There are errors in the Author Contributions. The correct contributions are: Conceived and designed the experiments: GL LX TF YC GJ HZ JW XC CW. Performed the experiments: GL LX TF JW. Analyzed the data: GL LX TF. Contributed reagents/materials/analysis tools: GL. Wrote the paper: GL.
Additionally, there is an error in the legends for Fig. 1 and Fig. 2. “Table 1” should say “Table 5.” Please view the complete, correct Fig. 1 and Fig. 2 legends below.
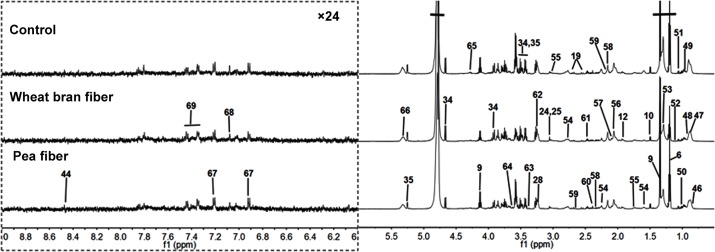
Fig 1. Representative one-dimensional 1H NMR spectra urine metabolites obtained from the (A) control, (B) pea fiber, and (C) wheat bran fiber groups.
The region of δ6.2–9.5 was magnified 16 times compared with corresponding region of δ0.5–6.2 for the purpose of clarity. Metabolite keys are given in Table 5.
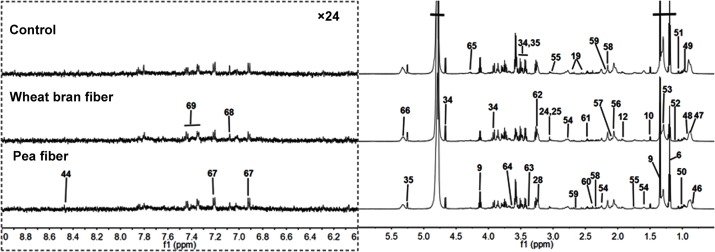
Fig 2. Typical 600 MHz 1H NMR spectra of plasma metabolites obtained from the (A) control, (B) pea fiber, and (C) wheat bran fiber groups.
The region of δ6.0–9.0 was magnified 24 times compared with corresponding region of δ0.5–6.0 for the purpose of clarity. Metabolite keys are given in Table 5.
Table 5. 1H NMR data for metabolites in rat urine and plasma.
| keys | Metabolites | moieties | δ 1H (ppm) and multiplicity | samples a |
|---|---|---|---|---|
| 1 | bile acids | CH3 | 0.62(m), 0.75(m) | U |
| 2 | Butyrate | CH3 | 0.9(t) | U |
| 3 | α-hydroxybutyrate | CH3 | 0.94(t) | U |
| 4 | α-hydroxy-iso-valerate | δCH3 | 0.97(d) | U |
| 5 | isobutyrate | CH3 | 1.14(d) | U, P |
| 6 | Ethanol | CH3, CH2 | 1.19(t), 3.66(q) | U, P |
| 7 | methylmalonate | CH3, CH | 1.26(d), 3.76(m) | U |
| 8 | α-hydroxy-n-valerate | CH3, γCH2 | 0.89(t), 1.31(m) | U |
| 9 | lactate | αCH, βCH3 | 4.13(q), 1.33(d) | U, P |
| 10 | alanine | αCH, βCH3 | 3.77(q), 1.48(d) | U, P |
| 11 | citrulline | γCH2, βCH2 | 1.56(m), 1.82(m) | U |
| 12 | acetate | CH3 | 1.92(s) | U, P |
| 13 | acetamide | CH3 | 1.99(s) | U |
| 14 | N-acetylglutamate | βCH2, γCH2, CH3 | 2.07(m), 1.88(m), 2.04(s) | U |
| 15 | acetone | CH3 | 2.25(s) | U, P |
| 16 | acetoacetate | CH3 | 2.3(s) | U |
| 17 | succinate | CH2 | 2.41(s) | U |
| 18 | α-ketoglutarate | βCH2, γCH2 | 2.45(t), 3.01(t) | U |
| 19 | citrate | CH2 | 2.55(d), 2.68(d) | U, P |
| 20 | methylamine | CH3 | 2.62(s) | U |
| 21 | dimethylamine | CH3 | 2.73(s) | U |
| 22 | trimethylamine | CH3 | 2.88(s) | U |
| 23 | dimethylglycine | CH3 | 2.93(s) | U |
| 24 | creatine | CH3, CH2 | 3.04(s), 3.93(s) | U, P |
| 25 | creatinine | CH3, CH2 | 3.04(s), 4.05(s) | U, P |
| 26 | ethanolamine | CH2 | 3.13(t) | U |
| 27 | malonate | CH2 | 3.16(s) | U |
| 28 | choline | OCH2, NCH2, N(CH3)3 | 4.07(t), 3.53(t), 3.20(s) | U, P |
| 29 | taurine | -CH2-S, -CH2-NH2 | 3.26(t), 3.43(t) | U |
| 30 | glycine | CH2 | 3.57(s) | U |
| 31 | phenylacetyglycine | 2,6-CH, 3,5-CH, 7-CH, 10-CH | 7.31(t), 7.37(m), 7.42(m), 3.68(s) | U |
| 32 | hippurate | CH2, 3,5-CH, 4-CH, 2,6-CH | 3.97(d), 7.57(t), 7.65(t), 7.84(d) | U |
| 33 | N-methylnicotinamide | CH3, 5-CH, 4-CH, 6-CH, CH2 | 4.44(s), 8.18(d), 8.89(d), 8.96(d), 9.26(s) | U |
| 34 | β-glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 4.65(d), 3.25(dd), 3.49(t), 3.41(dd), 3.46(m), 3.73(dd), 3.90(dd) | U, P |
| 35 | α-glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 5.24(d), 3.54(dd), 3.71(dd), 3.42(dd), 3.84(m), 3.78(m) | U, P |
| 36 | allantoin | CH | 5.40(s) | U, P |
| 37 | urea | NH2 | 5.82(s) | U |
| 38 | homogentisate | 6-CH, 5-CH | 6.7(d), 6.76(d), | U |
| 39 | p-hydroxyphenylacetate | 6-CH, 2-CH, 3,5-CH | 3.6(s), 6.87(d), 7.15(d) | U |
| 40 | m-hydroxyphenylacetate | 6-CH, 4-CH, 3-CH | 6.92(m), 7.04(d), 7.26(t) | U |
| 41 | nicotinate | 2,6-CH, 4-CH, 5-CH | 8.62(d), 8.25(d), 7.5(dd) | U |
| 42 | 4-aminohippurate | CH2 | 7.71(d) | U |
| 43 | trigonelline | 2-CH, 4-CH, 6-CH, 5-CH, CH3 | 9.12(s), 8.85(m), 8.83(dd), 8.19(m), 4.44(s) | U |
| 44 | formate | CH | 8.46(s) | U |
| 45 | unknown | 8.54(s) | U | |
| 46 | HDL* | CH3(CH2)n | 0.84(m) | P |
| 47 | LDL* | CH3(CH2)n | 0.87(m) | P |
| 48 | VLDL* | CH3CH2CH2C = | 0.89(t) | P |
| 49 | isoleucine | αCH, βCH, βCH3, γCH2, δCH3 | 3.68(d), 1.99(m), 1.01(d), 1.26(m), 1.47(m), 0.94(t) | P |
| 50 | leucine | αCH, βCH2, γCH, δCH3 | 3.73(t), 1.72(m), 1.72(m), 0.96(d), 0.97(d) | P |
| 51 | valine | αCH, βCH, γCH3 | 3.62(d), 2.28(m), 0.99(d), 1.04(d) | P |
| 52 | propionate | CH3, CH2 | 1.08(t), 2.18(q) | P |
| 53 | 3-hydroxybutyrate | αCH2, βCH, γCH3 | 2.28(dd), 2.42(dd), 4.16(m), 1.20(d) | P |
| 54 | lipids (triglycerids and fatty acids) | (CH2)n, CH2CH2CO, CH2C = C, CH2CO,C = CCH2C = C | 1.28(m),1.58(m), 2.01(m), 2.24(m), 2.76(m) | P |
| 55 | lysine | αCH, βCH2, γCH2, εCH2 | 3.76(t), 1.91(m), 1.48(m), 1.72(m), 3.01(t) | P |
| 56 | N-acetyl glycoprotein | CH3 | 2.04(s) | P |
| 57 | O-acetyl glycoprotein | CH3 | 2.08(s) | P |
| 58 | glutamate | αCH, βCH2, γCH2 | 3.75(m), 2.12(m), 2.35(m) | P |
| 59 | methionine | αCH, βCH2, γCH2, S-CH3 | 3.87(t), 2.16(m), 2.65(t), 2.14(s) | P |
| 60 | pyruvate | CH3 | 2.37(s) | P |
| 61 | glutamine | αCH, βCH2, γCH2 | 3.78(m), 2.14(m), 2.45(m) | P |
| 62 | glycerolphosphocholine | CH3, βCH2, αCH2 | 3.22(s), 3.69(t), 4.33(t) | P |
| 63 | phosphorylcholine | N(CH3)3, OCH2, NCH2 | 3.22(s), 4.21(t), 3.61(t) | P |
| 64 | myo-inositol | 1,3-CH, 2-CH, 5-CH, 4,6-CH | 3.60(dd), 4.06(t), 3.30(t), 3.63(t) | P |
| 65 | threonine | αCH, βCH, γCH3 | 3.58(d), 4.24(m), 1.32(d) | P |
| 66 | unsaturated lipids | = CH-CH2C =, -CH = CH- | 5.19 (m), 5.30(m) | P |
| 67 | tyrosine | 2,6-CH, 3,5-CH | 7.20(dd), 6.91(d) | P |
| 68 | 1-methylhistidine | 4-CH, 2-CH | 7.05(s), 7.78(s) | P |
| 69 | phenylalanine | 2,6-CH, 3,5-CH, 4-CH | 7.32(m), 7.42(m), 7.37(m) | P |
| 70 | 3-methylhistidine | 4-CH, 2-CH | 7.07(s), 7.67(s) | P |
a U, urine; P, plasma;
* HDL, high density lipoprotein; LDL, low density lipoprotein; VLDL, very low density lipoprotein; s, singlet; d, doublet; t, triplet; q, quartet; dd, doublet of doublets; m, multiplet
Further, there is an error in the footnotes for Table 5. VLDL stands for “very low density lipoprotein,” not “low density lipoprotein.” Please view the complete, correct Table 5 footnotes below.
Reference
- 1. Liu G, Xiao L, Fang T, Cai Y, Jia G, Zhao H, et al. (2014) Pea Fiber and Wheat Bran Fiber Show Distinct Metabolic Profiles in Rats as Investigated by a 1H NMR-Based Metabolomic Approach. PLoS ONE 9(12): e115561 doi:10.1371/journal.pone.0115561 [DOI] [PMC free article] [PubMed] [Google Scholar]