Abstract
Dysfunction of Müller cells has been implicated in the pathogenesis of several retinal diseases. In order to understand the potential contribution of Müller cells to retinal disease better, we have developed a transgenic model in which foci of Müller cell ablation can be selectively induced. MicroRNAs (miRNAs), small non-coding RNAs that are involved in post-transcriptional modulation, have critical functions in various biological processes. The aim of this study was to profile differential expression of miRNAs and to examine changes in their target genes 2 weeks after Müller cell ablation. We identified 20 miRNAs using the miScript HC PCR array. Data analysis using two target gene prediction databases (TargetScan and mirTarBase) revealed 78 overlapping target genes. DAVID and KEGG pathway analysis suggested that the target genes were generally involved in cell apoptosis, p53, neurotrophin, calcium, chemokine and Jak-STAT signalling pathways. Changes in seven target genes including Cyclin D2, Caspase 9, insulin-like growth factor 1, IL-1 receptor-associated kinase (IRAK), calmodulin (CALM) and Janus kinase 2 (Jak2), were validated with qRT-PCR and western blots. The cellular localisation of cleaved-caspase 9, Cyclin D2, Jak2 and CALM was examined by immunofluorescence studies. We found that the transcription of some miRNAs was positively, rather than negatively, correlated with their target genes. After confirming that overexpressed miR-133a-3p was localised to the outer nuclear layer in the damaged retina, we validated the correlation between miR-133a-3p and one of its predicted target genes, cyclin D2, with a luciferase assay in 661 photoreceptor cells. Results revealed by miRNA profiling, target gene analysis and validation were generally consistent with our previous findings that selective Müller cell ablation causes photoreceptor degeneration and neuroinflammation. Our data on alterations of miRNAs and their target gene expression after Müller cell ablation provide further insights into the potential role of Müller cell dysfunction in retinal disease.
Introduction
Müller cells, the principal glial cells of the mammalian retina, are essential for retinal homeostasis. Spanning the entire thickness of the retina, they support the surrounding neurones, including photoreceptors [1, 2], and are involved in forming and maintaining the blood retinal barrier (BRB)[3]. Dysfunction of Müller cells has been implicated in many retinal diseases such as diabetic retinopathy and macular telangiectasia [4, 5]. To investigate the relationship between Müller cell dysfunction and retinal diseases, we have created a transgenic mouse model in which Müller cells can be selectively and inducibly ablated [6]. These mice develop photoreceptor degeneration, neuroinflammation, including activation of microglia and surviving Müller cells, and differential expression of mature and precursor forms of neurotrophic factors such as neurotrophin-3 within 2 weeks of Müller cell ablation [6, 7]. We have previously reported differential expression of genes that was consistent with many of these pathological processes in this model [2].
MicroRNAs (miRNAs) are small, non-coding RNA molecules which are ~22 nucleotide (nt) long which are in transcribed in the nucleus [8, 9]. They regulate post-transcriptional gene expression by binding to the complementary site of mRNA [10, 11]. The short hairpin structure of primary miRNAs are processed into pre-miRNA by RNase III-type enzyme, Drosha, in the nucleus and exported into the cytoplasm by exportin-5 [12]. Pre-miRNAs are catalyzed by Dicer, another kind of RNase III-type enzyme, in the cytoplasm to become ~22 nt mature miRNA [13]. Mature miRNAs are assembled into a ribonucleoprotein effector complex known as miRNA induced silencing complex and bind to proteins of the argonaute family and GW182 family [8, 10]. It was previously thought that the main function of miRNAs was to repress transcription and destabilize mRNA, however, recent studies have proposed that miRNAs can also stimulate target mRNA expression [14–17].
The role of miRNAs in diseases of the vertebrate retina is an area of considerable potential significance [18, 19]. Many studies have been focused on the pathophysiological role of miRNAs in the pathogenesis of retinal diseases such as diabetic retinopathy and retinitis pigmentosa [8, 20–23]. There are few studies, however, of differential expression of miRNAs in retinal diseases in which Müller cell dysfunction is suspected to play a role. The transgenic model described above, in which Müller cells can be selectively ablated, provides a unique opportunity to study alterations of retinal miRNAs after induced Müller cell ablation. The aims of this study were to profile the differential expression of miRNAs and their target genes, and to investigate changes in target gene expression 2 weeks after selective Müller cell ablation, which is the peak time of photoreceptor apoptosis in this model [6].
Materials and Methods
Animals
This study was performed in accordance with the Association for Research in Vision and Ophthalmology guidelines for animal research and approved by The University of Sydney Animal Ethics Committee. Rlbp1-CreER mice were crossed with Rosa-DTA176 mice to produce Rlbp-CreER-DTA176 transgenic mice. Müller cell ablation was induced by daily intraperitoneal injection of tamoxifen (TMX) for 4 consecutive days at 8–10 weeks of age [6]. Wild type mice receiving TMX in the same way were used as controls. A total of 12 mice, including 6 transgenic mice with induced Müller cell ablation and 6 wild type mice, were used in this study. Mice were euthanized 2 weeks after Müller cell ablation. Retinae were isolated, snap frozen in liquid nitrogen and stored at-80°C until use.
miRNA Polymerase Chain Reaction (PCR) Array
Retinae were thawed then total RNA, including miRNAs, was extracted with a miRNeasy Mini Kit (Qiagen, 217004) according to manufacturer’s instructions. The quantity and quality were assessed by an Experion Automated Electrophoresis System (Biorad, 701–7001). Six retinae from each group were reverse-transcribed with miScript II RT kit (Qiagen, 218160) and diluted with RNase free water. miRNA PCR array was performed with 384 well-plate miScript miRNA High Content PCR array (Qiagen, MIMM-3001Z) which contains miScript primers for 372 of the most well characterised miRNAs and duplicates of 6 internal reference miRNAs. The identities of 372 miRNAs are listed in Supporting Information (Table A in S1 File). The thermo-cycle of the PCR array consisted of denaturing at 94° for 15 seconds, annealing at 55° for 20 seconds and extension at 70° for 30 seconds. Relative quantification was performed by the ΔΔCT method recommended by the manufacturer (http://pcrdataanalysis.sabiosciences.com/mirna/arrayanalysis.php?target=analysis). A p-value <0.05 was considered statistically significant and the fold change cut-off criteria was ± 2.
Functional Analysis of miRNAs
We started with functional analysis of each differentially expressed miRNAs by performing a PubMed literature search to identify recent publications, which would shed light on their related functions (http://www.ncbi.nlm.nih.gov/pubmed). In addition, we used 2 target gene prediction databases: TargetScan (http://www.targetscan.org) and miRTarbase (http://mirtarbase.mbc.nctu.edu.tw). Overlapping target genes from these 2 databases were identified for further analysis. Functional analysis of the target genes was performed by Database for Annotation, Visualisation Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) and associated pathways were identified by KEGG pathway database (http://www.genome.jp/kegg/pathway.html). We also performed in silico study with mirTarBase to examine direct seed matching sequence alignments between the identified miRNAs and their target genes prior to further investigations (Table 1).
Table 1. Direct Sequence Alignment Between miRNAs and Their Target Genes 3’UTR.
| ID | Target Gene | Binding Sites | Position | Target Score |
|---|---|---|---|---|
| miR133a-3p | Caspase 9 | miRNA 3′ gucgACC—AACUU—-CCCCUGGUUu 5′ | 1706–1732 | 136.00 |
| ||| || || | ||||||: | ||||
| Target 5′ gtagTGGTCTTAAAAGTGTGGACCAGt 3′ | ||||
| miRNA 3′ gucGAC-CAACUUCC———-CCUGGUUu 5′ | 1416–1445 | 127.00 | ||
| : || ||| |: | ||:|||| | ||||
| Target 5′ gcaTTGAGTTCAGTGACTCTCAGGGCCAAg 3′ | ||||
| miRNA 3′ gucgACC-AACUUCCCCUGGUUu 5′ | 45–67 | 122.00 | ||
| ||| |||:|| |:|||| | ||||
| Target 5′ ctcaTGGCTTGGAGCTGGCCAAg 3′ | ||||
| miR133a-3p | Cyclin D2 | miRNA 3′ guCGACCAACUUCCCCUGGUuu 5′ | 3974–3990 | 133.00 |
| ||||| || |||||| | ||||
| Target 5′ ggGCTGG—GA—-GGACCAca 3′ | ||||
| miRNA 3′ guCGACCAACUUCCCCUGGUUu 5′ | 3865–3883 | 125.00 | ||
| ||| || ||||||| | | ||||
| Target 5′ taGCTCTTT—-GGGGACCCAc 3′ | ||||
| miRNA 3′ gucGACCA—ACUUCCCCUGGuuu 5′ | 2963–2985 | 124.00 | ||
| ||||| | || |||||| | ||||
| Target 5′ cttCTGGTCCTTAA-GGGACCcca 3′ | ||||
| miR133a-3p | Insulin like Growth Factor 1 Receptor | miRNA 3′ gucGACCAACUUC—CCCUGGUuu 5′ | 5357–5380 | 141.00 |
| |||| |||| |||||| | ||||
| Target 5′ ggcCTGGAAGAAGCATGGACCAta 3′ | ||||
| miRNA 3′ gucgaccaacuuccCCUGGUUu 5′ | 5988–6009 | 140.00 | ||
| ||||||| | ||||
| Target 5′ agaaattctccccaGGACCAAt 3′ | ||||
| miRNA 3′ guCGAC-CAACUUCC——CCUGGUUu 5′ | 5022–5048 | 138.00 | ||
| |||| |||:: | ||||||: | ||||
| Target 5′ caGCTGCATTGGGAGACCTGGACCAGa 3′ | ||||
| miR-146a-5p | Interleukin Receptor Associated Kinase 1 | miRNA 3′ uuGGGUACCUUAAG—UCAAGAGu 5′ | 29–50 | 154.00 |
| |:|| ||: || ||||||| | ||||
| Target 5′ gaCTCA—GAGGTCAAAGTTCTCa 3′ | ||||
| miRNA 3′ uuGGGUAC—-CUUAAGUCAAGAGU——— 5′ | 45–72 | 135.00 | ||
| |:|||| |:| |||||||| | ||||
| Target 5′ ttCTCATGCTTGGA—-AGTTCTCATAGTGT 3′ | ||||
| miRNA 3′ uugGGUACCU—UAAGUCAAGAGu 5′ | 133–156 | 125.00 | ||
| |: |||| | | |||||| | ||||
| Target 5′ gggCAGTGGACCCTGCTGTTCTCa 3′ | ||||
| miR1a-1 | Calmodulin | miRNA 3′ uaUGUAUGAAGAAAUGUAAGGu 5′ | 2743–2762 | 144.00 |
| ||||| |:||||:||||: | ||||
| Target 5′ gtACATA—TTTTTATATTCTc 3′ | ||||
| miRNA 3′ uaUG-UAUGAAG—AAAUGUAAGGu 5′ | 3350–3373 | 133.00 | ||
| ||: || ||| | |||||:| | ||||
| Target 5′ gcACTGTA-TTCAATAAACATTTCt 3′ | ||||
| miRNA 3′ uaUGUAUGAAG———AAAUGUAAGGu 5′ | 1234–1261 | 126.00 | ||
| ||||: ||: | |::||||| | ||||
| Target 5′ agACATGATTTAGTGTGTCTGTATTCCt 3′ | ||||
| miR-375–3p | Janus Kinase 2 | miRNA 3′ agugcgcUCGGCUUGCUUGUUu 5′ | 563–584 | 127.00 |
| ||| || |||||| | ||||
| Target 5′ ttttcaaAGCAAAAGGAACAAa 3′ | ||||
| miRNA 3′ agugcgcucggcuugCUUGUUu 5′ | 33–54 | 120.00 | ||
| |||||| | ||||
| Target 5′ agacttccagaaccaGAACAAa 3′ | ||||
| miRNA 3′ agUGCGCUCGGCUUGCUUGUuu 5′ | 1073–1092 | 104.00 | ||
| | ||| |:: ||| ||| | ||||
| Target 5′ gcAGGCGTGT—GACGCACAtc 3′ |
Quantitative RT-PCR (qRT-PCR)
qRT-PCR was performed as previously described [2]. In brief, an equal amount of total RNA was reverse-transcribed with SuperScript VILO cDNA Synthesis kit (Life Technologies, 11754050) according to manufacturer’s instruction, and diluted with RNase free water and Express SYBR GreenER qPCR Supermix (Life Technologies, 11784–200). Primers we used in this study are listed in Table 2. The relative quantitative analysis was performed by Relative Expression Software Tool (REST) 2009 [24]. Expression values were normalised to the geometric mean of 3 reference genes including 18SrRNA, GAPDH, β-Tubulin.
Table 2. Information on primers used for qRT-PCR validation.
| Primers | Forward (5′->3′) | Reverse (5′->3′) |
|---|---|---|
| Cyclin D2 | GAGTGGGAACTGGTAGTGTTG | CGCACAGAGCGATGAAGGT |
| Casp 9 | TCCTGGTACATCGAGACCTTG | AAGTCCCTTTCGCAGAAACAG |
| Igf | CTGGACCAGAGACCCTTTGC | GGACGGGGACTTCTGAGTCTT |
| Irak | AGCCGAGGTCTGCATTACATT | TGGCAGTCTGGATAACTGATGA |
| Calm1 | TGGGAATGGTTACATCAGTGC | CGCCATCAATATCTGCTTCTCT |
| Calm2 | ACGGGGATGGGACAATAACAA | TGCTGCACTAATATAGCCATTGC |
| Jak2 | GAACCTACAGATACGGAGTGTCC | CAAAATCATGCCGCCACT |
| Gs | TGAACAAAGGCATCAAGCAAATG | CAGTCCAGGGTACGGGTCTT |
| Gfap | TCCTTCCAAGGTTGTCCATC | CCAATCAGCCTCAGAGAAGG |
| 18srRNA | GCAATTATTCCCCATGAACG | GGGACTTAATCAACGCAAGC |
| Β-Tub | GATCGGTGCTAAGTTCTGGGA | AGGGACATACTTGCCACCTGT |
| Gapdh | AAGATGGTGATGGGCTTCCCG | TGGCAAAGTGGAGATTGTTGCC |
Western Blot
Western blot was performed as previously described [2]. Briefly, proteins were extracted from each retina then the quantity was assessed by QuantiPro BCA assay kit (Sigma-Aldrich, QPBCA). Equal amounts of protein were loaded into NuPage Bis-Tris gels (Life Technologies, NP3023BOX), and transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 5% BSA in TBST and the primary antibodies were incubated overnight at 4°C. Primary antibodies included cyclin D2 (cell signalling, #2924), cleaved caspase 9 (cell signalling, #9509S), calmodulin (CALM, abcam, ab45689), insulin-like growth factor 1 receptor (IGF1r, abcam, ab39398), glial fibrillary acidic protein (GFAP, abcam, ab53554) and janus kinase 2 (Jak2, abcam, ab39636). Protein bands were visualised after incubation with the corresponding secondary antibodies conjugated with horseradish peroxidase. GeneTool image scanning and analysis software package was used to perform protein bands densitometry and the results were normalised with loading control α/β tubulin (Cell Signalling, #2148).
Immunofluorescence
Immunofluorescence labelling was performed as previously described [2]. Briefly, eyes were enucleated and anterior segments were dissected immediately. The remaining eye cups were fixed with 4% paraformaldehyde in PBS for an hour at room temperature and cryo-protected with 20% sucrose in PBS for overnight at 4°C. The eyecups were embedded with Optimum Cutting Medium and sectioned at 12μm on Superfrost glass slides. Sections were washed with PBS 3 times for 5 minutes and blocked with normal goat serum for 1 hour at room temperature. After washing with PBS, sections were incubated with primary antibody for overnight at 4°C. The primary antibodies used in this study were cleaved caspase 9 (cell signalling, #9509S), cyclin D2 (cell signalling, #2924), JAK2 (abcam, ab39636) and calmodulin (abcam, ab45689). Secondary antibody incubation was followed on the next day for 2 hours at room temperature and nucleus staining was performed with Hoechst. Stained sections were examined by confocal microscopy as described previously [2, 6].
In Situ Hybridisation
In situ hybridisation was performed to localise miRNA expression in the retina using Dig-labelled miR-133a-3p miRCURY LNA probe (Exiqon, Cat#39270–01) according to the manufacturer’s instruction with minor modifications. Briefly, cryosections were thawed, air-dried and washed with PBS for 3 times. Sections were treated with proteinase K (100ng in 200ml water) in a slide wash container for 5mins followed by acetylation treatment (2.3ml of triethanolamine and 500μl acetic anhydride in 200ml water) for 10mins. A hybridisation buffer containing 25nM of probe was pre-warmed for 5min at 65°C and then applied onto sections. Sections were cover-slipped and incubated in a chamber humidified with 50% formamide in saline sodium citrate (SSC) solution overnight at 53°C. On the next day, sections were washed, blocked with 10% normal goat serum and incubated with an anti-Dig antibody (Roche, 11093274910) overnight at 4°C. NBT/BCIP alkaline phosphatase colour reaction (Vector laboratories, SK-5400) was performed to visualise the cellular localisation of miR-133a expression according to the manufacturer’s instruction.
Luciferase Assay
Luciferase assay was performed to validate the direct correlation between miRNA and its predicted target gene expression. We chose to validate miR-133a-3p and one of its target genes, cyclin D2, since cyclin D2 was the most differentially expressed gene after Muller cell ablation. Briefly, 3’UTR of cyclin D2 was amplified by PCR, digested with XhoI and NotI restriction enzymes (New England Biolabs, R0146S and R3189S) and cloned into a multiple cloning site located downstream of the renilla translational stop codon within the psiCHECKTM-2 vector (Promega, C8021). 661w photoreceptor cells were seeded in 12-well plates (50,000 cells per well) and cultured for 2 days. Once they reached 70% confluence, the cells were transfected with the vector alone, or co-transfected with a miRNA-133a-3p mimic (Ambion, 4464067) or a control miRNA (Ambion; 4464058) using Lipofectamin 3000 (Life Technologies, L3000001) for 48 hrs. Luciferase assay was performed using the Dual-Glo Luciferase Assay System (Promega, E2920) and the luciferase activity was measured by Safire2TM Tecan plate reader (Tecan Austria, Safire-BASIC). Firefly luciferase activity was normalised to the renilla luciferase activity.
Statistic Analysis
Results are expressed as mean ± SEM. Data were analysed using unpaired Student t-test. A p value <0.05 was regarded as statistically significant.
Results
miRNA PCR Array
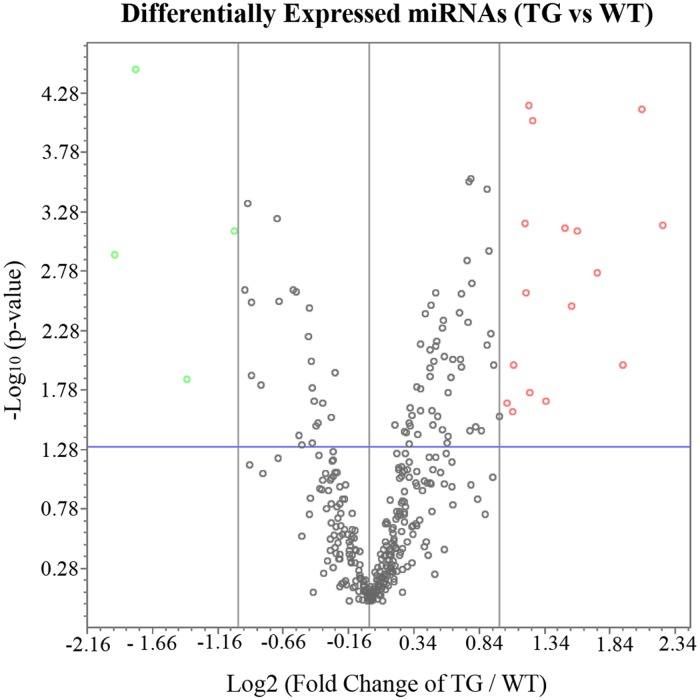
miRNA PCR array analysis revealed 20 differentially expressed miRNAs with greater than 2 fold change (P < 0.05) 2 weeks after Müller cell ablation. Of these, 16 were upregulated (miR-1196–5p, miR-133a-3p, miR-142–3p, miR-142–5p, miR-146a-5p, miR-146b-3p, miR-146b-5p, miR-190a-5p, miR-1a-1–5p, miR-1a-3p, miR-200a-3p, miR-222–5p, miR-29b-3p, miR-335–3p, miR-335–5p, and miR-653–5p) and 4 were downregulated (miR-1983, miR-375–3p, miR-376c-3p, and miR-542–5p) (Table 3 and Fig. 1). A literature search based functional analysis revealed that the potential functions of the differentially expressed miRNAs were generally associated with pathways contributing to neuronal damage, such as neuronal inflammation and neuronal apoptosis (Table 3). Some of the miRNAs that we found to be differentially regulated after Müller cell ablation have been reported in retinal diseases such as diabetic retinopathy and retinitis pigmentosa (Table 3). In general, the results we obtained from this miRNA PCR array were consistent with our previous studies which identified retinal morphological changes, microarray profiling, neuroinflammation and differential expression of neurotrophins after selective Müller cell ablation [2, 6, 7].
Table 3. List of Differentially Expressed miRNAs 2 weeks after induced Müller Cell Ablation.
| miRNA | Fold Change | P-value | Potential Functions | References |
|---|---|---|---|---|
| mmu-miR-1196–5p | 2.14 | 0.0255 | Not well characterised | N/A |
| mmu-miR-133a-3p | 2.15 | 0.0103 | Inflammation, Retinitis pigmentosa, Retinal degeneration | [21, 23, 25] |
| mmu-miR-142–3p | 4.26 | 0.00072 | Neuroinflammation, Neurodegeneration, Retinitis Pigmentosa, Diabetes, Retinal degeneration | [21–23, 27, 52, 53] |
| mmu-miR-142–5p | 4.75 | 0.000685 | Neuroinflammation, Autoimmune uvities, Retinitis pigmentosa, Retinal degeneration | [21, 23, 27, 53, 54] |
| mmu-miR-146a-5p | 3.02 | 0.000072 | Neuroinflammation, Diabetes, Neurodegeneration | [22, 26–28, 53, 55, 56] |
| mmu-miR-146b-3p | 3.84 | 0.01025 | Neuroinflammation, Neuronal diseases | [27, 28] |
| mmu-miR-146b-5p | 2.83 | 0.000728 | Neuroinflammation, Neuronal diseases | [27, 28] |
| mmu-miR-190a-5p | 2.08 | 0.02156 | Not well characterised | N/A |
| mmu-miR-1983 | -3.46 | 0.000033 | Not well characterised | N/A |
| mmu-miR-1a-1–5p | 3.35 | 0.001722 | Retinitis pigmentosa, Retinal degeneration | [21, 23] |
| mmu-miR-1a-3p | 2.29 | 0.00252 | Retinitis pigmentosa, Retinal degeneration | [21, 23] |
| mmu-miR-200a-3p | 2.55 | 0.020964 | Neuroinflammation, Neurodegeneration, Diabetes | [22, 25, 26, 28, 29, 52] |
| mmu-miR-222–5p | 2.35 | 0.017673 | Neurnal diseases | [28] |
| mmu-miR-29b-3p | 2.38 | 0.000091 | Diabetes, Neuroinflammation, Neurodegeneration, Neuronal development, Neuronal apoptosis | [18, 26, 28, 55, 57] |
| mmu-miR-335–3p | 2.33 | 0.000068 | Neuroinflammation | [53, 58] |
| mmu-miR-335–5p | 2.29 | 0.000664 | Neuroinflammation | [53, 58] |
| mmu-miR-375–3p | -2.04 | 0.000766 | Diabetes | [59] |
| mmu-miR-376c-3p | -2.63 | 0.01363 | Neuroinflammation, Neuronal development | [53] |
| mmu-miR-542–5p | -3.86 | 0.001219 | Pro-Apoptotic | [60] |
| mmu-miR-653–5p | 2.92 | 0.003296 | Not well characterised | N/A |
Fig 1. Volcano Plot of Differentially Expressed miRNA in after Selective Müller cell ablation.
The red circles represent upregulated miRNAs and the green circles indicate downregulated miRNAs (2 fold-change). The blue line indicates p-value of 0.05.
Target Gene Analysis and qRT-PCR Validation
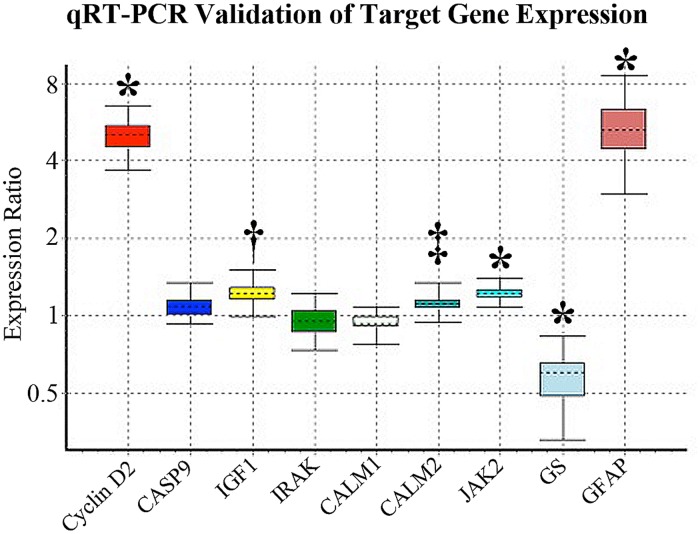
We used 2 different databases, TargetScan and mirTarBase, to analyse genes which could be potentially targeted by the differentially expressed miRNAs. We identified 78 overlapping genes that could be potentially targeted by the 20 differentially expressed miRNAs. Of these, 72 corresponded to upregulated miRNAs and 6 to downregulated miRNAs (Table B and Table C in S1 File). DAVID and KEGG pathway analysis revealed that 41 target genes had significant biological functions (Table D in S1 File). Based on their potential roles in retinal pathology as previously reported [2, 6], we chose 7 genes for qRT-PCR validation, including Cyclin D2, Caspase 9, IGF1, IRAK, CALM1, CALM2, and Jak2 (Table 3). Prior to further validation with qRT-PCR, we performed in silico study with mirTarBAse which revealed seed sequence alignments between miRNAs and their target genes (Table 1). Glutamine synthetase (GS) and GFAP were used as Müller cell markers to correlate changes in these 7 genes with Müller cell disruption. Consistent with our previous studies [6, 7], Müller cell ablation resulted in downregulation of GS and upregulation of GFAP (Table 3). qRT-PCR analysis revealed that Cyclin D2, IGF1, CALM2 and Jak2 were significantly upregulated 2 weeks after Müller cell ablation (p<0.05) (Table 4 and Fig. 2). The KEGG pathway analysis suggested that these genes were involved in cell apoptosis, p53, neurotrophin, calcium, chemokine and Jak-STAT signalling pathways (Table 4).
Table 4. Summary of some miRNA target genes and signalling pathways that might be involved in the retinal pathology caused by selective Müller cell ablation.
| Gene Name | KEGG Pathways/Markers | qRT-PCR Expression | P-value | qRT-PCR Result |
|---|---|---|---|---|
| CyclinD2 | p53 Signalling pathway, | 5.047 | 0.001 | UP |
| Casp9 | Apoptosis, p53 Signalling pathway, | 1.081 | 0.101 | - |
| Igf1 | p53 Signalling pathway | 1.220 | 0.003 | UP |
| Irak | Apoptosis, Neurotrophin pathway | 0.950 | 0.426 | - |
| Calm1 | Calcium signalling pathway, Neurotrophin pathway | 0.932 | 0.110 | - |
| Calm2 | Calcium signalling pathway, Neurotrophin pathway | 1.109 | 0.012 | UP |
| Jak2 | Chemokine Signalling pathway, Jak-STAT Signalling Pathway | 1.222 | 0.001 | UP |
| GS | Müller Cell Marker | 0.561 | 0.000 | DOWN |
| Gfap | Gliosis Marker | 5.277 | 0.001 | UP |
Fig 2. qRT-PCR validation of target gene expression.
The boxes represent the interquartile range, and the dotted line represents the median gene expression. Whiskers indicate the maximum and minimum values. * indicates p<0.001, † represents p<0.01 and ‡ shows p<0.05.
Western Blot Analysis
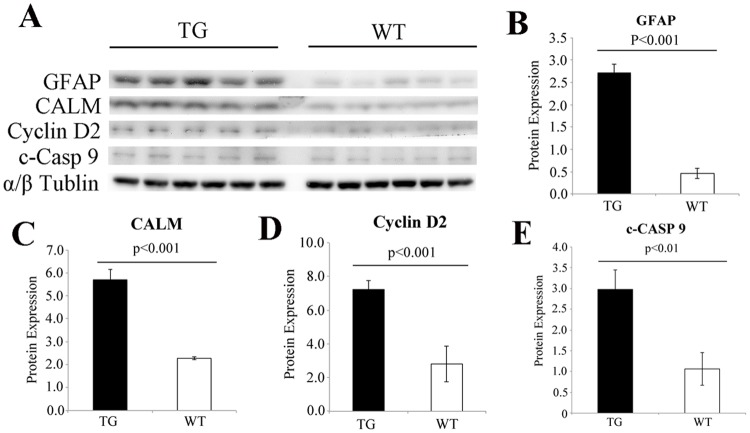
Western blot analysis after Müller cell ablation found changes that were consistent with target gene analysis revealed by qRT-PCR. Genes involved in the p53 signalling pathway, cell apoptosis including cleaved-caspase 9 and cyclin D2 and calmodulin which plays a role in calcium signalling and the neurotrophin pathway, were all significantly upregulated after Müller cell ablation (Fig. 3). Jak2 tended to be upregulated but the change was not statistical significant (P = 0.2).
Fig 3. Western blot validation of target gene expression.
(A-E) Western blot analysis revealed significant upregulation of proteins related to gliosis, calcium signalling pathway and p53 signalling pathways. The results were consistent with the qRT-PCR analysis. GFAP; glia fibrillary acidic protein, CALM; calmodulin, c-CASP 9; cleaved caspase 9.
Immunofluorescence
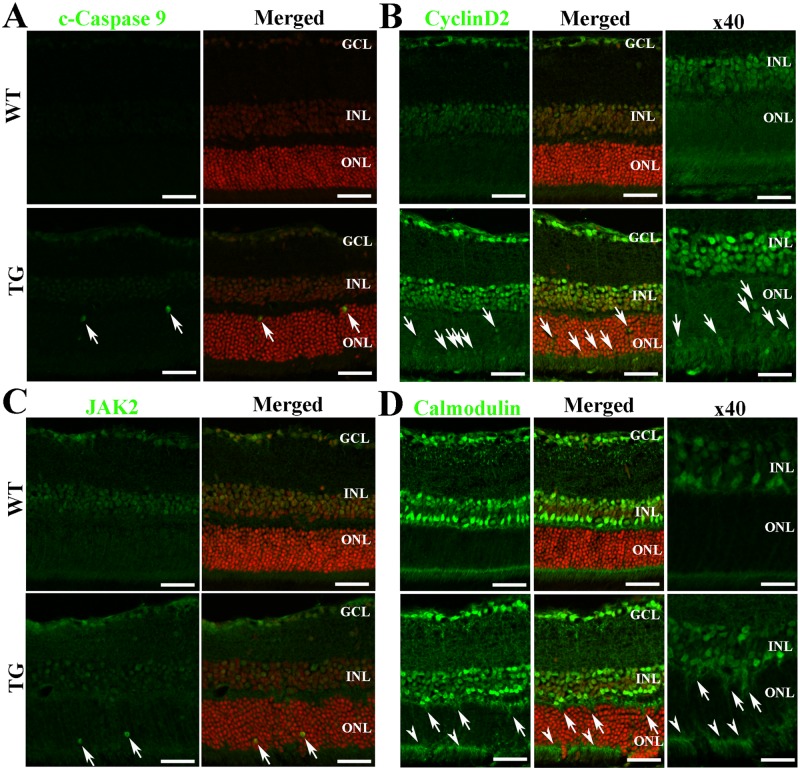
We performed immunofluorescence with antibodies against four proteins encoded by target genes including cleaved-caspase 9, cyclin D2, Jak2 and calmodulin to examine their cellular specific expression (Fig. 4). We found that cleaved-caspase9 (Fig. 4A) and Jak2 (Fig. 4C) were mainly expressed in the outer nuclear layer (ONL) in areas corresponding with patches of Müller cell ablation. Upregulation of cyclin D2 was observed in not only the inner nuclear layer (INL) and the ONL but also in some cell bodies of photoreceptors near the outer limiting membrane after Müller cell ablation compared with controls (Fig. 4B). Calmodulin was mainly expressed in the INL and weakly expressed in photoreceptor inner segments in the normal retinae. It was markedly upregulated in the outer plexiform layer, ONL and photoreceptor inner segments in areas of Müller cell ablation (Fig. 4D).
Fig 4. Immunofluorescent staining for proteins encoded by genes targeted by differentially expressed miRNAs after Müller cell ablation.
The genes encoding C-caspase 9 and CyclinD2 are targeted by miR133a-3p. “Merged” includes Hoechst stain. Jak2 is encoded by a gene that is targeted by miR-375–3p. Calmodulin is a gene targeted by miR1a-1. Cleaved-caspase 9 (A, arrows) and Jak2 (C, arrows) were only observed after Müller cell ablation (A and C). Increased immunoreactivity for CyclinD2 was observed in the ganglion cell layer (GCL), inner nuclear layer (INL) and outer nuclear layer (ONL) after Müller cell ablation compared with controls (B). Calmodulin was mainly expressed in the GCL and retinal neurons in the INL in the normal retina but increased expression (arrows) was observed in the outer plexiform layer, ONL and photoreceptor segments (arrow heads) in areas of Müller cell ablation, which are marked by focal areas of degeneration in the ONL (D). Scale bars represent 50μm. Scale bars in figures with high magnification (x40 in B and D) represent 25μm.
Validation of the Correlation of miR-133a-3p with Its Target Gene Cyclin D2 Expression Using Luciferase Assay
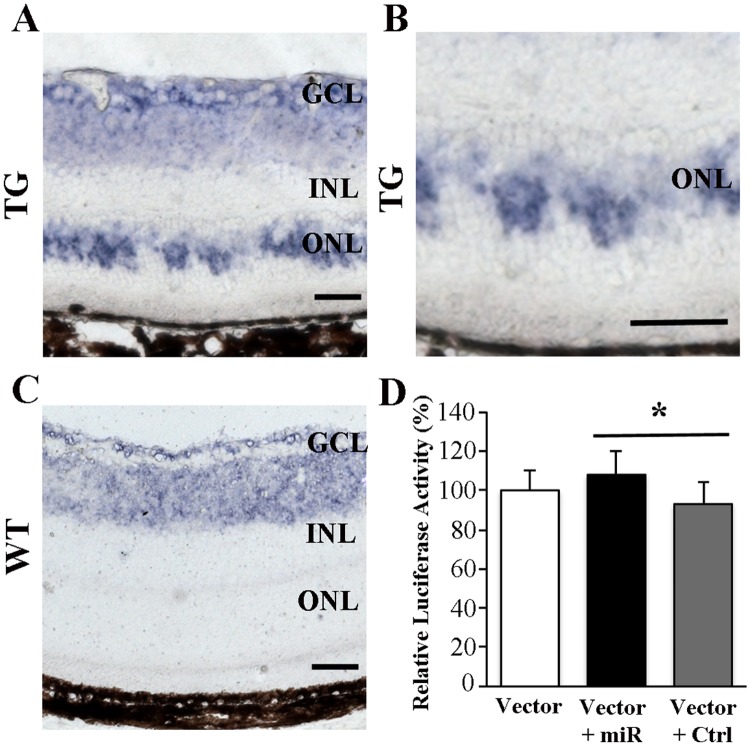
miRNA PCR array showed 2.15-fold increase in miR-133a-3p expression (Table 3) while qRT-PCR analysis showed 5-fold increase in cyclin D2 expression after Muller cell ablation (Table 4). As cyclin D2 is one of the genes targeted by miR-133a-3p, our data indicate a positive correlation between miR-133a-3p and cyclin D2 expression. We performed in situ hybridisation and found that the increased miR-133a-3p expression was mainly localised to the outer nuclear layer in the retina after Muller cell ablation (Fig. 5A-C). In order to validate the functional correlation between miR-133a-3p and cyclin D2 expression, we cloned the 3’UTR region of cyclin D2 gene into a vector and conducted luciferase assay in 661w photoreceptor cells. We found that co-transfection of 661w cells with a miR-133a-3p mimic along with the vector resulted in significant increase in the normalised luciferase activity compared with cells co-transfected with a control miRNA and the vector (P<0.05, Fig. 5D), thus confirming the positive correlation between miR-133a-3p and cyclin D2 expression.
Fig 5. (A-C), in situ hybridisation for miR-133a-3p expression in normal (C) and diseased (A and B) retinas. (D), luciferase assay to study the correlation of miR-133a-3p with cyclin D2 expression in 661 photoreceptor cells.
The expression of miR-133a-3p in the outer nuclear layer (ONL) was increased after induced Müller cell ablation. Co-transfection of 661w cells using a vector containing 3’UTR region of the cyclin D2 gene along with a miR-133a-3p mimic (Vector + miR) induced a significant increase in the normalised firefly luciferase activity compared with cells co-transfected using the vector with a control miRNA (Vector + Ctrl) (*P<0.05, n = 6/group). Data is presented as Mean + SEM. Scale bars in A-C: 50μm. GCL = ganglion cell layer. INL = inner nuclear layer. ONL = outer nuclear layer.
Discussion
We have identified 20 miRNAs that were differentially expressed during the photoreceptor degeneration that occurred after selective Müller cell ablation. We then used 2 different databases, TargetScan and mirTarBase, to identify a number of genes that could be potentially targeted by these miRNAs. Of 72 target genes that we identified, 41 were found, by DAVID and KEGG pathway analysis, to be involved in several biological functions. Based on our previous studies, we chose 7 target genes which were involved in apoptosis, p53, chemokine, Jak-STAT, calcium and neurotrophin signalling pathways for further validation. Upregulation of the chosen target genes was confirmed with qRT-PCR and validated by western blot analysis. Immunofluorescence studies revealed that the upregulation of these miRNA-target genes were mainly observed in the ONL where photoreceptor degeneration occurred after selective Müller cell ablation. We conducted in situ hybridisation for miR-133a-3p and found that the increased expression of miR-133a-3p was mainly localised in the ONL. Furthermore, we performed luciferase assay to validate the positive correlation between miR133a-3p and one of its predicted targets genes, cyclin D2 in 661 photoreceptor cells. The results were generally consistent with our previous findings that selective Müller cell ablation causes photoreceptor degeneration [2, 6].
Neuronal damage in this transgenic model is characterized by photoreceptor degeneration caused by selective Müller cell ablation [6]. MiRNA profiling conducted 2 weeks after Müller cell ablation identified a number of differentially expressed miRNAs whose target genes were associated with cell apoptosis and neurodegeneration in the central nervous system including retinal degeneration [21, 23, 25–29]. For instance, one of the genes targeted by miR-133a-3p is caspase-9, which can form an apoptosome with cytochrome-c released from mitochondria and lead to activation of other caspases and cell death [30]. Calmodulin, which is targeted by mir-1a-3, has been reported that calmodulin plays a role in neurotransmitter release and trafficking in synapsis [31, 32]. However, overexpression of calmodulin has been reported to be involved in neuronal apoptosis by activating calcium/calmodulin-dependent protein kinase II [33, 34]. Inhibition of calmodulin activity has been reported to prevent retinal neuronal apoptosis [35–38].
Our previous studies found that the microarray profiling in which we performed 1 week after Müller cell ablation, also revealed upregulation of genes involved in apoptosis [2], and the characterization study showed photoreceptor apoptosis was peaked at 2 weeks after Müller cell ablation [6]. We did not observe any significant differences in the number of TUNNEL positive cells the retina between 1 week and 2 weeks after Müller cell ablation, thus we believe the current study is generally supported by our previously observations.
Interestingly, miR-133a-3p also targets an anti-apoptotic gene. Cyclin D2, which has been reported to play a role in anti-apoptosis by phosphorylating retinoblastoma protein, cell cycle arrest and neurogenesis[39–42], was significantly upregulated after Müller cell ablation. After confirmation that the increased miR-133a-3p expression was mainly localised to the outer nuclear layer after Muller cell ablation, we conducted luciferase assay in 661w photoreceptors to validate the positive correlation between miR-133a-3p and cyclin D2 expression. We found that co-transfection of 661w cells using the vector with a miR-133a mimic induced significant increase in luciferase activity compared with cells co-transfected using the vector with a control miRNA (Fig. 5D). A previous study reported a negative correlation between miR-133a and cyclin D2 expression in cardiac muscle cells and cos-1 cells [41]. The discrepancy between that study and the results we present here could be due to the diverse roles of miR-133a and cyclin D2 in different tissues and biological models. Cyclin D family has been reported to play a neuroprotective role in brain injury and retinal degeneration [41, 43]. Moreover, cyclin D2 is involved in neurogenesis [40]. It is unclear whether the overexpressed miR-133a and cyclin D2 are due to a defensive response to the retinal stress caused by Muller cell ablation. Future studies are warranted to investigate the effects of intervention of miR-133a and cyclin D2 overexpression on photoreceptor degeneration in our transgenic mice.
Neuroinflammation was also shown in the transgenic model of selective Müller cell ablation [2, 6, 7]. Mir-146a-5p, which has been reported to play a role in neuroinflammation and neurodegeneration [22, 27], was upregulated after induced Müller cell ablation. It has been reported that its target genes, such as IRAK1, IRAK2, and TRAF6, are involved in cell survival and neuroprotection. IRAK recruits the p75 neurotrophin receptor and activates nuclear factor keppa B (NF-kB) [44, 45]. We recently showed that the p75 neurotrophin receptor was profoundly upregulated in activated surviving Müller cells and that photoreceptor degeneration occurred concurrently with activation of microglial cells in this model. Linking the result of upregulation of mir-146a-5p with features of photoreceptor degeneration and neuroinflammation caused by Müller cell ablation indicates a potential positive correlation between mir-146a-5p expression and IRAK expression, although our qRT-PCR analysis of the change in IRAK transcription was not statistically significant between transgenic and control mice. Further analysis on the relationship between miR-146a-5p upregulation of changes in different isoforms of IRAKs and their downstream effectors is warranted to provide a better understanding of the role of miRNA-146 in photoreceptor degeneration and neuroinflammation caused by Müller cell dysfunction.
Interestingly, miR-375–3p was downregulated after Müller cell ablation, and Jak2, one of its target genes, was upregulated. The negative correlation between miR-375 and Jak2 is consistent with a recent report in which target mutation of a predicted miR-375-binding site abolished the activity of a luciferase reporter carrying the 3’ untranslated region of Jak2 [46]. Jak2 has been reported to be involved in activation of immune response/chemokine signalling and the Jak/STAT pathways. Inflammation induced by lipopolysachrides in the rat retina caused activation of the Jak2/STAT3 pathway and neuronal gliosis [47]. Our recent studies found that selective Müller cell ablation resulted in profound activation of surviving Müller cells and microglia and overexpression of TNFα. It is plausible that the downregulation of miR-375–3p may trigger activation of the Jak2/STAT3 pathway and chemokine signalling, both of which can damage photoreceptors.
We conducted immunostaining to study the cellular localization of a number of target genes in normal and degenerating retinae. Cleaved-caspase 9, cyclin D2 and JAK2, were mainly expressed in the ONL in areas corresponding to Müller cell ablation, whereas upregulation of calmodulin was observed in the outer plexiform layer, ONL and photoreceptor segments after Müller cell ablation. Calmodulin is a highly conserved regulatory protein found in all eukaryotic organisms which mediates a variety of calcium ion-dependent signaling pathways [31]. A recent study showed that over-activation of calcium/calmodulin-activated protein kinase II is involved in photoreceptor degeneration in the rd1 mouse [48]. Our observation that calmodulin was upregulated after Müller cell ablation suggests that activation of the calcium/calmodulin signalling may contribute to photoreceptor degeneration. Interestingly, cyclin D2 was highly upregulated in the INL of retinae with Müller cell ablation compared with the normal retina in controls. As cyclin D2 is anti-apoptosis [39], this observation may partially explain the phenomenon that neuronal damage is not obvious in the INL following Müller cell ablation in this transgenic model [6].
One of the interesting findings we found in this study is that a number of differentially expressed miRNAs appear to be positively correlated with their target gene expression after selective Müller cell ablation. While the precise molecular mechanisms of which miRNA control gene expression are still largely obscure, there has been a growing number of reports of a positive correlation between miRNA and mRNA expression [17, 49–51]. Some hypothetical and in silico studies of the mechanism of which miRNA promotes gene expression have recently been reported [14–17, 50].
We acknowledge that this profiling study only covered 372 miRNAs rather than an unbiased genome-wide screen. We cannot rule out the possibility that the screening of a limited number of miRNAs might have missed some miRNAs which make important contributions towards photoreceptor degeneration and neuroinflammation in retinal disease.
We report here that miRNAs and their target genes that are differentially expressed soon after selective Müller ablation are mainly involved in neuronal apoptosis, inflammation and neurodegenration. Müller cell dysfunction has been implicated in the pathogenesis of many retinal diseases but its precise contribution to the photoreceptor degeneration, which ultimately results in loss of vision, remains poorly understood. The results of miRNA profiling after Müller cell ablation that we present here provide further insights into the pathogenesis of photoreceptor degeneration associated with Müller cell dysfunction.
Supporting Information
Table A. The list of 372 miRNA IDs used in Qiagen HC PCR array. Table B. Target gene analysis from the upregulated miRNAs. The list shows the overlapping target genes from TargetScan and miTarBase. Table C. Target gene analysis from the dwonregulated miRNAs. The list is the overlapping target genes from TargetScan and miTarBase. Table D. Functional analysis of the target genes performed by DAVID and KEGG.
(DOCX)
Acknowledgments
The authors thank Drs. Natalie Castrechini, Teresa Lyons and Fanfan Zhu for their technical advice.
Funding Statement
This study was supported by grants from Lowy Medical Research Institute, National Health and Medical Research Council (NHMRC, Australia; APP1028393) and Ophthalmic Research Institute of Australia. Mark Gillies is a fellow of Sydney Medical School Foundation and supported by a NHMRC Practitioner Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jablonski MM, Iannaccone A (2000) Targeted disruption of Muller cell metabolism induces photoreceptor dysmorphogenesis. Glia 32: 192–204. [DOI] [PubMed] [Google Scholar]
- 2. Chung SH, Shen W, Gillies MC (2013) Laser capture microdissection-directed profiling of glycolytic and mTOR pathways in areas of selectively ablated Muller cells in the murine retina. Investigative ophthalmology & visual science 54: 6578–6585. [DOI] [PubMed] [Google Scholar]
- 3. Tout S, Chan-Ling T, Hollander H, Stone J (1993) The role of Muller cells in the formation of the blood-retinal barrier. Neuroscience 55: 291–301. [DOI] [PubMed] [Google Scholar]
- 4. Powner MB, Gillies MC, Tretiach M, Scott A, Guymer RH, Hageman GS, et al. (2010) Perifoveal muller cell depletion in a case of macular telangiectasia type 2. Ophthalmology 117: 2407–2416. 10.1016/j.ophtha.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rungger-Brandle E, Dosso AA PM (2000) Glial reactivity, an early feature of diabetic retinopathy. Investigative ophthalmology & visual science 41: 1971–1980. [PubMed] [Google Scholar]
- 6. Shen W, Fruttiger M, Zhu L, Chung SH, Barnett NL, Kirk JK, et al. (2012) Conditional Muller Cell Ablation Causes Independent Neuronal and Vascular Pathologies in a Novel Transgenic Model. The Journal of neuroscience: the official journal of the Society for Neuroscience 32: 15715–15727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen W, Zhu L, Lee SR, Chung SH, Gillies MC (2013) Involvement of NT3 and P75NTR in photoreceptor degeneration following selective Muller cell ablation. Journal of neuroinflammation 10: 137 10.1186/1742-2094-10-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundermeier TR, Palczewski K (2012) The physiological impact of microRNA gene regulation in the retina. Cellular and molecular life sciences: CMLS 69: 2739–2750. 10.1007/s00018-012-0976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 10. Huntzinger E E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews Genetics 12: 99–110. 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 11. Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry 79: 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 12. Fabian MR, Sundermeier TR, Sonenberg N (2010) Understanding how miRNAs post-transcriptionally regulate gene expression. Progress in molecular and subcellular biology 50: 1–20. 10.1007/978-3-642-03103-8_1 [DOI] [PubMed] [Google Scholar]
- 13. Siomi H, Siomi MC (2010) Posttranscriptional regulation of microRNA biogenesis in animals. Molecular cell 38: 323–332. 10.1016/j.molcel.2010.03.013 [DOI] [PubMed] [Google Scholar]
- 14. Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- 15. Vasudevan S (2012) Posttranscriptional upregulation by microRNAs. Wiley interdisciplinary reviews RNA 3: 311–330. 10.1002/wrna.121 [DOI] [PubMed] [Google Scholar]
- 16. Lee S, Vasudevan S (2013) Post-transcriptional stimulation of gene expression by microRNAs. Advances in experimental medicine and biology 768: 97–126. 10.1007/978-1-4614-5107-5_7 [DOI] [PubMed] [Google Scholar]
- 17. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America 105: 1608–1613. 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hackler L Jr, Wan J, Swaroop A, Qian J, Zack DJ (2010) MicroRNA profile of the developing mouse retina. Investigative ophthalmology & visual science 51: 1823–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, et al. (2010) miRNeye: a microRNA expression atlas of the mouse eye. BMC genomics 11: 715 10.1186/1471-2164-11-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, et al. (2010) Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141: 618–631. 10.1016/j.cell.2010.03.039 [DOI] [PubMed] [Google Scholar]
- 21. Loscher CJ, Hokamp K, Wilson JH, Li T, Humphries P, Farrar GJ, et al. (2008) A common microRNA signature in mouse models of retinal degeneration. Experimental eye research 87: 529–534. 10.1016/j.exer.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kovacs B, Lumayag S, Cowan C, Xu S (2011) MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Investigative ophthalmology & visual science 52: 4402–4409. [DOI] [PubMed] [Google Scholar]
- 23. Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, et al. (2007) Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome biology 8: R248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfaffl MW, Horgan GW L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freilich RW, Woodbury ME, Ikezu T (2013) Integrated Expression Profiles of mRNA and miRNA in Polarized Primary Murine Microglia. PloS one 8: e79416 10.1371/journal.pone.0079416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ (2013) Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Frontiers in cellular neuroscience 7: 178 10.3389/fncel.2013.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ksiazek-Winiarek DJ, Kacperska MJ, Glabinski A (2013) MicroRNAs as novel regulators of neuroinflammation. Mediators of inflammation 2013: 172351 10.1155/2013/172351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maes OC, Chertkow HM, Wang E, Schipper HM (2009) MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Current genomics 10: 154–168. 10.2174/138920209788185252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S (2011) MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 60: 1314–1323. 10.2337/db10-1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fridman J, SLowe SW (2003) Control of apoptosis by p53. Oncogene 22: 9030–9040. [DOI] [PubMed] [Google Scholar]
- 31. DeLorenzo RJ (1982) Calmodulin in neurotransmitter release and synaptic function. Federation proceedings 41: 2265–2272. [PubMed] [Google Scholar]
- 32. Hinds HL, Goussakov I, Nakazawa K, Tonegawa S, Bolshakov VY (2003) Essential function of alpha-calcium/calmodulin-dependent protein kinase II in neurotransmitter release at a glutamatergic central synapse. Proceedings of the National Academy of Sciences of the United States of America 100: 4275–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ui-Tei K, Nagano M, Sato S, Miyata Y (2000) Calmodulin-dependent and-independent apoptosis in cell of a Drosophila neuronal cell line. Apoptosis: an international journal on programmed cell death 5: 133–140. [DOI] [PubMed] [Google Scholar]
- 34. Yu W, Niwa T, Miura Y, Horio F, Teradaira S, Ribar TJ, et al. (2002) Calmodulin overexpression causes Ca(2+)-dependent apoptosis of pancreatic beta cells, which can be prevented by inhibition of nitric oxide synthase. Laboratory investigation; a journal of technical methods and pathology 82: 1229–1239. [DOI] [PubMed] [Google Scholar]
- 35. Goebel DJ (2009) Selective blockade of CaMKII-alpha inhibits NMDA-induced caspase-3-dependent cell death but does not arrest PARP-1 activation or loss of plasma membrane selectivity in rat retinal neurons. Brain research 1256: 190–204. 10.1016/j.brainres.2008.12.051 [DOI] [PubMed] [Google Scholar]
- 36. Fan W, Agarwal N, Kumar MD, Cooper NG (2005) Retinal ganglion cell death and neuroprotection: Involvement of the CaMKIIalpha gene. Brain research Molecular brain research 139: 306–316. [DOI] [PubMed] [Google Scholar]
- 37. Fan W, Agarwal N, Cooper NG (2006) The role of CaMKII in BDNF-mediated neuroprotection of retinal ganglion cells (RGC-5). Brain research 1067: 48–57. [DOI] [PubMed] [Google Scholar]
- 38. Fan W, Cooper NG (2009) Glutamate-induced NFkappaB activation in the retina. Investigative ophthalmology & visual science 50: 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bai M, Tsanou E, Skyrlas A, Sainis I, Agnantis N, Kanavaros P (2007) Alterations of the p53, Rb and p27 tumor suppressor pathways in diffuse large B-cell lymphomas. Anticancer research 27: 2345–2352. [PubMed] [Google Scholar]
- 40. Kowalczyk A, Filipkowski RK, Rylski M, Wilczynski GM, Konopacki FA, Jaworski J, et al. (2004) The critical role of cyclin D2 in adult neurogenesis. The Journal of cell biology 167: 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes & development 22: 3242–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urbach A, Robakiewicz I, Baum E, Kaczmarek L, Witte OW, Filipkowski RK (2013) Cyclin D2 knockout mice with depleted adult neurogenesis learn Barnes maze task. Behavioral neuroscience 127: 1–8. 10.1037/a0031222 [DOI] [PubMed] [Google Scholar]
- 43. Ma C, Papermaster D, Cepko CL (1998) A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proceedings of the National Academy of Sciences of the United States of America 95: 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cao Z, Henzel W, JGao X (1996) IRAK: a kinase associated with the interleukin-1 receptor. Science 271: 1128–1131. [DOI] [PubMed] [Google Scholar]
- 45. Mamidipudi V, Li X, Wooten MW (2002) Identification of interleukin 1 receptor-associated kinase as a conserved component in the p75-neurotrophin receptor activation of nuclear factor-kappa B. The Journal of biological chemistry 277: 28010–28018. [DOI] [PubMed] [Google Scholar]
- 46. Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, et al. (2010) MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell research 20: 784–793. 10.1038/cr.2010.79 [DOI] [PubMed] [Google Scholar]
- 47. Jang S, Lee JH, Choi KR, Kim D, Yoo H, SOh S (2007) Cytochemical alterations in the rat retina by LPS administration. Neurochemical research 32: 1–10. [DOI] [PubMed] [Google Scholar]
- 48. Hauck SM, Ekstrom PA, Ahuja-Jensen P, Suppmann S, Paquet-Durand F, van Veen T, et al. (2006) Differential modification of phosducin protein in degenerating rd1 retina is associated with constitutively active Ca2+/calmodulin kinase II in rod outer segments. Molecular & cellular proteomics: MCP 5: 324–336. [DOI] [PubMed] [Google Scholar]
- 49. Mortensen RD, Serra M, Steitz JA, Vasudevan S (2011) Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proceedings of the National Academy of Sciences of the United States of America 108: 8281–8286. 10.1073/pnas.1105401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, et al. (2013) Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC genomics 14: 725 10.1186/1471-2164-14-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nunez-Iglesias J, Liu CC, Morgan TE, Finch C, EZhou XJ (2010) Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PloS one 5: e8898 10.1371/journal.pone.0008898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lau P, Bossers K, Janky R, Salta E, Frigerio CS, Barbash S, et al. (2013) Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO molecular medicine 5: 1613–1634. 10.1002/emmm.201201974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. (2013) Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. The Journal of neuroscience: the official journal of the Society for Neuroscience 33: 5127–5137. 10.1523/JNEUROSCI.0600-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishida W, Fukuda K, Higuchi T, Kajisako M, Sakamoto S, Fukushima A (2011) Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Investigative ophthalmology & visual science 52: 611–617. [DOI] [PubMed] [Google Scholar]
- 55. Omran A, Elimam D, Yin F (2013) MicroRNAs: new insights into chronic childhood diseases. BioMed research international 2013: 291826 10.1155/2013/291826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Feng B, Chen S, McArthur K, Wu Y, Sen S, Ding Q, et al. (2011) miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 60: 2975–2984. 10.2337/db11-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, Reis RI, et al. (2011) Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Molecular vision 17: 2228–2240. [PMC free article] [PubMed] [Google Scholar]
- 58. Cossetti C, Smith JA, Iraci N, Leonardi T, Alfaro-Cervello C, Pluchino S (2012) Extracellular membrane vesicles and immune regulation in the brain. Frontiers in physiology 3: 117 10.3389/fphys.2012.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230. [DOI] [PubMed] [Google Scholar]
- 60. Bray I, Tivnan A, Bryan K, Foley NH, Watters KM, Tracey L, et al. (2011) MicroRNA-542–5p as a novel tumor suppressor in neuroblastoma. Cancer letters 303: 56–64. 10.1016/j.canlet.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. The list of 372 miRNA IDs used in Qiagen HC PCR array. Table B. Target gene analysis from the upregulated miRNAs. The list shows the overlapping target genes from TargetScan and miTarBase. Table C. Target gene analysis from the dwonregulated miRNAs. The list is the overlapping target genes from TargetScan and miTarBase. Table D. Functional analysis of the target genes performed by DAVID and KEGG.
(DOCX)