 ± 4.3 | <0.001 |
| Smoking status, N (%) | |||
| Non-smokers | 72 (17.3) | 157 (37.7) | 1.00 (ref.) |
| Ex-smokers | 185 (44.5) | 142 (34.1) | 5.14 (3.19, 8.28) a |
| Current smokers | 159 (38.2) | 117 (28.1) | 5.60 (3.45, 9.09) a |
| Cumulative smoking amounts, pack-years, mean ± SD | 39.1 ± 29.1 | 19.9 ± 23.6 | <0.001 |
| Cumulative smoking amounts, N (%) | |||
| 0 pack-years (Non-smokers) | 72 (17.3) | 157 (38.1) | 1.00 (ref.) |
| <30 pack-years | 81 (19.5) | 127 (30.8) | 2.84 (1.73, 4.68) a |
| ≥30 pack-years | 263 (63.2) | 128 (31.1) | 11.34 (6.80, 18.90) a |
| Alcohol consumption, N (%) | |||
| Non-drinker | 185 (44.6) | 167 (40.6) | 1.00 (ref.) |
| Drinker | 230 (55.4) | 244 (59.4) | 0.84 (0.62, 1.14) b |
| 8-OHdG, μg/g creatinine, GM (95% CI) | 5.06 (4.55, 5.62) | 4.81 (4.43, 5.22) | 0.457 |
| TBARS, μmol/g creatinine, GM (95% CI) | 0.88 (0.79, 0.98) | 0.84 (0.77, 0.92) | 0.486 |
TBARS: thiobarbituric acid reactive substances; 8-OHdG: 8-hydroxydeoxyguanosine; GM: geometric mean; CI: confidence intervals.
aAdjusted for age and sex.
bAdjusted for age, sex and smoking status.
cIndividuals who have work experience in occupations related lung cancer risk, such as petrochemicals, construction, mining, asbestos or rockwool production, welding, electrical manufacture, plastic or rubber manufacture, smelting, and asphalt.
dReference category is all other occupations.
The distributions of the seven SNPs (rs662, rs13306698, rs854572, rs854573, rs854552, rs854565, and rs854568) in the PON1 gene among lung cancer cases and controls are shown in Table 2. The rs662 AA (192QQ) genotype showed a significantly lower risk for lung cancer than the GG (192RR) genotype (OR = 0.60, 95% CI: 0.36–0.99). The rs854565 GA genotype also showed a marginal association with reduced lung cancer risk when compared with GG genotype (OR = 0.77, 95% CI: 0.54–1.04). However, neither of those associations were statistically significant after controlling for the FDR.
Table 2. Associations between seven genetic polymorphisms of PON1 and the risk of lung cancer.
| SNP ID | Genotypes | Cases, N (%) | Controls, N (%) | OR a (95% CI) | P-value | FDR |
|---|---|---|---|---|---|---|
| rs13306698 | AA | 354 (85.1) | 357 (85.8) | 1.00 (ref.) | ||
| AG | 60 (14.4) | 58 (13.9) | 1.02 (0.68, 1.53) | 0.944 | 0.944 | |
| GG | 2 (0.5) | 1 (0.2) | 2.07 (0.19, 23.02) | 0.553 | 0.863 | |
| AG+GG | 62 (14.9) | 59 (14.2) | 1.03 (0.69, 1.55) | 0.872 | 0.872 | |
| rs662 | GG | 209 (50.2) | 180 (43.3) | 1.00 (ref.) | ||
| GA | 170 (40.9) | 188 (45.2) | 0.77 (0.57, 1.04) | 0.087 | 0.312 | |
| AA | 37 (8.9) | 48 (11.5) | 0.60 (0.36, 0.99) | 0.044 | 0.308 | |
| GA+AA | 207 (49.8) | 236 (56.7) | 0.73 (0.55, 0.98) | 0.034 | 0.238 | |
| rs854552 | TT | 234 (56.3) | 237 (57) | 1.00 (ref.) | ||
| TC | 147 (35.3) | 149 (35.8) | 1.04 (0.76, 1.41) | 0.808 | 0.943 | |
| CC | 35 (8.4) | 30 (7.2) | 1.14 (0.66, 1.97) | 0.637 | 0.863 | |
| TC+CC | 182 (43.8) | 179 (43.0) | 1.06 (0.79, 1.41) | 0.709 | 0.872 | |
| rs854565 | GG | 236 (56.7) | 215 (51.7) | 1.00 (ref.) | ||
| GA | 152 (36.5) | 172 (41.4) | 0.77 (0.57, 1.04) | 0.089 | 0.312 | |
| AA | 28 (6.7) | 29 (7.0) | 0.82 (0.45, 1.47) | 0.500 | 0.863 | |
| GA+AA | 180 (43.3) | 201 (48.3) | 0.78 (0.58, 1.04) | 0.086 | 0.301 | |
| rs854568 | AA | 190 (45.7) | 198 (47.6) | 1.00 (ref.) | ||
| AG | 182 (43.8) | 178 (42.8) | 1.10 (0.81, 1.48) | 0.553 | 0.774 | |
| GG | 44 (10.6) | 40 (9.6) | 1.03 (0.63, 1.69) | 0.903 | 0.903 | |
| AG+GG | 226 (54.3) | 218 (52.4) | 1.08 (0.81, 1.44) | 0.586 | 0.820 | |
| rs854572 | CC | 103 (24.8) | 124 (29.8) | 1.00 (ref.) | ||
| CG | 208 (50.0) | 201 (48.3) | 1.23 (0.87, 1.73) | 0.244 | 0.569 | |
| GG | 105 (25.2) | 91 (21.9) | 1.36 (0.91, 2.03) | 0.139 | 0.487 | |
| CG+GG | 313 (75.2) | 292 (70.2) | 1.27 (0.92, 1.75) | 0.150 | 0.350 | |
| rs854573 | TT | 329 (79.1) | 322 (77.4) | 1.00 (ref.) | ||
| TC | 82 (19.7) | 89 (21.4) | 0.89 (0.62, 1.27) | 0.519 | 0.774 | |
| CC | 5 (1.2) | 5 (1.2) | 0.80 (0.22, 2.92) | 0.740 | 0.863 | |
| TC+CC | 87 (20.9) | 94 (22.6) | 0.89 (0.63, 1.25) | 0.488 | 0.820 |
aAdjusted for age, sex, smoking status, and occupational history.
bFalse discovery rate adjusted using the Benjamini-Hochberg procedure[26].
Table 3 shows the effect of PON1 SNPs on lung cancer risk according to smoking status. In non-smokers, the rs662 AA (192QQ) genotype exhibited a significantly reduced lung cancer risk (OR = 0.25, 95% CI: 0.06–0.98), and the lung cancer risk significantly decreased as the number of rs662 A alleles increased (p = 0.047). In current and ex-smokers, however, no statistically significant association was found. When stratified according to smoking status, the rs13306698, rs854552, rs854565 and rs854568 genotypes were not associated with lung cancer risk for any smoking status. In addition, no interaction between PON1 SNPs and smoking status was significantly associated with lung cancer risk.
Table 3. Associations between seven genetic polymorphisms of PON1 and the risk of lung cancer, according to smoking status.
| SNP ID | Genotypes | Non-smokers | Current or ex-smokers | P for interaction | ||
|---|---|---|---|---|---|---|
| Cases/controls | OR (95% CI) | Cases/controls | OR (95% CI) | |||
| rs13306698 | AA | 60/138 | 1.00 (ref.) | 294/219 | 1.00 (ref.) | 0.451 |
| AG | 11/19 | 1.44 (0.59, 3.50) | 49/39 | 0.95 (0.60, 1.51) | ||
| GG | 1/0 | - | 1/1 | 1.04 (0.07, 16.84) | ||
| AG+GG | 12/19 | 1.56 (0.65, 3.72) | 50/40 | 0.95 (0.60, 1.51) | ||
| P trend b | 0.241 | 0.839 | ||||
| rs662 | GG (RR) | 39/69 | 1.00 (ref.) | 170/111 | 1.00 (ref.) | 0.454 |
| GA (QR) | 30/70 | 0.73 (0.39, 1.37) | 140/118 | 0.78 (0.55, 1.10) | ||
| AA (QQ) | 3/18 | 0.25 (0.06, 0.98) | 34/30 | 0.72 (0.41, 1.26) | ||
| GA+AA | 33/88 | 0.63 (0.34, 1.15) | 174/148 | 0.77 (0.55, 1.07) | ||
| P trend b | 0.047 | 0.125 | ||||
| rs854552 | TT | 35/89 | 1.00 (ref.) | 199/148 | 1.00 (ref.) | 0.253 |
| TC | 30/59 | 1.49 (0.78, 2.86) | 117/90 | 0.98 (0.68, 1.39) | ||
| CC | 7/9 | 2.11 (0.65, 6.81) | 28/21 | 1.03 (0.56, 1.91) | ||
| TC+CC | 37/68 | 1.58 (0.85, 2.93) | 145/111 | 0.99 (0.71, 1.37) | ||
| P trend b | 0.122 | 0.986 | ||||
| rs854565 | GG | 45/87 | 1.00 (ref.) | 191/128 | 1.00 (ref.) | 0.443 |
| GA | 26/57 | 0.88 (0.46, 1.67) | 126/115 | 0.74 (0.52, 1.04) | ||
| AA | 1/13 | 0.12 (0.01, 1.09) | 27/16 | 1.15 (0.59, 2.24) | ||
| GA+AA | 27/70 | 0.74 (0.40, 1.38) | 153/131 | 0.79 (0.57, 1.10) | ||
| P trend b | 0.117 | 0.430 | ||||
| rs854568 | AA | 27/77 | 1.00 (ref.) | 163/121 | 1.00 (ref.) | 0.390 |
| AG | 39/68 | 1.51 (0.80, 2.85) | 143/110 | 0.98 (0.69, 1.39) | ||
| GG | 6/12 | 0.95 (0.30, 3.00) | 38/28 | 1.00 (0.58, 1.74) | ||
| AG+GG | 45/80 | 1.41 (0.76, 2.61) | 181/138 | 0.99 (0.71, 1.37) | ||
| P trend b | 0.523 | 0.968 | ||||
| rs854572 | CC | 16/46 | 1.00 (ref.) | 87/78 | 1.00 (ref.) | 0.857 |
| CG | 39/78 | 1.35 (0.64, 2.83) | 169/123 | 1.16 (0.79, 1.72) | ||
| GG | 17/33 | 1.49 (0.61, 3.63) | 88/58 | 1.30 (0.82, 2.06) | ||
| CG+GG | 56/111 | 1.39 (0.69, 2.81) | 257/181 | 1.21 (0.84, 1.74) | ||
| P trend | 0.366 | 0.262 | ||||
| rs854573 | TT | 60/123 | 1.00 (ref.) | 269/199 | 1.00 (ref.) | 0.392 |
| TC | 12/32 | 0.72 (0.32, 1.61) | 70/57 | 0.90 (0.60, 1.35) | ||
| CC | 0/2 | - | 5/3 | 1.15 (0.27, 4.91) | ||
| TC+CC | 12/34 | 0.64 (0.29, 1.43) | 75/60 | 0.92 (0.62, 1.36) | ||
| P trend | 0.195 | 0.733 | ||||
aAdjusted for age, sex, smoking status, and occupational history.
bFor the linear trend across all three genotypes.
The rs662, rs13306698, and rs854565 genotypes showed a strong LD to one another (D’ >0.9). The strongest LD was found in two non-synonymous SNPs, rs13306698 and rs662 (D’ = 0.998), which were then selected for haplotype analysis (S1 Fig.). After adjusting for age, sex, smoking status, and occupational history, the A-A haplotype was significantly associated with lung cancer risk compared with the A-G haplotype in the additive genetic model (OR = 0.77, 95% CI: 0.61–0.96). However, after stratifying by smoking status, there were no haplotypes associated with the risk of lung cancer (Table 4).
Table 4. Associations between the haplotypes of two non-synonymous SNPs of PON1 and the risk of lung cancer, according to smoking status.
| Haplotypes (rs1330669, rs662) | All subjects | Non-smokers | Current or ex-smokers | P for interaction | |||
|---|---|---|---|---|---|---|---|
| Frequency of cases/controls | OR (95% CI) a | Frequency of cases/controls | OR (95% CI) b | Frequency of cases/controls | OR (95% CI) b | ||
| A-G | 0.59/0.61 | 1.00 (ref.) | 0.66/0.60 | 1.00 (ref.) | 0.62/0.58 | 1.00 (ref.) | 0.570 |
| A-A | 0.34/0.32 | 0.77 (0.61–0.96) | 0.25/0.34 | 0.63 (0.38–1.05) | 0.30/0.34 | 0.81 (0.63–1.05) | |
| G-G | 0.07/0.07 | 0.95 (0.64–1.42) | 0.09/0.06 | 1.35 (0.57–3.18) | 0.07/0.08 | 0.89 (0.57–1.40) | |
aAdjusted for age, sex, smoking status, and occupational history.
bAdjusted for age, sex, and occupational history.
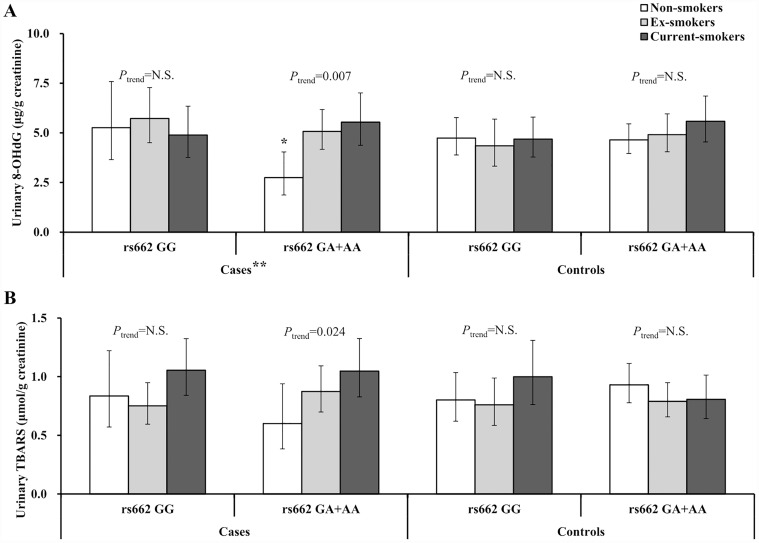
Fig. 1 presents the levels of oxidative stress biomarkers in the lung cancer patients and controls, according to PON1 rs662 SNP and smoking status. In lung cancer patients with one or two PON1 rs662 A (192Q) alleles, 8-OHdG and TBARS levels showed a significant positive exposure-response trend for smoking status (P trend = 0.007 and 0.024, respectively), but this trend was not found in lung cancer patients with the PON1 rs662 GG (192RR) genotype and controls. Among non-smoker patients, urinary 8-OHdG level was significantly lower in individuals with the rs662 GA (192RQ) and AA (192QQ) genotypes than in those with the GG (192RR) genotype. We found a significant interaction effect between PON1 rs662 SNP and smoking status on the urinary 8-OHdG level in lung cancer patients (P interaction = 0.025). These significant interactions were not identified for PON1 rs854572, rs854573, rs854552, rs854565, or rs854568 SNPs (data not shown).
Fig 1. Geometric means and 95% confidence intervals for urinary oxidative stress markers (A: 8-hydroxydeoxyguanosine, B: thiobarbituric acid reactive substances) according to smoking status and PON1 rs662 SNP in lung cancer patients and controls.
*Significant difference between the genotypes in non-smokers (P = 0.017), ** Significant interaction (SNP×smoking)(P = 0.025).
In addition, we tested the influence of the PON1 SNPs on lung cancer risk after stratification according to histological type. The odds ratio of the rs662 GA + AA (192RQ + QQ) genotype in the adenocarcinoma group (OR = 0.59, p = 0.053) was marginally significant and lower than for other histological types (squamous cell carcinoma OR = 0.76, p = 0.246; other non-small cell carcinoma OR = 0.90, p = 0.658) (data not shown).
Discussion
This case-control study found that the PON1 rs662 (R192Q) polymorphism is associated with a risk of lung cancer, especially among non-smokers. Furthermore, we observed a significant interaction between the PON1 rs662 SNP and smoking on oxidative stress in lung cancer patients.
The results of this study indicate that the individuals with one or two rs662 A (192Q) alleles of the PON1 rs662 polymorphism showed a reduced risk for lung cancer. Until now, several epidemiological studies have reported that the PON1 genetic polymorphism is associated with lung cancer risk [19–21] or reduction of lung function [29]. Among them, the two most recent studies suggested that the 192Q allele of the PON1 is a potential protective factor against lung cancer [20,21], which is in concordance with this present study. Similarly, the PON1 192Q allele has been reported to reduce the risk of bladder cancer [30], ovarian cancer [31], and B-cell lymphoma [32]; in contrast, it has been reported to increase the risk of lung [19], breast [33] and prostate cancers [34].
Cigarette smoking decreases serum PON1 activity in humans [12,17], and cigarette smoke extracts inhibit serum PON1 activity through the modification of the free thiol residue at amino acid 283 in PON [35]. In addition, a recent study reported that myeloperoxidase (MPO), a heme enzyme abundantly produced by neutrophils, inactivates PON1 functioning [36]. The number of neutrophils was found to be higher in current or ex-smokers than in non-smokers [37], with greater MPO activity in smokers than in non-smokers [38]. These results indicate that the serum PON1 activity of individuals exposed to tobacco smoke would be inhibited regardless of their PON1 genotype. This supports our data which indicate that the PON1 rs662 polymorphism was significantly associated with lung cancer risk in non-smokers, but not in ex- or current smokers. Furthermore, a similar association was also observed between the PON1 rs662 polymorphism and oxidative stress level in lung cancer patients. The PON1 rs662 A (192Q) allele was associated with a significant reduction in the urinary 8-OHdG level of non-smoking lung cancer patients, but this protective allelic effect was not observed in current or ex-smokers with lung cancer.
The effect of the PON1 rs662 polymorphism on the risk of lung cancer differed according to the smoking status. Particularly, non-smokers with PON1 rs662 AA (192QQ) genotype had a significant 75% decrease in lung cancer risk (OR = 0.25, 95% CI [0.06–0.98]), but this reduction was not observed in current or ex-smokers (OR = 0.72, 95% CI [0.41–1.26]). However, the interaction between the PON1 rs662 polymorphism and smoking habits on lung cancer risk was not statistically significant, probably owing to the relatively small sample size. Nevertheless, our results showed a clear difference in the magnitude of lung cancer risk across PON1 rs662 genotypes according to the smoking status. This is suggestive of a possible gene-environment interaction that influences lung cancer risk.
The protective effect of the PON1 rs662 SNP on 8-OHdG which could be modified by smoking habits was statistically significant only in lung cancer patients, but not in controls. This finding suggests the possibility that the PON1 192Q allele reduces the lung cancer risk of non-smoking individuals by protecting against oxidative stress [7]. However, it is not certain that the protective effect of the PON1 rs662 SNP against oxidative stress exists in the body of lung cancer patients before the start of carcinogenesis. We cannot rule out the possibility that lung cancer can change the action of the PON1 192Q enzyme on oxidative stress. These results, therefore, need to be interpreted with caution and should be further investigated with prospective studies.
PON1 is an antioxidant enzyme that can act as a scavenger for systemic oxidative stress [10–13]. Previous studies have reported associations between PON1 genetic polymorphisms or PON1 enzyme activity and oxidative stress, but the results are still inconclusive. Serum PON1 activity has been negatively correlated with urinary 8-OHdG levels in patients with Alzheimer’s disease [39] and laryngeal squamous cell carcinoma [40]. Bhattacharyya et al. found that the PON1 192 RR genotype was associated with a lower level of oxidative stress [13], and Ji et al. similarly reported that carriers of the PON1 rs662 Q allele have a significantly higher level of 8-OHdG in sperm DNA than R allele carriers [41]. In contrast, Min et al. reported that individuals carrying the PON1 192RR genotypes showed a higher level of urinary 8-OHdG than those with other genotypes [42]. Our study found that non-smoker lung cancer patients with the PON1 192RR genotype showed a significantly higher level of urinary 8-OHdG than those with the 192RQ and QQ genotypes.
The PON1 rs662 (R192Q) polymorphism significantly modifies the catalytic efficiency of PON1 in a substrate-dependent manner [12]. The PON1 192R allele hydrolyzes paraoxon and chlorpyrifos oxon more efficiently than the PON1 192Q allele in vitro, while diazoxon, sarin, and soman are hydrolyzed more rapidly by the PON1 192Q allele than the PON1 192R allele [17,43]. The PON1 192Q-alloenzyme protects low-density lipoproteins from oxidative modification more effectively when compared to the R-alloenzyme [44], and arylesterase activity of PON1 is associated with antioxidant capacity to a greater degree than with paraoxonase activity [45,46]. There is substantial evidence that the PON1 R192Q polymorphism plays an important role in enzyme activity; specifically, this genetic polymorphism contributes to HDL binding and stability of PON1 [47]. In this study, the PON1 192Q allele was associated with lower oxidative stress in non-smoking lung cancer patients, and with an overall reduction in lung cancer risk. Our previous study showed that PON1-paraoxonase activity in Koreans with the PON1 192RR genotype was higher than in those with the QQ genotype, and that PON1-arylesterase activity in those with the PON1 192QQ genotype was higher than in those with the 192RR genotype [17]. Therefore, our present findings suggest that the PON1 192Q allele might reduce lung cancer risk or oxidative stress through increased PON1-arylesterase activity.
Previous studies reported that distributions of SNPs and serum PON1 activity were not different between histological types of lung cancer [21,48]. However, in our stratified analyses according to the histological types, differing associations were observed between PON1 rs662 SNP and lung cancer risk for different histological types, although statistical significance was not reached, probably owing to the small sample size. PON1 rs662 SNP was marginally associated with lung cancer risk solely in the adenocarcinoma group, which was more common in non-smokers. Concordantly, our findings showed that the PON1 rs662 SNP was associated with a significant reduction in the lung cancer risk of non-smokers. These facts suggest that PON1 rs662 SNP may be considered as a genetic susceptibility marker for lung cancer in non-smokers.
In conclusion, the results of this study suggest that functional polymorphisms of PON1 may be associated with the risk of lung cancer and that the effect of PON1 polymorphisms on lung carcinogenesis and oxidative stress may be modulated by tobacco smoking.
Supporting Information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1120330).
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2. Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG, Lee JS (2013) Cancer statistics in korea: Incidence, mortality, survival and prevalence in 2010. Cancer Res Treat 45: 1–14. 10.4143/crt.2013.45.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahsan H, Thomas DC (2004) Lung cancer etiology: Independent and joint effects of genetics, tobacco, and arsenic. JAMA 292: 3026–3029. [DOI] [PubMed] [Google Scholar]
- 4. Alberg A, Brock M, Ford J, Samet J, Spivack S (2013) Epidemiology of lung cancer. Chest 143: e1S–e29S. 10.1378/chest.12-2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. In KH, Kwon YS, Oh IJ, Kim KS, Jung MH, Lee KH, et al. (2009) Lung cancer patients who are asymptomatic at diagnosis show favorable prognosis: A korean lung cancer registry study. Lung Cancer 64: 232–237. 10.1016/j.lungcan.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 6. Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91: 1194–1210. [DOI] [PubMed] [Google Scholar]
- 7. Valko M, Rhodes C, Moncol J, Izakovic M, Mazur M (2006) Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 160: 1–40. [DOI] [PubMed] [Google Scholar]
- 8. Halliwell B, Gutteridge JM, editors (2007) Free radicals in biology and medicine.: Oxford University Press. [Google Scholar]
- 9. Crawford A, Fassett RG, Geraghty DP, Kunde DA, Ball MJ, Robertson IK, et al. (2012) Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 501: 89–103. 10.1016/j.gene.2012.04.011 [DOI] [PubMed] [Google Scholar]
- 10. Rozenberg O, Rosenblat M, Coleman R, Shih DM, Aviram M (2003) Paraoxonase (PON1) deficiency is associated with increased macrophage oxidative stress: Studies in PON1-knockout mice. Free Radic Biol Med 34: 774–784. [DOI] [PubMed] [Google Scholar]
- 11. Costa LG, Richter RJ, Li WF, Cole T, Guizzetti M, Furlong CE (2003) Paraoxonase (PON 1) as a biomarker of susceptibility for organophosphate toxicity. Biomarkers 8: 1–12. [DOI] [PubMed] [Google Scholar]
- 12. Costa LG, Vitalone A, Cole TB, Furlong CE (2005) Modulation of paraoxonase (PON1) activity. Biochem Pharmacol 69: 541–550. [DOI] [PubMed] [Google Scholar]
- 13. Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, et al. (2008) Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA 299: 1265–1276. 10.1001/jama.299.11.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackness B, Beltran-Debon R, Aragones G, Joven J, Camps J, Mackness M (2010) Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 62: 480–482. 10.1002/iub.347 [DOI] [PubMed] [Google Scholar]
- 15. Rodrigo L, Hernandez AF, Lopez-Caballero JJ, Gil F, Pla A (2001) Immunohistochemical evidence for the expression and induction of paraoxonase in rat liver, kidney, lung and brain tissue. implications for its physiological role. Chem Biol Interact 137: 123–137. [DOI] [PubMed] [Google Scholar]
- 16. Rainwater DL, Rutherford S, Dyer TD, Rainwater ED, Cole SA, Vandeberg JL, et al. (2009) Determinants of variation in human serum paraoxonase activity. Heredity (Edinb) 102: 147–154. 10.1038/hdy.2008.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eom SY, Kim YS, Lee CJ, Lee CH, Kim YD, Kim H (2011) Effects of intronic and exonic polymorphisms of paraoxonase 1 (PON1) gene on serum PON1 activity in a korean population. J Korean Med Sci 26: 720–725. 10.3346/jkms.2011.26.6.720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang DH, Fan CH, Ji Q, Qi BX, Li J, Wang L (2012) Differential effects of paraoxonase 1 (PON1) polymorphisms on cancer risk: Evidence from 25 published studies. Mol Biol Rep 39: 6801–6809. 10.1007/s11033-012-1505-3 [DOI] [PubMed] [Google Scholar]
- 19. Lee CH, Lee KY, Choe KH, Hong YC, Kim YD, Kang JW, et al. (2005) Effects of oxidative DNA damage induced by polycyclic aromatic hydrocarbons and genetic polymorphism of the paraoxonase-1 (PON1) gene on lung cancer. J Prev Med Public Health 38: 345–350. [PubMed] [Google Scholar]
- 20. Aksoy-Sagirli P, Cakmakoglu B, Isbir T, Kaytan-Saglam E, Kizir A, Topuz E, et al. (2011) Paraoxonase-1 192/55 polymorphisms and the risk of lung cancer in a turkish population. Anticancer Res 31: 2225–2229. [PubMed] [Google Scholar]
- 21. Wang H, Li L, Ding L, Zhang Z, Pu C (2012) Association of genetic polymorphisms in the paraoxonase 1 gene with the risk and prognosis of non-small cell lung cancer in chinese han population. J Investig Med 60: 592–597. 10.231/JIM.0b013e318245d557 [DOI] [PubMed] [Google Scholar]
- 22. Harik-Khan RI, Wise RA, Fozard JL (1998) Determinants of maximal inspiratory pressure. the baltimore longitudinal study of aging. Am J Respir Crit Care Med 158: 1459–1464. [DOI] [PubMed] [Google Scholar]
- 23. Kang HJ, Choi KO, Kim BD, Kim S, Kim YJ (2005) FESD: A functional element SNPs database in human. Nucleic Acids Res 33: D518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agarwal R, Chase SD (2002) Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci 775: 121–126. [DOI] [PubMed] [Google Scholar]
- 25. Purcell S, Cherny SS, Sham PC (2003) Genetic power calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150. [DOI] [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol: 289–300. [Google Scholar]
- 27. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 28. Sole X, Guino E, Valls J, Iniesta R, Moreno V (2006) SNPStats: A web tool for the analysis of association studies. Bioinformatics 22: 1928–1929. [DOI] [PubMed] [Google Scholar]
- 29. Seo T, Pahwa P, McDuffie HH, Nakada N, Goto S, Ghosh S, et al. (2008) Interactive effect of paraoxonase-1 Q192R polymorphism and smoking history on the lung function decline in grain workers. Ann Epidemiol 18: 330–334. [DOI] [PubMed] [Google Scholar]
- 30. Ozturk O, Kagnici OF, Ozturk T, Durak H, Tuzuner BM, Kisakesen HI, et al. (2009) 192R allele of paraoxanase 1 (PON1) gene as a new marker for susceptibility to bladder cancer. Anticancer Res 29: 4041–4046. [PubMed] [Google Scholar]
- 31. Lurie G, Wilkens LR, Thompson PJ, McDuffie KE, Carney ME, Terada KY, et al. (2008) Genetic polymorphisms in the paraoxonase 1 gene and risk of ovarian epithelial carcinoma. Cancer Epidemiol Biomarkers Prev 17: 2070–2077. 10.1158/1055-9965.EPI-08-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conesa-Zamora P, Ruiz-Cosano J, Torres-Moreno D, Español I, Gutiérrez-Meca MD, Trujillo-Santos J, et al. (2013) Polymorphisms in xenobiotic metabolizing genes (EPHX1, NQO1 and PON1) in lymphoma susceptibility: A case control study. BMC Cancer 13: 228 10.1186/1471-2407-13-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saadat M (2012) Paraoxonase 1 genetic polymorphisms and susceptibility to breast cancer: A meta-analysis. Cancer epidemiol 36: e101–e103. 10.1016/j.canep.2011.10.015 [DOI] [PubMed] [Google Scholar]
- 34. Antognelli C, Mearini L, Talesa VN, Giannantoni A, Mearini E (2005) Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. Prostate 63: 240–251. [DOI] [PubMed] [Google Scholar]
- 35. Nishio E, Watanabe Y (1997) Cigarette smoke extract inhibits plasma paraoxonase activity by modification of the enzyme’s free thiols. Biochem Biophys Res Commun 236: 289–293. [DOI] [PubMed] [Google Scholar]
- 36. Huang Y, Wu Z, Riwanto M, Gao S, Levison BS, Gu X, et al. (2013) Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J Clin Invest 123: 3815–3828. 10.1172/JCI67478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parry H, Cohen S, Schlarb JE, Tyrrell DA, Fisher A, Russell MA, et al. (1997) Smoking, alcohol consumption, and leukocyte counts. Am J Clin Pathol 107: 64–67. [DOI] [PubMed] [Google Scholar]
- 38. Loke WM, Lam KM, Chong WL, Chew SE, Quek AM, Lim ECh, et al. (2012) Products of 5-lipoxygenase and myeloperoxidase activities are increased in young male cigarette smokers. Free Radic Res 46: 1230–1237. 10.3109/10715762.2012.701291 [DOI] [PubMed] [Google Scholar]
- 39. Zengi O, Karakas A, Ergun U, Senes M, Inan L, Yucel D (2012) Urinary 8-hydroxy-2′-deoxyguanosine level and plasma paraoxonase 1 activity with alzheimer’s disease. Clin Chem Lab Med 50: 529–534. 10.1515/CCLM.2011.792 [DOI] [PubMed] [Google Scholar]
- 40. Karaman E, Uzun H, Papila I, Balci H, Ozdilek A, Genc H, et al. (2010) Serum paraoxonase activity and oxidative DNA damage in patients with laryngeal squamous cell carcinoma. J Craniofac Surg 21: 1745–1749. 10.1097/SCS.0b013e3181f4040a [DOI] [PubMed] [Google Scholar]
- 41. Ji G, Gu A, Wang Y, Huang C, Hu F, Zhou Y, et al. (2012) Genetic variants in antioxidant genes are associated with sperm DNA damage and risk of male infertility in a chinese population. Free Radic Biol Med 52: 775–780. 10.1016/j.freeradbiomed.2011.11.032 [DOI] [PubMed] [Google Scholar]
- 42. Min J, Park H, Park B, Kim YJ, Park J, Lee H, et al. (2006) Paraoxonase gene polymorphism and vitamin levels during pregnancy: Relationship with maternal oxidative stress and neonatal birthweights. Reprod Toxicol 22: 418–424. [DOI] [PubMed] [Google Scholar]
- 43. Costa LG, Richter RJ, Li WF, Cole T, Guizzetti M, Furlong CE (2003) Paraoxonase (PON 1) as a biomarker of susceptibility for organophosphate toxicity. Biomarkers 8: 1–12. [DOI] [PubMed] [Google Scholar]
- 44. Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN (1998) Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modification. FEBS Lett 423: 57–60. [DOI] [PubMed] [Google Scholar]
- 45. Aydin O, Yacinkaya S, Eren E, Ergin M, Eroglu M, Yilmaz N (2013) Diminished arylesterase enzyme activity and total thiol levels in bladder cancer patients. Clin Lab 59:1231–1237. [DOI] [PubMed] [Google Scholar]
- 46. Kurtul N, Söylemez S, Çelik M (2014) Plasma paraoxonase and arylesterase activities in smokers and smokeless tobacco users as maras powder. Inhal Toxicol 26: 235–239. 10.3109/08958378.2013.878007 [DOI] [PubMed] [Google Scholar]
- 47. Gaidukov L, Rosenblat M, Aviram M, Tawfik DS (2006) The 192R/Q polymorphs of serum paraoxonase PON1 differ in HDL binding, lipolactonase stimulation, and cholesterol efflux. J Lipid Res 47: 2492–2502. [DOI] [PubMed] [Google Scholar]
- 48. Elkiran ET, Mar N, Aygen B, Gursu F, Karaoglu A, Koca S (2007) Serum paraoxonase and arylesterase activities in patients with lung cancer in a turkish population. BMC Cancer 7: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.