Abstract
Background/Aims
This study was conducted to identify microRNAs (miRNAs) that are differentially expressed in Helicobacter pylori-infected patients with an intestinal type of gastric cancer using miRNA microarray and to confirm the candidate miRNA expression levels.
Methods
Total RNA was extracted from the cancerous and noncancerous regions of formalin-fixed, paraffin-embedded tissues of H. pylori-positive (n=8) or H. pylori-negative (n=8) patients with an intestinal type of gastric cancer. RNA expression was analyzed using a 3,523 miRNA profiling microarray based on the Sanger miRBase. Validation analysis was performed using TaqMan miRNA assays.
Results
A total of 219 miRNAs in the aberrant miRNA profiles across the miRNA microarray showed at least a 2-fold change differential expression in H. pylori-positive and H. pylori-negative cancer tissues. After candidate miRNAs were selected using online miRNA databases, TaqMan miRNA assays confirmed that three miRNAs (miR-99b-3p, miR-564, and miR-638) were significantly increased in three H. pylori-positive cancer tissues compared to the H. pylori-negative cancer tissues. Additionally, four miRNAs (miR-204-5p, miR-338-5p, miR-375, and miR-548c-3p) were significantly increased in H. pylori-negative cancer tissues compared to H. pylori-positive cancer tissues.
Conclusions
miRNA expression in the intestinal type of H. pylori infection-dependent gastric cancer suggests that different gastric cancer pathogenesis mechanisms could exist between H. pylori-positive and H. pylori-negative gastric cancer. Additional functional studies are required.
Keywords: Gastric carcinogenesis, Helicobacter pylori, Microarray, MicroRNAs, TaqMan miRNA assays
INTRODUCTION
About 20 length nucleotides of nonprotein coding microRNA (miRNA) regulates gene expression by hybridizing to the 3′ untranslated region of specific messenger RNA (mRNA) targets. Up to now, approximately 1,600 human miRNAs have been studied and the relationship of miRNAs with human diseases is being intensively studied.1,2
Microarray-based hybridization profiling is a powerful technique for screening and use of fresh tissue specimens recommended to reduce collateral damage of miRNAs. However, it is hard to get fresh tissue specimens right after operations. Therefore, it is helpful to use the miRNA preserving formalin-fixed paraffin-embedded (FFPE) tissue for miRNA screening analysis. It has been used for extraction of miRNAs and a few studies have examined the correlation with clinical data.3,4 In addition, a strong correlation in global miRNA expression between fresh frozen and FFPE human cancer samples has been revealed.5
Epidemiological studies have implicated that colonization of the stomach by Helicobacter pylori is a risk for various development of gastric diseases including gastric cancer.6 Given the possible pathogenesis of gastric cancer, abnormal miRNA expression after H. pylori infection might cause unregulated inflammation response and immune system disruption.7
The occurrence of gastric cancer still remains very high in much of Asia and especially, à Laurens’ classification divided gastric cancer into two histological main types; intestinal-type and diffuse-type,8,9 in which the number of intestinal-type gastric cancer patients in South Korea is more prevalent than the other type.10
On the other hand, 5.3% of gastric cancer patients were not infected with H. pylori in Korea.11 Few articles noted that different genetic background in gastric mucosa depending on H. pylori infection and a recent study has reported that miRNA expression patterns in H. pylori-infected and -uninfected gastric normal mucosa were different.12–14 Thus different miRNAs could be involved in gastric carcinogenesis depending on H. pylori as well. However, no study has ever reported regarding distinct miRNA profiles in intestinal type of gastric cancer FFPE specimens depending on H. pylori infection. Furthermore, it has not been determined whether miRNA expression patterns which are different in this gastric cancer depending on H. pylori infection are different or not in normal gastric mucosa.
From this background, the present study was undertaken to identify whether miRNA expression profiles differ between H. pylori-positive and -negative FFPE specimens in the intestinal type of gastric cancer patients using miRNA microarray. The microarray results were confirmed by TaqMan miRNA assays.
MATERIALS AND METHODS
1. Subjects
Sixteen gastric cancer patients matched for age, sex, and H. pylori status, who received curative operation at Seoul National University Bundang Hospital, were included for microarray study. Table 1 shows the baseline characteristics of the study subjects. All subjects of this study received gastroscopy for gastric cancer screening and conformation of histological gastric adenocarcinoma diagnosis. For TaqMan miRNA assays, gastric cancer tissue was retrieved from gastric cancer patients who received endoscopic submucosal dissection with current H. pylori (n=28) or without any H. pylori infection evidence at all (n=24), including 16 patients in whom microarray experiments were performed. In addition, gastric body tissue was obtained from controls with current H. pylori (n=24) or without any H. pylori infection evidence at all (n=24). The subjects who underwent gastroscopy and H. pylori gastric cancer screening but did not show any significant gastroduodenal diseases, such as gastric cancer, dysplasia, mucosa-associated lymphoid tissue lymphoma, esophageal cancer or peptic ulcer disease were enrolled into the control group. The study protocol was approved by the Ethics Committee of Seoul National University Bundang Hospital (Institutional Review Board number: B-1301-186-111). All participants provided their written informed consent to participate in this study.
Table 1.
Characteristics of the Subjects Used in the MicroRNA Microarray and TaqMan Assay Analyses
| miRNA microarray | TaqMan miRNA assay | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Hp- GC (n=8) | Hp+ GC (n=8) | Hp- Cont. (n=24) | Hp+ Cont. (n=24) | Hp- GC (n=24) | Hp+ GC (n=28) | |
| Age, yr | 69.3±1.7 | 67.8±3.4 | 60.1±11.0 | 61.4±8.5 | 66.5±8.8 | 67.3±8.1 |
| Male sex | 6 (75) | 5 (62.5) | 17 (70.8) | 15 (62.5) | 19 (79.2) | 18 (64.3) |
| Intestinal type histology | 8 (100.0) | 8 (100.0) | - | - | 24 (100.0) | 28 (100.0) |
Data are presented as mean±standard error or number (%). Hp+, current active Helicobacter pylori infection if any one of these endoscopy-based tests are positive (Campylobacter-like organism [CLO] test, culture, and histology); Hp-, the absence of H. pylori by both endoscopy-based tests and serological testing.
miRNA, microRNA; Hp, Helicobacter pylori; GC, gastric cancer group; Cont., noncancer controls.
2. H.pylori testing
Three types of H. pylori testing (histology, Campylobacter-like organism [CLO]-test, and culture) were conducted in both the antrum and the body as previously described.15 If one of the three examinations was positive, the patients were H. pylori-positive. If all three tests were negative then H. pylori serology test was performed using anti-H. pylori immunoglobulin G in an enzyme-linked immunosorbant assay (Green Cross Medical Science, Eumsung, Korea). Intestinal metaplasia (IM) was graded according to the modified Sydney system in hematoxylin and eosin stained tissue.16 All of the 48 H. pylori-negative cases (24 controls and 24 patients with gastric cancer) were sero-negative and IM grade was absent.
3. RNA isolation and miRNA microarray analysis
After manual dissection under microscopic guidance avoiding the contamination of inflammatory cells and stromal cells, hematoxylin and eosin stained sections 50 μm in thickness from cancerous and noncancerous regions of intestinal type of gastric cancer FFPE samples were reviewed by one pathologist (H.S.L.). Each section was incubated in xylene and total RNA was extracted using a RecoverAll™ Total Nucleic Acid Isolation kit (Life Technologies, Carlsbad, CA, USA). Each 400 ng RNA was dephosphorylated with 15 units of calf intestine alkaline phosphatase, followed by RNA denaturation with 40% dimethylsulfoxide. Dephosphorylated RNA was ligated with pCp-Cy3 mononucleotide and resuspended in Gene Expression Blocking Reagent and Hi-RPM Hybridization buffer. The denatured, labeled samples were pipetted onto assembled Agilent Human miRNA microarray Release 16.0 platform and hybridized at 55°C for 20 hours at 20 rpm. The hybridization images were analyzed using a DNA microarray scanner (Agilent Technologies, Palo Alto, CA, USA). The average fluorescence intensity for each spot was calculated and local background was subtracted. Data visualization and analysis were performed with GeneSpring GX 7.3 software (Agilent Technologies). Signal cutoff measurements were <0.01.
4. Selection of miRNA candidates
The microarray showed 219 miRNAs which were at least 2-fold change between the H. pylori-positive and -negative cancer. Only 37 miRNAs were statistical significance (p<0.05) and those miRNAs were compared with miRNAs of Validated Targets associated with gastric cancer in miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk)17 and HMDD v2.0 (http://202.38.126.151/hmdd/tools/hmdd2.html).18 Specifically, 19 miRNAs which were highly expressed in H. pylori-negative gastric cancer and 18 miRNAs which were highly expressed in H. pylori-positive gastric cancer were compared with target miRNAs of “stomach neoplasms” in each site. At last, 2-fold changed and statistically significant seven miRNAs which have been reported to be associated with gastric cancer were selected for next validation assay.
5. TaqMan miRNA validation assay
miRNA was extracted from the frozen gastric cancer tissue in gastric cancer patients and gastric body in control cases, which had been obtained during gastroscopy and had been kept at −80°C, with a mirVana™ miRNA Isolation Kit (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed using 5 μL of miRNA and TaqMan MicroRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied Biosystems, Foster City, CA, USA). The assays were carried in duplicate. The 20 μL reaction mixture contained reverse transcription reaction product, TaqMan Universal PCR Master Mix without uracil-N-glycosylase, TaqMan miRNA assay mix, and nuclease-free water. Amplification was performed using the 7500/7500 Fast PCR system (Applied Biosystems) at 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Amplification signals were computed with 7500 software v2.0.6 (Applied Biosystems). Relative miRNA expression levels are presented as 2−ΔΔCt method.
6. Statistical analysis
No interarray normalization was applied on the array, because the similarity between matched normal and cancer sample arrays was unknown. To identify distinct miRNAs hybridization signals, one-way analysis of variance (ANOVA) (p<0.05) and multiple testing correction (Benjamini and Hochberg false discovery rate) were employed for microarray clustering analysis. Relationship between two assays was evaluated by calculation of Spearman correlation tests. All statistical analyses were performed using the SPSS version 13.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
1. miRNA microarray data analysis
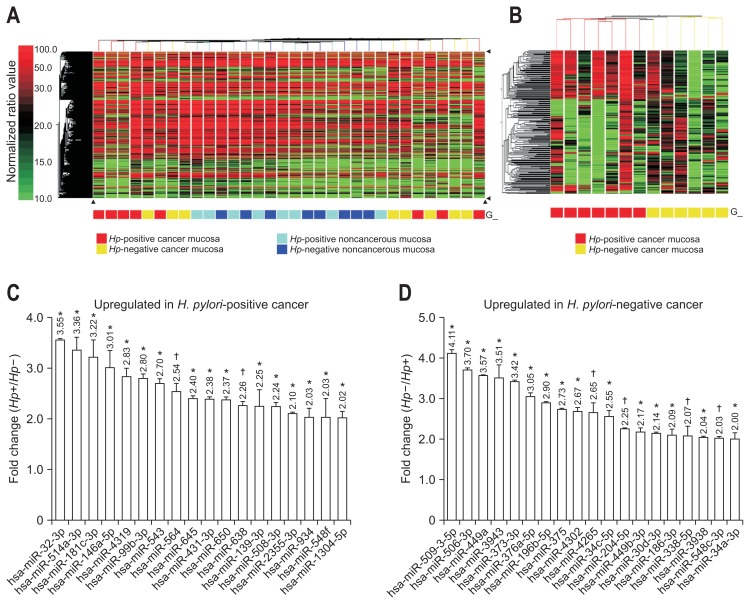
In all FFPE individual samples, the low intensity hybridization signals (<0.01) between miRNAs and probes were filtered out and 1,781 of 3,523 (50.55%) miRNA probes remained as the dataset and were used for further analysis. Among the four subdivided groups (current H. pylori-positive cancer or noncancerous region, and H. pylori-negative cancer or noncancerous region), unsupervised hierarchical clustering of hybridization values showed clustering trends of the cancerous and noncancerous groups (Fig. 1A). Especially, no interarray normalization on normalized ratio and multiple testing corrections were used for getting rid of discordant three samples’ hybridization signals and finally, clustering result of Fig. 1B shows 219 miRNAs which displayed at least a 2-fold changed expression between H. pylori-positive and -negative cancerous tissues. Among them, one-way ANOVA showed 19 miRNAs which were upregulated in H. pylori-negative cancerous tissue (p<0.05) and 18 miRNAs which were upregulated in H. pylori-positive cancerous tissue (p<0.05) (Table 2, Fig. 1C and D).
Fig. 1.
Unsupervised hierarchical clustering analysis of gastric cancer formalin-fixed paraffin-embedded tissues. (A) A total of 1,781 microRNA (miRNA) probes were identified as differentially expressed between the cancerous and noncancerous regions of patients with gastric cancer. The higher normalized ratio value denotes higher expression levels of the miRNAs. (B) Two subclasses, Helicobacter pylori (Hp)-positive (n=7) and H. pylori-negative (n=6), showed clustering results of 219 miRNAs that exhibited 2-fold changes in gastric cancer samples. (C) A fold-change graph of 18 miRNAs upregulated in H. pylori-positive cancer. (D) A fold-change graph of 19 miRNAs upregulated in H. pylori-negative cancer.
*p<0.05; †p<0.01.
Table 2.
Differential Expression of 37 MicroRNAs between the H. pylori-Positive and H. pylori-Negative Gastric Cancer Tissue Samples
| miRNA | Fold change* | p-value† | miRNA | Fold change | p-value | ||
|---|---|---|---|---|---|---|---|
| Up in Hp+ GC‡ | hsa-miR-32-3p | 3.55 | <0.0213 | Up in Hp- GC§ | hsa-miR-509-3-5p | 4.11 | <0.0102 |
| hsa-miR-514a-3p | 3.36 | <0.0193 | hsa-miR-506-3p | 3.70 | <0.0193 | ||
| hsa-miR-181c-3p | 3.22 | <0.0168 | hsa-miR-449a | 3.57 | <0.015 | ||
| hsa-miR-146a-5p | 3.01 | <0.0279 | hsa-miR-3943 | 3.51 | <0.0458 | ||
| hsa-miR-4319 | 2.83 | <0.0236 | hsa-miR-373-3p | 3.42 | <0.0085 | ||
| hsa-miR-99b-3p | 2.80 | <0.0446 | hsa-miR-376a-5p | 3.05 | <0.0244 | ||
| hsa-miR-543 | 2.70 | <0.0227 | hsa-miR-196b-5p | 2.90 | <0.0169 | ||
| hsa-miR-564 | 2.54 | <0.0055 | hsa-miR-375 | 2.73 | <0.0182 | ||
| hsa-miR-645 | 2.40 | <0.0322 | hsa-miR-4302 | 2.67 | <0.0406 | ||
| hsa-miR-431-3p | 2.38 | <0.0268 | hsa-miR-4265 | 2.65 | <0.0368 | ||
| hsa-miR-650 | 2.37 | <0.0312 | hsa-miR-34c-5p | 2.55 | <0.0049 | ||
| hsa-miR-638 | 2.26 | <0.006 | hsa-miR-204-5p | 2.25 | <0.0076 | ||
| hsa-miR-139-3p | 2.25 | <0.0273 | hsa-miR-449b-3p | 2.17 | <0.024 | ||
| hsa-miR-508-3p | 2.24 | <0.0309 | hsa-miR-30d-3p | 2.14 | <0.0366 | ||
| hsa-miR-2355-3p | 2.10 | <0.0394 | hsa-miR-186-3p | 2.09 | <0.0233 | ||
| hsa-miR-934 | 2.03 | <0.021 | hsa-miR-338-5p | 2.07 | <0.0445 | ||
| hsa-miR-548f | 2.03 | <0.0292 | hsa-miR-3938 | 2.04 | <0.0237 | ||
| hsa-miR-1304-5p | 2.02 | <0.0363 | hsa-miR-548c-3p | 2.03 | <0.0078 | ||
| hsa-miR-34a-3p | 2.00 | <0.0428 |
miRNA, microRNA; Hp, Helicobacter pylori; GC, gastric cancer group.
Fold change denotes reciprocal ratio, Hp+ GC/Hp- GC and Hp- GC/Hp+ GC. More than 2-fold changed hybridization signals were reserved for candidate miRNAs;
Differences in fold-change were considered statistically significant if the p-value of the one-way analysis of variance was <0.05;
Up in Hp+ GC, upregulated in the cancerous regions of H. pylori-positive cancer;
Up in Hp- GC, upregulated in the cancerous regions of H. pylori-negative cancer.
2. Selection of promising candidate miRNAs
Next, miRNA candidates which might be associated with gastric cancer were selected for validation assay. When 18 highly expressed miRNAs in H. pylori-positive cancer were compared with “disease target miRNAs,” 6 and 5 miRNAs were in agreement with target miRNAs of “stomach neoplasms” in miRWalk and HMDD, respectively. Then duplicated three miRNAs; hsa-miR-99b-3p, hsa-miR-564, and hsa-miR-638, were conserved (Table 3). In the same manner, among 19 highly expressed miRNAs in H. pylori-negative cancer, 8 and 9 miRNAs were matched with target miRNAs of “stomach neoplasms” in miR-Walk and HMDD v2.0. Overlapped four miRNAs; hsa-miR-204-5p, hsa-miR-338-5p, hsa-miR-375, and hsa-miR-548c-3p, were conserved.
Table 3.
List of the Validation Assay Targets
| Stomach neoplasms* | Assay ID‡ | Sequence | |||
|---|---|---|---|---|---|
|
| |||||
| miRWalk | HMDD | Conserved miRNAs† | |||
| Up in Hp+ GC | hsa-miR-32-3p | hsa-miR-99b-3p | hsa-miR-99b-3p | 002196 | CAAGCUCGUGUCUGUGGGUCCG |
| hsa-miR-99b-3p | hsa-miR-146a-5p | hsa-miR-564 | 001531 | AGGCACGGUGUCAGCAGGC | |
| hsa-miR-139-3p | hsa-miR-181c-3p | hsa-miR-638 | 001582 | AGGGAUCGCGGGCGGGUGGCGGCCU | |
| hsa-miR-564 | hsa-miR-564 | ||||
| hsa-miR-638 | hsa-miR-638 | ||||
| hsa-miR-650 | |||||
| Up in Hp- GC | hsa-miR-34a-3p | hsa-miR-30d-3p | hsa-miR-204-5p | 000508 | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-34c-5p | hsa-miR-186-3p | hsa-miR-338-5p | 002658 | AACAAUAUCCUGGUGCUGAGUG | |
| hsa-miR-196b-5p | hsa-miR-204-5p | hsa-miR-375 | 000564 | UUUGUUCGUUCGGCUCGCGUGA | |
| hsa-miR-204-5p | hsa-miR-338-5p | hsa-miR-548c-3p | 0001590 | CAAAAAUCUCAAUUACUUUUGC | |
| hsa-miR-338-5p | hsa-miR-375 | ||||
| hsa-miR-373-3p | hsa-miR-376a-5p | ||||
| hsa-miR-375 | hsa-miR-449a | ||||
| hsa-miR-548c-3p | hsa-miR-449b-3p | ||||
| hsa-miR-548c-3p | |||||
miRNA, microRNA; Hp, Helicobacter pylori; GC, gastric cancer group.
The miRNA list of stomach neoplasms denotes the validated targets of two databases, miRWalk and HMDD. The validation study was conducted using seven conserved miRNAs;
Conserved miRNAs denotes overlapping miRNAs in the screened 37 miRNAs and the miRNA list from miRWalk and HMDD;
ID of TaqMan miRNA assays in miRBase version 19.0. RNU6B was used as an endogenous control.
Finally, we used seven commercially available primers in TaqMan miRNA assays to confirm the miRNA microarray results (Table 3).
3. Correlation of candidate miRNAs and validation study by TaqMan miRNA assay
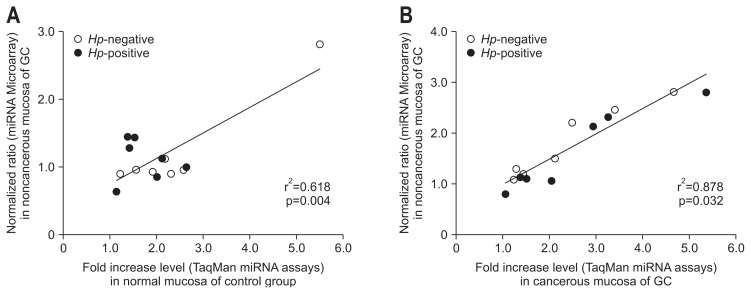
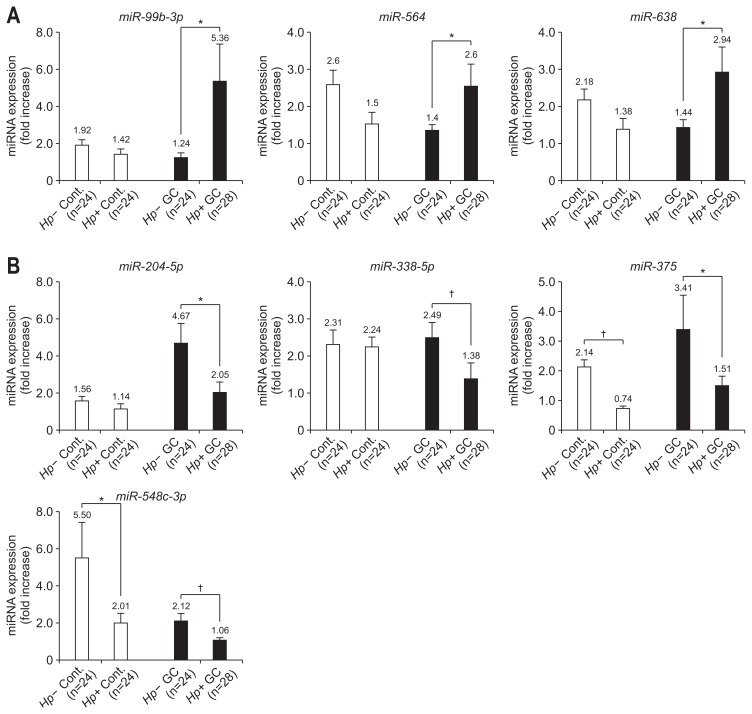
All TaqMan miRNA assays examined the fold-change of absolute expression levels of candidate miRNAs of each sample. To identify the correlation between the two assays, we compared the normalized ratio of the miRNA microarray hybridization signal and the fold-increase levels of the TaqMan miRNA assay. Although these two assays showed a low correlation (r2=0.618, p=0.004) (Fig. 2A) with H. pylori-positive and -negative control groups, there was a considerably high correlation (r2=0.878, p=0.032) (Fig. 2B) in cancer groups regardless of H. pylori-positive or -negative status. The miRNA expression in control subjects was not significantly different, except for miR-375 and miR-548c-3p, irrespective of H. pylori infection. However, the expression level of miR-99b-3p, miR-564, and miR-638 in the H. pylori-positive cancer increased 4.32-, 2.53-, and 2.04-fold compared to -negative cancer, respectively (Fig. 3A). Also, the expression level of miR-204-5p, miR-338-5p, miR-375, and miR-548-3p increased 2.28-, 1.81-, 2.25-, and 2.00-fold, respectively, in the H. pylori-negative cancer group compared to the- positive cancer group (Fig. 3B).
Fig. 2.
Correlation analysis representing the corresponding fold-increase levels from the TaqMan miRNA assays for the same microRNAs (miRNAs) in the miRNA microarray. (A) Between noncancerous mucosa from Helicobacter pylori (Hp)-positive and H. pylori-negative gastric cancer groups and normal mucosa of H. pylori-positive and H. pylori-negative control groups. (B) Between cancerous mucosa of H. pylori-positive and H. pylori-negative cancer groups. r2, Spearman correlation coefficient; p, value of paired Student t-test.
GC, gastric cancer group.
Fig. 3.
MicroRNA (miRNA) expression levels of seven miRNAs in the cancer and control groups. (A) The miRNA expression levels of three miRNAs overexpressed in the Helicobacter pylori-positive cancer group. (B) The miRNA expression levels of four miRNAs overexpressed in the H. pylori-negative cancer group.
Cont., noncancer control groups; GC, gastric cancer group; Hp-, H. pylori-negative; Hp+, H. pylori-positive. *p<0.05; †p<0.01.
DISCUSSION
There are some concerns about the integrity of miRNA from FFPE specimens and its suitability in the microarray assay because RNA in the FFPE tissue is fragmented and might be modified in the chemical reaction.19 However, it is reported that miR-NA is so small (about 20 nucleotides) that it cannot be degraded in FFPE preparation.3,4 And commercially available microarray platforms help to profile miRNA expression. Furthermore, there were a report which has shown a strong correlation in global miRNA expression between fresh frozen and FFPE human cancer samples using miRNA microarray platforms.3–5 Supporting miRNA preserving characteristics of FFPE samples, our FFPE samples showed biologically-useful RNA integrity and optical density value (RIN=2.26±0.04 and OD260/280=1.99±0.01, data not shown) in all eight paired RNAs. We found different result of cancerous region from noncancerous regions in the FFPE samples, and screened 219 miRNAs depending on H. pylori infection. For the clear results, multiple testing corrections were employed to minimize the probability of happening of these clustering results by chance originated from FFPE samples.
The miRWalk and HMDD v2.0 were employed to select candidate RNAs from 2-fold changed statistically significant 37 miRNAs. These two databases were created to offer a platform to scrutinize the mechanisms of miRNAs in disease.17,18 miRWalk has a comprehensive database of miRNAs on experimentally validated miRNA targets associated with 549 diseases.17 Also HMDD v2.0 provides experimentally supported miRNA and 397 diseases association data.18 Because the aim of our study was screening and selecting specific miRNAs in gastric cancer before functional study, we found seven candidate miRNAs after comparing screened results with validated miRNA lists of “stomach neoplasms.”
These seven miRNAs were presently validated with a good correlation (r2=0.878, p=0.032) between the microarray and TaqMan miRNA assays. Similarly, a few studies also reported similar correlation tendency between Agilent miRNA microarrays and TaqMan miRNA assays, with r2>0.9 and r2=0.83.20,21 Interestingly, this good correlation between microarray assay and TaqMan assay was not observed in normal mucosa of control group. The miRNA profile of noncancerous mucosa in cancer patients was not same as that of control patients22,23 and we also got different hybridization signal between cancerous and noncancerous mucosa in the cancer patients. This less correlation in the normal mucosa of controls may be attributed to different molecular background. Anyway, in contrast to the different expression of microRNA between cancer tissue of H. pylori-positive and -negative cancer patients the miRNA expression in control subjects was not different, except for miR-375 and miR-548c-3p, irrespective of H. pylori infection. These results might support the specificity of the different expression of seven miRNAs in the cancer tissue depending on H. pylori infection.
All of the seven miRNA candidates have been previously identified as potential regulator in gastric cancer aside from H. pylori infection. Interestingly, we found that three miRNAs (miR-99b-3p, miR-564, and miR-638) are strongly expressed in the H. pylori-positive gastric cancer tissue than H. pylori-negative. miR-99b-3p has been reported to be overexpressed in gastric cancer and a possible link between miR-99b-3p and the epithelial-to-mesenchymal transition (EMT) induced by transforming growth factor β has been suggested.24,25 As this miRNA was enhanced, especially, in the H. pylori-positive gastric cancer tissue in the present study miR-99b-3p might play a role in the H. pylori-positive gastric cancer tissue in terms of metastasis of gastric cancer by means of EMT. Now, we are investigating the role of miR-99b-3p on the EMT in the H. pylori-positive gastric cancer. Another candidate miR-564 was reported to control malignant phenotypes of gastric cancer cells.26 And its significantly suppressed expression levels in tumor material and in blood samples of gastric cancer patients has suggested that dysfunction of this miRNA may lead to gastric cancer.27,28 Our result that miR-564 has been overexpressed especially in the H. pylori-infected gastric cancer suggests that H. pylori infection somehow provokes miR-564 resulting to gastric cancer. In case of miR-638 its relationship with gastric cancer was not certain, so far. That is, the miR-638 expression level of gastric cancer mucosa was lower than normal mucosa.29 However, miR-638 has been reported to be a specific oncomir in gastric cancer.30 The present study result strongly supports a relationship between miR-638 and H. pylori-positive gastric cancer. Further functional analyses are necessary to uncover the relationships between these miRNAs and gastric carcinogenesis depending on H. pylori infection.
Other four miRNAs (miR-204-5p, miR-338-5p, miR-375, and miR-548c-3p) were strongly expressed in the H. pylori-negative gastric cancer tissue than H. pylori-positive. In case of miR-204-5p its downregulated expression has caused to overexpression of ezrin target and Ras activation, which promoted development of gastric cancer.31 Interestingly, it increased in intestinal metaplastic gland tissue after H. pylori eradication, suggesting a negative relationship with H. pylori infection.32 And our study result could support this report. However, it also needs functional study. Recent study has identified a miRNA signature of miR-338-5p for overall survival and relapse-free survival of gastric cancer patients.33 Deregulated miR-338 in drug resistant cell and its higher expression in gastric cancer patients show a possible involvement of miR-338 in gastric cancer pathogenesis.29,34 miR-338-5p overexpression could be related with poor prognosis of H. pylori-negative cancer than H. pylori-positive although it needs further study.11,35
Especially, a significant inverse correlation between miR-375 expression and JAK2 protein level in gastric cancer and a suppression of potent antiapoptotic 14-3-3ζ, PDK1 and Akt phosphorylation in miR-375 transfected cell were reported.36,37 These data suggest that it may function as a tumor suppressor. Unexpectedly, as miR-375 has been overexpressed in the H. pylori-negative gastric cancer in the present study it is rather puzzling and challenging. In the future it also needs functional study. Especially, miR-548c-3p overexpression was reported in blood of gastric cancer patients.24,38,39 However, this report did not classify this result depending on H. pylori-positivity. According to our study, this higher expression of miR-548c-3p might be more definite in case of H. pylori-negative gastric cancer. Taken together all of these results suggest that miRNAs might be differently involved in gastric carcinogenesis depending on H. pylori infection. Also, to the best of our knowledge, the present study is the first to address the different expression patterns of these seven miRNAs in terms of H. pylori infection in gastric cancer and noncancer control patients.
This study has a few limitations. First, microarray experiments were performed in the only 16 cancer patients. However, our data showed good correlation between the microarray and TaqMan miRNA assay. In addition, we tried to leave little sample-to-sample variation in this validation assay by distinguishing the patients according not only to the H. pylori infection but also in an intestinal metaplasia and all cases were intestinal type of cancer. Secondly, normal control groups were not included in the miRNA microarray in this study. The main reason was the difficulty of FFPE gastric tissue of normal control. Instead, we screened miRNAs displaying more than 2-fold altered expression level between cancerous and noncancerous tissues in the same host. Then, we included normal gastric mucosa in the validation study step to reinforce microarray results, in comparison to miRNA expression patterns with cancer tissue and normal mucosa of the control group depending on H. pylori infection. Interestingly, there was a significant difference in cancer tissue between H. pylori-positive and -negative, but not in the normal mucosa, with the exception of miR-375 and miR-548c-3p.
In conclusion, with the findings of microRNA microarray using FFPE specimens, the results of this study demonstrated that there are somewhat different miRNA expression patterns in cancerous region of intestinal type gastric cancer depending on H. pylori infection. And validation assay with selected seven candidate miRNAs confirmed the different expression of miR-NAs. Functional study is in progress to identify how these seven miRNAs could lead to different gastric carcinogenesis between H. pylori-positive and -negative cancer and what target genes would be regulated.
ACKNOWLEDGEMENTS
This work was supported by the Global Core Research Center (GCRC) grant (2011-0030001) from the National Research Foundation (NRF), Ministry of Education, Science and Technology (MEST), Republic of Korea.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 2.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glud M, Klausen M, Gniadecki R, et al. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129:1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomura AM, Perez-Perez GI, Lee J, Stemmermann G, Blaser MJ. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am J Epidemiol. 2002;155:1054–1059. doi: 10.1093/aje/155.11.1054. [DOI] [PubMed] [Google Scholar]
- 7.Belair C, Darfeuille F, Staedel C. Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin Microbiol Infect. 2009;15:806–812. doi: 10.1111/j.1469-0691.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- 8.Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 9.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 10.Jeong O, Park YK. Clinicopathological features and surgical treatment of gastric cancer in South Korea: the results of 2009 nationwide survey on surgically treated gastric cancer patients. J Gastric Cancer. 2011;11:69–77. doi: 10.5230/jgc.2011.11.2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon H, Kim N, Lee HS, et al. Helicobacter pylori-negative gastric cancer in South Korea: incidence and clinicopathologic characteristics. Helicobacter. 2011;16:382–388. doi: 10.1111/j.1523-5378.2011.00859.x. [DOI] [PubMed] [Google Scholar]
- 12.Ruzzo A, Graziano F, Pizzagalli F, et al. Interleukin 1B gene (IL-1B) and interleukin 1 receptor antagonist gene (IL-1RN) polymorphisms in Helicobacter pylori-negative gastric cancer of intestinal and diffuse histotype. Ann Oncol. 2005;16:887–892. doi: 10.1093/annonc/mdi184. [DOI] [PubMed] [Google Scholar]
- 13.Ye H, Liu H, Raderer M, et al. High incidence of t(11;18)(q21;q21) in Helicobacter pylori-negative gastric MALT lymphoma. Blood. 2003;101:2547–2550. doi: 10.1182/blood-2002-10-3167. [DOI] [PubMed] [Google Scholar]
- 14.Matsushima K, Isomoto H, Inoue N, et al. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 15.Shin CM, Kim N, Jung Y, et al. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci. 2010;101:1337–1346. doi: 10.1111/j.1349-7006.2010.01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N, Park RY, Cho SI, et al. Helicobacter pylori infection and development of gastric cancer in Korea: long-term follow-up. J Clin Gastroenterol. 2008;42:448–454. doi: 10.1097/MCG.0b013e318046eac3. [DOI] [PubMed] [Google Scholar]
- 17.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Lu M, Zhang Q, Deng M, et al. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu A, Xu X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol Biol. 2011;724:259–267. doi: 10.1007/978-1-61779-055-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008;8:69. doi: 10.1186/1472-6750-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando T, Yoshida T, Enomoto S, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 24.Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 25.Turcatel G, Rubin N, El-Hashash A, Warburton D. MIR-99a and MIR-99b modulate TGF-β induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One. 2012;7:e31032. doi: 10.1371/journal.pone.0031032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH, Kim HK, Rettig RL, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah AA, Leidinger P, Backes C, et al. A set of specific miRNAs is connected with murine and human gastric cancer. Genes Chromosomes Cancer. 2013;52:237–249. doi: 10.1002/gcc.22024. [DOI] [PubMed] [Google Scholar]
- 29.Yao Y, Suo AL, Li ZF, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 30.Krutovskikh VA, Herceg Z. Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. Bioessays. 2010;32:894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 31.Lam EK, Wang X, Shin VY, et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3:209–218. [PMC free article] [PubMed] [Google Scholar]
- 32.Shiotani A, Iishi H, Uedo N, et al. Evidence that loss of sonic hedgehog is an indicator of Helicobater pylori-induced atrophic gastritis progressing to gastric cancer. Am J Gastroenterol. 2005;100:581–587. doi: 10.1111/j.1572-0241.2005.41001.x. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 34.Wu XM, Shao XQ, Meng XX, et al. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxyc-amptothecin-resistant gastric cancer cells. Acta Pharmacol Sin. 2011;32:259–269. doi: 10.1038/aps.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marrelli D, Pedrazzani C, Berardi A, et al. Negative Helicobacter pylori status is associated with poor prognosis in patients with gastric cancer. Cancer. 2009;115:2071–2080. doi: 10.1002/cncr.24253. [DOI] [PubMed] [Google Scholar]
- 36.Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 38.Kozomara A, Griffiths-Jones S. miRBase: integrating microR-NA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Xie J, Xu X, et al. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-lambda1. Protein Cell. 2013;4:130–141. doi: 10.1007/s13238-012-2081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]