Abstract
Background/Aims
Hematological abnormalities during hepatitis C virus (HCV) combination therapy with pegylated interferon α and ribavirin often necessitate dose reduction. Variants of the ITPA gene have been reported to protect against anemia during the early stages of HCV combination treatments but have also been associated with larger decreases in platelet counts. We aimed to identify the association between specific ITPA gene polymorphisms and hematological abnormalities in patients undergoing HCV combination therapy.
Methods
In this retrospective study, 175 patients treated with HCV combination therapy were enrolled at St. Martin De Porres Hospital in Taiwan between 2006 and 2012. Two single nucleotide polymorphisms (SNP) within or adjacent to the ITPA gene (rs1127354, rs6051702) were genotyped. We investigated the effect of ITPA gene variants on hematological abnormalities during the therapy.
Results
The ITPA rs1127354 minor variants were significantly associated with protection against anemia at week 4 (p=1.86×10−6) and with more severe decreases in platelet counts during HCV combination therapy. SNP rs6051702 was not associated with the hemoglobin decline to >3 g/dL at week 4 in our study (p=0.055).
Conclusions
The ITPA SNP rs1127354 is a useful predictor of ribavirin-induced anemia in Taiwanese patients and may be related to more severe decreases in platelet counts during the early stage of HCV combination therapy.
Keywords: Hematologic abnormalities, Ribavirin, Chronic hepatitis C, Polymorphism, ITPA
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is a major health problem affecting 170 million people worldwide.1 Standard treatment for chronic hepatitis C (CHC) is a course of pegylated interferon α (PEG-IFN-α) combined with ribavirin (RBV) therapy for 24 or 48 weeks.2–4 The goal of such a treatment is to reach the sustained virological response (SVR), defined as undetectable HCV RNA in the serum after 24 weeks of posttreatment follow-up. The achievement of SVR after therapy can prevent liver-related complications and improves survival.5–8 However, less than 50% of patients infected with HCV genotype 1 achieve SVR or are cured of the infection following this type of therapy.9–11 A number of host and viral factors have been linked to the response to therapy.12,13 Recent studies have revealed a genetic polymorphism in the region of the interleukin 28B gene (IL28B) encoding IFN-lambda that is strongly associated with viral clearance following PEG-IFN-α/RBV therapy.9,10,14 In addition, treatment is often poorly tolerated due to side effects, particularly RBV-induced hemolytic anemia;15,16 some patients are thus prevented from completing therapy or require RBV dose reduction that increases the risk of treatment failure.17
Variation in the degree of RBV-induced hemolysis and anemia between individuals is likely affected by both clinical and genetic factors. A recent genome-wide association study (GWAS) identified a single nucleotide polymorphism (SNP) in C20orf194, rs6051702, as significantly associated with decreased hemoglobin (Hb) in European-American HCV-genotype-1 patients receiving PEG-IFN-α plus RBV therapy.18 The gene C20orf194, rs6051702, locates on chromosome 20p13 in a 1.8 Mb region encodes an uncharacterized protein and is associated with two genetic variants (rs1127354 and rs7270101) in the IPTA gene as determined using HapMap data from Utah residents with northern and western European ancestry.19 Variations within the ITPA gene are responsible for the protection against hemolytic anemia in HCV-infected patients in the original GWAS.18 The ITPA gene encodes inosine triphosphatase (ITPase), a protein that hydrolyses inosine triphosphate (ITP). ITPase deficiency results in accumulation of ITP in erythrocytes and increased toxicity of purine analogue drugs.20,21 Reduced ITPase activity can be caused by a missense variant in exon 2 (rs1127354, resulting in a proline-to-threonine substitution denoted P32T) and a splice-modifying SNP located in the second intron (rs7270101).22–25 Variations within the ITPA gene (rs1127354 and rs7270101) lead to ITPase deficiency and protect against hemolytic anemia in HCV-infected patients during early stages of treatment;18 however, the effect of these ITPA gene variants on the outcome of therapy was inconsistent.18,26,27 In studies of the Asian population, rs7270101 is excluded because no such variants are reported in the international HapMap project database.
In addition to RBV-related hemolytic anemia, bone marrow suppression is an important adverse effect of PEG-IFN therapy. The resulting neutropenia and thrombocytopenia2 leads to a decrease in medication dose or premature withdrawal from therapy in 10% to 14% of patients.28 The decline in platelet counts that occurs during antiviral therapy is less pronounced when IFN is combined with RBV than when administered alone, suggesting RBV may also play a role in thrombocytopenia.29,30 A genome-wide association study in Japanese HCV patients found that the IPTA SNP (rs1127354) CA/AA genotype was significantly associated with a lower absolute decrease in Hb levels, especially during the early weeks of therapy, but independently associated with a greater decrease in platelet counts.31,32
ITPA gene variants that protect against anemia have been established among Caucasian, Hispanic, African-American, and Japanese populations.11,33 The prevalence of HCV infection has been reported as high as 10.2% in southern Taiwan;34 one study reported that 37.7% of HCV patients develop anemia during combination therapy.35 HCV infection is also closely associated with thrombocytopenia in Taiwanese patients.36 Therefore, it is important to evaluate the association between IPTA gene polymorphisms and hematological abnormalities in HCV-infected Taiwanese patients receiving HCV combination therapy. Our findings reveal predictive biomarkers that could be used for effective pretreatment screening and could substantially reduce the cost of treatment.
MATERIALS AND METHODS
1. Patients
In this retrospective, cross-sectional study, patients with CHC treated with PEG-IFN-α/RBV therapy were enrolled from November 2006 to March 2012 at St. Martin De Porres Hospital in Taiwan and informed consent was obtained for the analysis of peripheral blood for DNA extraction and genetic testing. All patients had abnormal levels of serum alanine transaminase (ALT) for more than 6 months and were positive for both anti-HCV antibody and serum HCV RNA. All patients were also negative for hepatitis B surface antigen and HIV and had no evidence of other liver diseases, including alcoholic hepatitis, hemochromatosis, Wilson’s disease, and autoimmune hepatitis and had not received immunosuppressive therapy before enrollment in the study. All subjects gave written informed consent to participate in the study according to the process approved by the Ethics Committee of St. Martin De Porres Hospital.
Patients received weekly injections of PEG-IFN-α-2b (1.5 μg/kg body weight) or PEG-IFN-α-2a (180 μg) plus oral administration of RBV (800 to 1,200 mg daily depending on body weight) for 24 or 48 weeks. On-treatment dose reduction and discontinuation of PEG-IFN-α or RBV were chosen based on the recommendations stated in package inserts or clinical circumstances of individual patients to avoid possible side-effects.
2. Patient evaluation
The following factors were analyzed to determine whether they were related to the efficacy of combination therapy: age, sex, body weight, height, BMI, and pretreatment biochemical parameters such as, Hb, platelet counts, white cell counts, ALT level, thyroid function test, serum creatinine, estimated glomerular filtration rate as calculated by the modification of diet in renal disease (MDRD) equation, HCV genotype, and serum HCV RNA level (log IU/mL). Hb values, white cell counts, and platelet counts were measured at baseline and every 4 weeks until 24 weeks. A decrease in Hb of more than 3 g/dL or Hb levels of less than 10 g/dL were considered clinically relevant anemia.
3. HCV RNA levels and genotypes
Plasma HCV-RNA virus load and genotypes were determined using real-time polymerase chain reaction (PCR) (COBAS TaqMan; Roche Molecular System, South Branchburg, NJ, USA) with a lower limit of detection of 25 IU/mL. All values are reported as log IU/mL. HCV RNA levels were determined at weeks 4, 12, the end of treatment, and 24 weeks after the completion of treatment. Patients were classified as having achieved rapid virological response (RVR) if HCV RNA was undetectable (<25 IU/mL) in serum at treatment week 4 and as having SVR if HCV RNA was undetectable in serum 24 weeks after the completion of therapy.
4. DNA extraction and TaqMan SNP assay
Genomic DNA was extracted from the whole blood of each patient using a genomic DNA extraction kit (GMbioslab Co., Taipei, Taiwan). Genetic polymorphisms for rs1127354 in ITPA, rs6051702 in C20orf194, and rs8099917 in IL28B were determined by TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, USA) using real-time PCR.37 In the present study, we categorized IL28B rs8099917 TT as the major variant and TG/GG as minor variants, ITPA rs1127354 CC as the major variant and CA/AA as minor variants, and C20orf194 rs6051702 AA as the major variant and AC/CC as minor variants.
5. Statistical analysis
For categorical data, proportions were compared between groups using chi-square test, while Student t-test was used for continuous data. Univariate and multivariate logistic regression was performed for Hb declines of more than 3 g/dL at week 4 and platelet count decreases ≥20 (109/L) at week 4 as dependent variables. Sex, age, weight, body mass index, rs1127354, rs8099917, rs6051702, viral genotype, viral load, serum creatinine, MDRD, baseline Hb, baseline platelet counts, and RBV start dose were considered as potential explanatory variables. The multivariate logistic regression models were constructed sequentially with variables entered one at a time; the significance, colinearity, and variance contributing to the model was assessed at each step. A significance level of 0.05 was used to eliminate variables from the model.
We considered p-values less than 0.05 to indicate significance for all statistical tests, and odds ratios are presented with 95% confidence intervals. Statistical analyses were conducted using SPSS version 14.0 (SPSS Inc., Chicago, IL, USA) software or Microsoft Excel 2007 (Microsoft Corp., Redmond, WA, USA).
RESULTS
1. Patient characteristics
The clinical characteristics and SNP genotypes of the 175 patients included in our cohort are summarized in Table 1. The SNPs of ITPA rs1127354, C20orf194 rs6051702, and IL28B rs8099917 were successfully genotyped in 100%, 98.9%, and 99.4%, respectively. The minor allele frequencies at rs1127354 (A-allele=0.17) and rs6051702 (C-allele=0.15) were similar to those found in HapMap individuals of Northern European descent (CEU population, http://www.hapmap.org) (Table 1). Next, we compared the baseline clinical and host genetic characteristics of patient groups according to ITPA rs1127354 and C20orf194 rs6051702 genotypes (Table 2). There were no significant differences in age, sex, blood cell counts, ALT levels, or serum viral loads between the two groups based on the SNP of either ITPA rs1127354 or C20orf194 rs6051702. The SNP of ITPA rs1127354 did not show significant linkage with that of IL28B rs8099917 (p=0.544). In contrast to a previous study,18 the SNP of ITPA rs1127354 did not show significant linkage with the SNP of C20orf194 rs6051702 in our study (p=0.712) (Table 2).
Table 1.
Baseline Patient Characteristics
| Characteristic | Value |
|---|---|
| No. of patients | 175 |
| Age, yr* | 55.53±11.50 |
| Sex, male/female | 116/59 |
| SNP genotype | |
| ITPA, rs1127354 CC/CA/AA | 127/41/7 |
| C20orf194, rs6051702 AA/AC/CC | 122/44/7 |
| IL28B, rs8099917 TT/TG/GG | 149/24/1 |
| MAF of ITPA, rs1127354, % | 16.6 |
| MAF of C20orf194, rs6051702, % | 15.1 |
| Body weight, kg* | 64.67±10.69 |
| BMI, kg/m2* | 24.66±3.13 |
| HCV genotypes | |
| 1/non-1 | 111/62 |
| 1, high viral load†/others | 99/74 |
| Baseline white blood cell value,/mm3* | 5.75±1.78 |
| Baseline Hb value, g/dL* | 14.38±1.51 |
| Baseline platelet count, 109/L* | 170.50±57.45 |
| Serum creatinine, mg/dL* | 0.94±0.89 |
| MDRD* | 95.45±26.30 |
| ALT, IU/L* | 105.45±72.00 |
| Serum HCV RNA, log IU/mL* | 6.02±1.08 |
| PEG IFN, PEG-IFN-α-2a/PEG-IFN-α-2b | 79/96 |
| Hb decrease at week 4, g/dL* | 2.21±1.55 |
| RBV dose reduction or discontinuation at week 4, n (%) | 96 (56.5) |
| Hb decline >3 g/dL, n (%) | 50 (28.6) |
SNP, single-nucleotide polymorphism; ITPA, inosine triphosphatase; IL28B, interleukin-28B; MAF, minor allele frequency; BMI, body mass index; HCV, hepatitis C virus; Hb, hemoglobin; MDRD, modification of diet in renal disease; ALT, alanine transaminase; PEG IFN, pegylated interferon; RVB, ribavirin.
Mean±SD;
High viral load, HCV RNA >5 log IU/mL.
Table 2.
Clinical and Genetic Characteristics of Patients with Respect to IPTA rs1127354 and C20orf194 rs6051702
| ITPA rs1127354 | C20orf194 rs6051702 | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| CC (n=127) | CA+AA (n=48) | p-value | AA (n=122) | AC+CC (n=51) | p-value | |
| Age, yr* | 55.77±11.76 | 54.90±10.88 | 0.654 | 55.40±12.12 | 56.00±9.89 | 0.756 |
| Sex, male/female | 87/40 | 29/19 | 0.313 | 77/45 | 37/14 | 0.233 |
| Weight, kg* | 64.10±10.35 | 66.17±11.54 | 0.254 | 63.85±10.28 | 66.77±11.61 | 0.103 |
| BMI, kg/m2* | 24.45±2.96 | 25.21±3.52 | 0.152 | 24.65±3.13 | 24.76±3.18 | 0.821 |
| Serum creatinine, mg/dL* | 0.92±0.83 | 1.01±1.02 | 0.566 | 0.98±1.05 | 0.85±0.21 | 0.381 |
| MDRD* | 96.89±26.97 | 91.63±24.29 | 0.239 | 94.96±28.41 | 97.14±20.89 | 0.621 |
| ALT, IU/L* | 105.30±75.18 | 105.88±63.59 | 0.962 | 106.66±74.37 | 101.52±67.78 | 0.672 |
| Serum HCV RNA, log IU/mL* | 5.96±1.11 | 6.20±0.98 | 0.182 | 6.06±1.06 | 5.90±1.14 | 0.361 |
| White blood cells,/mm3* | 5.80±1.85 | 5.61±1.57 | 0.531 | 5.73±1.66 | 5.72±2.06 | 0.959 |
| Hb, g/dL* | 14.31±1.52 | 14.56±1.48 | 0.340 | 14.30±1.57 | 14.56±1.38 | 0.304 |
| Platelet count, ×109/L* | 170.83±57.08 | 169.63±59.02 | 0.902 | 173.43±59.63 | 163.47±50.65 | 0.297 |
| PEG IFN, 2a/2b | 58/69 | 21/27 | 0.820 | 52/70 | 25/26 | 0.440 |
| C20orf194, rs6051702, AA/AC/CC | 91/30/5 | 31/14/2 | 0.712 | - | - | - |
| IL28B, rs8099917, TT/TG | 110/17 | 39/8 | 0.544 | 106/15 | 41/10 | 0.220 |
| Hb decline >3 g/dL at week 4, n (%) | 49 (38.6) | 1 (2.1) | 1.86×10−6 | 30 (24.6) | 20 (39.2) | 0.055 |
p-values were calculated using Student t-test or by chi-square analysis. ITPA rs1127354, major allele-C and minor allele-A; C20orf194 SNP, major allele-A and minor allele-C; IL28B rs8099917, major allele-T and minor allele-G.
ITPA, inosine triphosphatase; BMI, body mass index; MDRD, modification of diet in renal disease; ALT, alanine transaminase; HCV, hepatitis C virus; Hb, hemoglobin; PEG IFN, pegylated interferon; IL28B, interleukin-28B.
Mean±SD.
2. Association of ITPA variants with Hb decline during the course of anti-HCV therapy
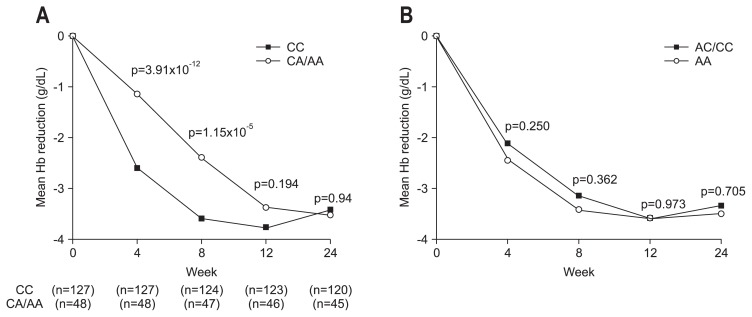
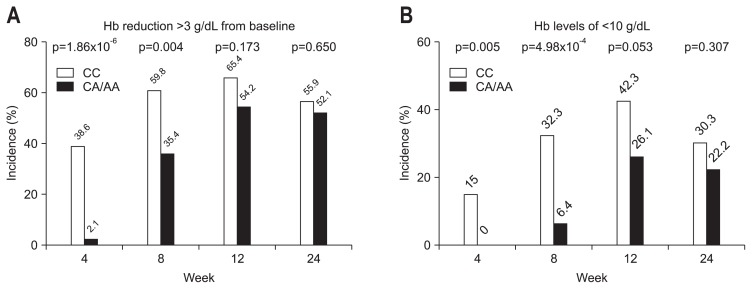
Because the decrease in Hb during the course of treatment is most clinically relevant, we analyzed the association of the SNPs of ITPA rs1127354 and C20orf194 rs6051702 with Hb decline at weeks 4, 8, 12, and 24 of PEG-IFN-α/RBV treatment (Fig. 1). The SNP of C20orf194 rs6051702 did not correlate with the decline in Hb at any time point (Fig. 1B). In contrast, patients with the minor allele ITPA rs1127354 had a smaller decrease in Hb after 4 and 8 weeks of treatment (Fig. 1A). The rs1127354 SNP of the ITPA gene showed a significant association with Hb decline at week 4 (p=3.91×10−12) and week 8 (p=1.15×10−5) (Fig. 1A); after week 8, no difference in Hb decline between IPTA major and minor variants was observed (Fig. 1A). Significant anemia is defined clinically as a decline in Hb levels of more than 3 g/dL or Hb levels of less than 10 g/dL at week 4, which is the threshold at which RBV dose reduction is recommended. To assess the clinical involvement of these SNPs, we analyzed the proportion of patients suffering from clinically relevant anemia. As shown in Fig. 2, 38.6% of ITPA-CC patients showed a decrease in Hb of more than 3 g/dL (Fig. 2A), and 15% of patients had Hb levels of less than 10 g/dL at week 4 (Fig. 2B). In contrast, only 2.1% patients with ITPA-CA/AA developed clinically relevant anemia (Fig. 2A). Further, SNP rs1127354 of the ITPA gene was significantly associated with clinically relevant anemia at weeks 4 and week 8 (p=1.86×10−6 and p=0.004, respectively, for Hb reduction levels >3 g/dL; p=0.005 and p=4.98×10−4, respectively, for Hb levels <10 g/dL) (Fig. 2A and B). However, SNP rs6051702 of C20orf194, which showed the strongest significant association with Hb decline in the European-American population,18 was not associated with Hb decline >3 g/dL at week 4 in our study (p=0.055) (Table 2). These results demonstrate that the ITPA CA/AA genotypes are significantly associated with a lower incidence of clinically relevant anemia, especially during the early weeks of therapy, and protect against the development of severe anemia.
Fig. 1.
The association between the single-nucleotide polymorphisms of IPTA rs1127354, C20orf194 rs6051702, and hemoglobin (Hb) decline. (A) Time-dependent Hb decline in ITPA major and minor variants. The ITPA minor variant was associated with smaller decreases in Hb levels at weeks 4 and 8. The numbers in parentheses denote the numbers of patients. (B) Time-dependent Hb declines in C20orf194, rs6051702 major and minor variants. The SNP of C20orf194 rs6051702 was not associated with a decrease in Hb at any time point.
Fig. 2.
Effects of ITPA single nucleotide polymorphisms on clinically relevant anemia induced by pegylated interferon plus ribavirin treatment. (A) Percentage of subjects with a hemoglobin (Hb) decrease >3 g/dL during the course of anti-hepatitis C virus (HCV) therapy. (B) Percentage of subjects with Hb concentrations <10 g/dL during the course of anti-HCV therapy.
3. Factors influencing hemoglobin decrease at week 4 during treatment
To determine the factors associated with decreased hemoglobin levels during the early stage of PEG-IFN/RBV therapy, univariate and multivariate logistic regression analysis was performed. The ITPA minor variant rs1127354 was the only factor that was strongly associated with Hb decrease >3 g/dL at week 4 in univariate and multivariate logistic regression analysis (Table 3), with this minor allele having a protective effect.
Table 3.
Univariate and Multivariate Logistic Regression Analyses of Patient Characteristics to Assess Baseline Predictors of Hemoglobin Decline >3 g/dL at Week 4
| p-value (univariate) | p-value (multivariate) | OR | 95% CI | |
|---|---|---|---|---|
| Sex, male vs female | 0.174 | - | - | - |
| Age | 0.145 | - | - | - |
| rs1127354, CC vs CA+AA | 0.001 | 0.001 | 0.034 | 0.01–0.26 |
| rs6051702, AA vs AC+CC | 0.055 | - | - | - |
| rs8099917, TT vs TG+GG | 0.301 | - | - | - |
| BW | 0.023 | 0.533 | 0.981 | 0.924–1.042 |
| BMI | 0.017 | 0.430 | 0.919 | 0.746–1.133 |
| MDRD | 0.997 | - | - | - |
| RVR, yes/no | 0.829 | - | - | - |
| SVR, yes/no | 0.461 | - | - | - |
| PEG IFN, 2a/2b | 0.063 | - | - | - |
| Serum creatinine | 0.637 | - | - | - |
| Virus load | 0.826 | - | - | - |
| Genotype, 1/other | 0.100 | - | - | - |
| Baseline Hb | 0.080 | - | - | - |
| RBV starting dose | 0.179 | - | - | - |
OR, odds ratio; CI, confidence interval; BW, body weight; BMI, body mass index; MDRD, modification of diet in renal disease; RVR, rapid virological response; SVR, sustained virological response; PEG IFN, pegylated interferon; Hb, hemoglobin; RBV, ribavirin.
4. Association of the predicted ITPase deficiency with clinically relevant anemia
SNP rs1127354 in the ITPA gene is a missense variant in exon 2 that leads to decreased ITPase activity. Two previous studies have reported that ITPase activity can be predicted and scored according to SNP genotype.25,38 The ITPase activities for genotypes CC, CA, and AA are 100%, 20%–30%, and less than 10%, respectively. Notably, patients with the rs1127354 genotype AA, predicted to yield severe ITPase deficiency, experienced smaller decreases in Hb than did those with the CC/CA genotype and showed no clinically relevant anemia at week 4 (Table 4). The predicted ITPase deficiency remained protective for clinically relevant anemia at week 4 (p=2.54×10−8).
Table 4.
Distribution of the Predicted Level of ITPase Deficiency and ITPase Activity according to ITPA Genotype
| rs1127354 genotype | Predicted ITPase deficiency | ITPase activity, % | Patients, no. | Population frequency, % | Hb decrease at week 4, mean±SD, g/dL | p-value for Hb decrease >3 g/dL at week 4 | Patients with clinically relevant anemia, no.* |
|---|---|---|---|---|---|---|---|
| CC | − | 100 | 127 | 72.6 | −2.61±1.55 | 2.54×10−8 | 49 |
| CA | ++ | 20–30 | 41 | 23.4 | −1.23±0.95 | − | 1 |
| AA | +++ | <10 | 7 | 4.0 | −0.64±0.74 | − | 0 |
ITPase, inosine triphosphatase; Hb, hemoglobin.
Clinically relevant anemia: defined as a decrease in Hb levels of more than 3 g/dL or Hb levels of less than 10 g/dL at week 4.
5. ITPA gene variant is associated with RBV dose reduction at week 4 but does not affect sustained viral response
Because of decreased Hb levels, RBV dose reduction was required at week 4 in 42% of patients with the ITPA major variant but only 11.8% of those with the ITPA minor variant in our study. The incidence of RBV dose reduction at week 4 differed significantly between patients with major and minor variants of rs1127354 (CC vs non-CC, p=0.019). Knowing that drug reduction occurred significantly less frequently in patients with the ITPA minor variant, we next investigated whether the ITPA gene variants affected final treatment outcomes; we found no associations between the ITPA gene variants and SVR (Table 5).
Table 5.
Sustained Viral Response Rates in Patients according to IPTA Gene Variant
| Total | HCV genotype 1, high viral load* | Others† | ||||
|---|---|---|---|---|---|---|
| IPTA SNP rs1127354 | CC | CA/AA | CC | CA/AA | CC | CA/AA |
| Patients, no. | 124 | 46 | 68 | 29 | 56 | 17 |
| Patients achieving SVR, no. | 92 | 34 | 44 | 18 | 48 | 16 |
| SVR rate, % | 74.2 | 73.9 | 64.7 | 62.1 | 85.7 | 94.1 |
| p-value | 0.970 | 0.804 | 0.356 | |||
All of the hepatitis C virus (HCV) patients received combination therapy and underwent viral load assessment after 24 weeks of treatment. p-values were calculated by the chi-square analysis of sustained viral response (SVR).
ITPA, inosine triphosphatase; SNP, single-nucleotide polymorphism.
High viral load, HCV RNA >5 log IU/mL;
Others: includes HCV genotype 1, serum HCV RNA <5 log IU/mL, and non-1 HCV genotypes.
6. Association of ITPA gene variants with decrease in platelets during anti-HCV therapy
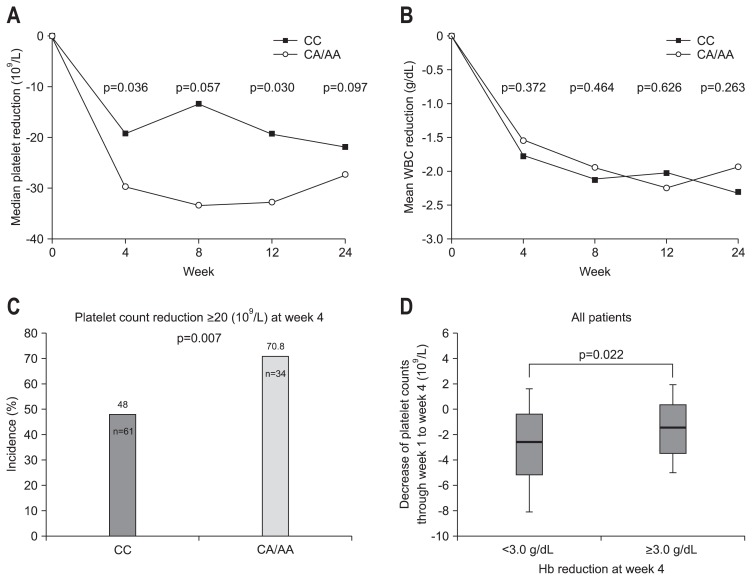
The median quantitative decrease in platelets from baseline according to ITPA rs1127354 genotype is shown in Fig. 3A. Following PEG-IFN-α/RBV therapy, patients with the ITPA CC genotype had smaller decreases in platelet counts compared to those with the ITPA CA/AA genotypes (Fig. 3A). The greatest differences in median platelet count decrease were found at weeks 4 and 12 (week 4, p=0.036; week 12, p=0.03). Leukocyte count decreases did not differ between patients with ITPA genotype CC and those with CA/AA (Fig. 3B). The fraction of patients with a platelet count decrease >20 (109/L) at week 4 was lower for ITPA genotype CC than CA/AA (Fig. 3C). Patients with clinically relevant anemia (Hb decrease ≥3.0 g/dL) at week 4 had a significantly smaller decrease in platelet count than did those without anemia (p=0.022) (Fig. 3D). Multivariate regression analysis for factors associated with a platelet decrease >20 (109/L) at week 4 showed that a lower platelet count at baseline and the rs1127354 genotypes CA/AA were independently associated with greater decreases in platelets (Table 6).
Fig. 3.
The association between ITPA rs1127354 and platelet or leukocyte count declines. (A) Time-dependent median platelet counts in ITPA major and minor variants (rs1127354). (B) Time-dependent mean leukocyte counts in ITPA major and minor variants (rs1127354). (C) The percentage of patients with a platelet count decrease >20 (109/L) from baseline at week 4 according to ITPA rs1127354 genotype. (D) Decrease in platelet counts over weeks 0–4 of pegylated interferon α/ribavirin therapy. Patients with anemia (hemoglobin [Hb] decrease ≥3.0 g/dL) at week 4 had a significantly lower decrease in platelet counts than those with smaller decreases in Hb (<3.0 g/dL).
WBC, white blood cell.
Table 6.
Univariate and Multivariate Logistic Regression Analyses of Patient Characteristics to Assess Baseline Predictors of Platelet Reduction ≥20 (109/L) at Week 4
| p-value (univariate) | p-value (multivariate) | OR | 95% CI | |
|---|---|---|---|---|
| Sex, male vs female | 0.673 | - | - | - |
| Age | 0.989 | - | - | - |
| rs1127354, CC vs CA+AA | 0.028 | 0.020 | 2.51 | 1.12–5.44 |
| rs6051702, AA vs AC+CC | 0.267 | - | - | - |
| rs8099917, TT vs TG+GG | 0.868 | - | - | - |
| BMI | 0.261 | - | - | - |
| MDRD | 0.351 | - | - | - |
| RVR, yes/no | 0.949 | - | - | - |
| SVR, yes/no | 0.498 | - | - | - |
| PEG IFN, 2a/2b | 0.186 | - | - | - |
| Serum creatinine | 0.354 | - | - | - |
| Virus load | 0.662 | - | - | - |
| HCV genotype (1, high virus load/other*) | 0.976 | - | - | - |
| Baseline Hb | 0.011 | 0.110 | 1.18 | 0.96–1.45 |
| Hb reduction ≥3.0 g/dL at week 4 | 0.096 | - | - | - |
| Baseline platelet counts | 0.002 | 0.012 | 1.01 | 1.00–1.02 |
OR, odds ratio; CI, confidence interval; BMI, body mass index; MDRD, modification of diet in renal disease; RVR, rapid virological response; SVR, sustained virological response; PEG IFN, pegylated interferon; HCV, hepatitis C virus; Hb, hemoglobin.
High viral load, HCV RNA >5 log IU/mL; other, including HCV genotype 1, serum HCV RNA <5 log IU/mL, and non-1 genotypes.
DISCUSSION
We observed that a functional SNP in the ITPA locus rs1127354, which confers decreased ITPase activity, protected patients from the development of anemia early in the treatment course (Table 4, Figs 1 and 2). Univariate and multivariate logistic regression analyses indicated that rs1127354 SNP was the most important factor associated with a decrease in Hb >3 g/dL at week 4 (Table 3). These data are consistent with previous studies in United States18 and Japanese26 populations. Although ITPA genetic variants protect against RBV-induced anemia, the difference in Hb decrease between ITPA major and minor variants did not persist to the end of therapy in our study. This result differs from other studies27,33 and may be due to our RBV dose adjustments after the onset of anemia or host compensation.
The precise mechanism whereby ITPA genetic variants protect against RBV-induced anemia is unclear. RBV enters erythrocytes, where it undergoes phosphorylation to form active RBV-phosphate conjugates. The RBV-phosphate conjugates are unable to cross the erythrocyte cell membrane and thus accumulate in the cell.39 Red blood cells with ITPase deficiency accumulate ITP, which may protect the cells from RBV-induced hemolysis by competing with RBV triphosphate.38,40 Another recent report proposes a mechanism whereby ITP confers protection against RBV-induced adenosine triphosphate (ATP) reduction by substituting for erythrocyte guanosine triphosphate, which is depleted by RBV, in the biosynthesis of ATP.41
Although ITPA genetic variants were observed to protect against RBV-induced anemia, 46% of the patients with the ITPA major variant (CC) did not experience a significant decrease in Hb at week 4 in our study (Fig. 2). This finding suggests the presence of other ITPA variants or polymorphisms in other enzymes affecting RBV-induced hemolysis. At least two other rare functional ITPA variants with reduced ITPase activity have been described,38,42 and their involvement still requires further investigation.
The SNP rs6051702 in C20orf194 showed a significant association with decreased Hb in the early stage of combination therapy for CHC in European-Americans.18 Our study demonstrated no significant association between this variant and decreased Hb in response to such therapy. This discrepancy may be due to the low linkage disequilibrium between this SNP and the ITPA gene in the Asian population as compared with the high linkage disequilibrium in white subjects. The association between the C20orf194 SNP and treatment-induced Hb decrease was reported to differ between races, including European-Americans (p=1.1×10−45), African-Americans (p=0.19) and Hispanics (p=9.5×10−3).18
Although the incidence of early RBV dose reduction was significantly lower in ITPA-minor (CA/AA) patients, these patients demonstrated no benefit in terms of treatment outcome in our study (Table 5). Several explanations for this observation are plausible. First, the incidence of anemia-protective (rs1127354 non-CC) genotypes is lower in Taiwanese (16.6%) than in Japanese patients (20%),43 making the power of our study too low to observe an effect of ITPA variants on treatment outcomes. Secondly, higher pretreatment levels of hemoglobin in Taiwanese patients compared with Japanese patients43 (14.4 g/dL vs 13.0 g/dL) might result in fewer RBV dose-reductions in Taiwanese patients, thereby decreasing the apparent effects of the ITPA SNP on treatment outcome. Thirdly, a previous study showed anemia was associated with higher rates of SVR in HCV-infected patients treated with PEG-IFN/RBV,44 suggesting an alternative hypothesis: the magnitude of Hb loss is a pharmacodynamic marker of RBV exposure, correlating more closely with antiviral effects than the administered RBV dose. Finally, reports suggest that the effect of RBV dose reduction on the likelihood of SVR may be negligible, as long as RBV is not discontinued,16,45 which is consistent with our results.
Thrombocytopenia is one of the major adverse effects of PEG-IFN-α. Additionaly, treatment-induced reductions in platelet counts were less marked following the addition of RBV to IFN monotherapy for CHC.29,30 The biological mechanism underlying this relationship between Hb levels and platelet counts is not clearly understood but may involve stimulation of the bipotent erythroid/megakaryocyte progenitor cell by erythropoietin.46,47 Our data demonstrate that patients with the ITPA rs1127354 major variant had smaller decreases in platelet counts during the early stage of therapy (Fig. 3A and C), supporting the results of a previous report.32 This data therefore presents an indirect genetic association where wild type ITPA activity is associated with more profound RBV-related anemia that in turn stimulates platelet production, manifesting as less severe IFN-induced thrombocytopenia. The ITPA genotype may thus provide a valuable pharmacogenetic diagnostic tool for the identification of patients at increased risk for significant anemia and thrombocytopenia before treatment initiation. Awareness of this risk will allow closer monitoring of Hb and platelet counts and early PEG-IFN/RBV dose decrements when necessary. In these patients, early intervention with RBV dose reduction and/or growth factor support could maximize safety and minimize premature discontinuation of RBV.
ACKNOWLEDGEMENTS
We would like to thank the St. Martin De Porres Hospital (grant number: P1101) for financially supporting this research. We thank Rui-Fang Huang at Department of Medical Research and Education, St. Martin De Porres Hospital for her analysis of data for the study.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/S0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Hsu CS, Kao JH. Peginterferon alfa-2b or alfa-2a with ribavirin for hepatitis C. N Engl J Med. 2009;361:1808–1809. doi: 10.1056/NEJMc091820. [DOI] [PubMed] [Google Scholar]
- 5.Imai Y, Kawata S, Tamura S, et al. Relation of interferon therapy and hepatocellular carcinoma in patients with chronic hepatitis C. Osaka Hepatocellular Carcinoma Prevention Study Group. Ann Intern Med. 1998;129:94–99. doi: 10.7326/0003-4819-129-2-199807150-00005. [DOI] [PubMed] [Google Scholar]
- 6.Shiratori Y, Imazeki F, Moriyama M, et al. Histologic improvement of fibrosis in patients with hepatitis C who have sustained response to interferon therapy. Ann Intern Med. 2000;132:517–524. doi: 10.7326/0003-4819-132-7-200004040-00002. [DOI] [PubMed] [Google Scholar]
- 7.Yu ML, Lin SM, Chuang WL, et al. A sustained virological response to interferon or interferon/ribavirin reduces hepatocellular carcinoma and improves survival in chronic hepatitis C: a nationwide, multicentre study in Taiwan. Antivir Ther. 2006;11:985–994. [PubMed] [Google Scholar]
- 8.Huang JF, Yu ML, Lee CM, et al. Sustained virological response to interferon reduces cirrhosis in chronic hepatitis C: a 1,386-patient study from Taiwan. Aliment Pharmacol Ther. 2007;25:1029–1037. doi: 10.1111/j.1365-2036.2007.03297.x. [DOI] [PubMed] [Google Scholar]
- 9.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto N, Tanaka Y, Nakagawa M, et al. ITPA gene variant protects against anemia induced by pegylated interferon-α and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res. 2010;40:1063–1071. doi: 10.1111/j.1872-034X.2010.00741.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee SS, Heathcote EJ, Reddy KR, et al. Prognostic factors and early predictability of sustained viral response with peginterferon alfa-2a (40KD) J Hepatol. 2002;37:500–506. doi: 10.1016/S0168-8278(02)00211-8. [DOI] [PubMed] [Google Scholar]
- 13.Asselah T, Estrabaud E, Bieche I, et al. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int. 2010;30:1259–1269. doi: 10.1111/j.1478-3231.2010.02283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 15.De Franceschi L, Fattovich G, Turrini F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 16.Reddy KR, Shiffman ML, Morgan TR, et al. Impact of ribavirin dose reductions in hepatitis C virus genotype 1 patients completing peginterferon alfa-2a/ribavirin treatment. Clin Gastroenterol Hepatol. 2007;5:124–129. doi: 10.1016/j.cgh.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 17.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 18.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bierau J, Lindhout M, Bakker JA. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics. 2007;8:1221–1228. doi: 10.2217/14622416.8.9.1221. [DOI] [PubMed] [Google Scholar]
- 21.Stocco G, Cheok MH, Crews KR, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mer-captopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85:164–172. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arenas M, Duley J, Sumi S, Sanderson J, Marinaki A. The ITPA c.94C>A and g.IVS2+21A>C sequence variants contribute to missplicing of the ITPA gene. Biochim Biophys Acta. 2007;1772:96–102. doi: 10.1016/j.bbadis.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Cao H, Hegele RA. DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J Hum Genet. 2002;47:620–622. doi: 10.1007/s100380200095. [DOI] [PubMed] [Google Scholar]
- 24.Stepchenkova EI, Tarakhovskaya ER, Spitler K, et al. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. J Mol Biol. 2009;392:602–613. doi: 10.1016/j.jmb.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumi S, Marinaki AM, Arenas M, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–367. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 26.Ochi H, Maekawa T, Abe H, et al. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy: a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010;139:1190–1197. doi: 10.1053/j.gastro.2010.06.071. [DOI] [PubMed] [Google Scholar]
- 27.Kurosaki M, Tanaka Y, Tanaka K, et al. Relationship between polymorphisms of the inosine triphosphatase gene and anaemia or outcome after treatment with pegylated interferon and ribavirin. Antivir Ther. 2011;16:685–694. doi: 10.3851/IMP1796. [DOI] [PubMed] [Google Scholar]
- 28.Afdhal N, McHutchison J, Brown R, et al. Thrombocytopenia associated with chronic liver disease. J Hepatol. 2008;48:1000–1007. doi: 10.1016/j.jhep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Poynard T, Marcellin P, Lee SS, et al. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT) Lancet. 1998;352:1426–1432. doi: 10.1016/S0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 30.Davis GL, Esteban-Mur R, Rustgi V, et al. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 31.Thompson AJ, Clark PJ, Singh A, et al. Genome-wide association study of interferon-related cytopenia in chronic hepatitis C patients. J Hepatol. 2012;56:313–319. doi: 10.1016/j.jhep.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y, Kurosaki M, Nishida N, et al. Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic hepatitis C. Hum Mol Genet. 2011;20:3507–3516. doi: 10.1093/hmg/ddr249. [DOI] [PubMed] [Google Scholar]
- 33.Thompson AJ, Fellay J, Patel K, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–1189. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai MC, Kee KM, Chen YD, et al. Excess mortality of hepatocellular carcinoma and morbidity of liver cirrhosis and hepatitis in HCV-endemic areas in an HBV-endemic country: geographic variations among 502 villages in southern Taiwan. J Gastroenterol Hepatol. 2007;22:92–98. doi: 10.1111/j.1440-1746.2006.04489.x. [DOI] [PubMed] [Google Scholar]
- 35.Tseng CW, Hsieh YH, Chang CK, et al. HLA-B*15:02 is associated with anemia in patients with chronic hepatitis C treated with pegylated interferon-α and ribavirin. Tissue Antigens. 2012;80:424–430. doi: 10.1111/j.1399-0039.2012.01956.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang CS, Yao WJ, Wang ST, Chang TT, Chou P. Strong association of hepatitis C virus (HCV) infection and thrombocytopenia: implications from a survey of a community with hyperendemic HCV infection. Clin Infect Dis. 2004;39:790–796. doi: 10.1086/423384. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high-throughput SNP typing system for genome-wide association studies. J Hum Genet. 2001;46:471–477. doi: 10.1007/s100380170047. [DOI] [PubMed] [Google Scholar]
- 38.Shipkova M, Lorenz K, Oellerich M, Wieland E, von Ahsen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52:240–247. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 39.Kowdley KV. Hematologic side effects of interferon and ribavirin therapy. J Clin Gastroenterol. 2005;39(1 Suppl):S3–S8. doi: 10.1097/01.mcg.0000145494.76305.11. [DOI] [PubMed] [Google Scholar]
- 40.Fraser JH, Meyers H, Henderson JF, Brox LW, McCoy EE. Individual variation in inosine triphosphate accumulation in human erythrocytes. Clin Biochem. 1975;8:353–364. doi: 10.1016/S0009-9120(75)93685-1. [DOI] [PubMed] [Google Scholar]
- 41.Hitomi Y, Cirulli ET, Fellay J, et al. Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adeny-losuccinate synthase function. Gastroenterology. 2011;140:1314–1321. doi: 10.1053/j.gastro.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 42.Atanasova S, Shipkova M, Svinarov D, et al. Analysis of ITPA phenotype-genotype correlation in the Bulgarian population revealed a novel gene variant in exon 6. Ther Drug Monit. 2007;29:6–10. doi: 10.1097/FTD.0b013e3180308554. [DOI] [PubMed] [Google Scholar]
- 43.Azakami T, Hayes CN, Sezaki H, et al. Common genetic polymorphism of ITPA gene affects ribavirin-induced anemia and effect of peg-interferon plus ribavirin therapy. J Med Virol. 2011;83:1048–1057. doi: 10.1002/jmv.22069. [DOI] [PubMed] [Google Scholar]
- 44.Sulkowski MS, Shiffman ML, Afdhal NH, et al. Hepatitis C virus treatment-related anemia is associated with higher sustained virologic response rate. Gastroenterology. 2010;139:1602–1611. 1611. e1. doi: 10.1053/j.gastro.2010.07.059. [DOI] [PubMed] [Google Scholar]
- 45.Shiffman ML, Ghany MG, Morgan TR, et al. Impact of reducing peginterferon alfa-2a and ribavirin dose during retreatment in patients with chronic hepatitis C. Gastroenterology. 2007;132:103–112. doi: 10.1053/j.gastro.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Cardier JE, Erickson-Miller CL, Murphy MJ., Jr Differential effect of erythropoietin and GM-CSF on megakaryocytopoiesis from primitive bone marrow cells in serum-free conditions. Stem Cells. 1997;15:286–290. doi: 10.1002/stem.150286. [DOI] [PubMed] [Google Scholar]
- 47.Broudy VC, Lin NL, Kaushansky K. Thrombopoietin (c-mpl ligand) acts synergistically with erythropoietin, stem cell factor, and interleukin-11 to enhance murine megakaryocyte colony growth and increases megakaryocyte ploidy in vitro. Blood. 1995;85:1719–1726. [PubMed] [Google Scholar]