Abstract
A flow-injection mass spectrometric metabolic fingerprinting method in combination with chemometrics was used to differentiate Aurantii Fructus Immaturus from its counterfeit Poniciri Trifoliatae Fructus Immaturus. Flow-injection mass spectrometric (FIMS) fingerprints of 9 Aurantii Fructus Immaturus samples and 12 Poniciri Trifoliatae Fructus Immaturus samples were acquired and analyzed using principal component analysis (PCA) and soft independent modeling of class analogy (SIMCA). The authentic herbs were differentiated from their counterfeits easily. Eight characteristic components which were responsible for the difference between the samples were tentatively identified. Furthermore, three out of the eight components, naringin, hesperidin, and neohesperidin, were quantified. The results are useful to help identify the authenticity of Aurantii Fructus Immaturus.
1. Introduction
Aurantii Fructus Immaturus (AFI) is mainly comprised of two different species: the immature fruits of Citrus aurantium L. (AFICA) and Citrus sinensis Osbeck (AFICS). They are usually collected from May to June. After removal of pollutants, they are cut in half through the middle and are dried in the shade. Clinically, they are used mainly for gastro-intestinal food retention, fullness and pain from distention of the stomach, and prolapse of the rectum and uterus [1]. Polymethoxylated flavones, coumarins, flavonoid glycosides and alkaloids were identified as the main chemical compounds which showed anticarcinogenic [2-4], antioxidant, antimicrobial [5], gastric mucosal protective [6], and neuroprotective [7] effects.
Another herbal medicine, Poniciri Trifoliatae Fructus Immaturus (PTFI), is the fruits of Poncirus trifolata Raf., which is usually misused as AFI in clinics. Before the Song dynasty of China, both AFI and PTFI were used interchangeably in folk medicine. Later, physicians realized that AFI and PTFI had different medical effects and stopped using the latter gradually [8]. By the Ming and Qing dynasties of China, PTFI was defined clearly as the counterfeit of AFI [8]. However, even today, PTFI is still often misused as AFI, either mistakenly or intentionally, due to their similar morphological appearances and their confusing Chinese names as PTFI is called “Lvyi Zhishi” that is similar in Chinese pronunciation with AFI (“Zhishi”).
Metabolic fingerprinting can be defined as high-throughput qualitative screening of the metabolic compositions of an organism or tissue with the primary aim of sample comparison and discrimination analysis. Generally, no attempt is initially made to identify the metabolites present. All steps from sample preparation, separation, and detection should be rapid and as simple as is feasible [9]. Flow-injection mass spectrometry (FIMS) is one of the easiest and fastest analytical tool for obtaining metabolic fingerprints of samples. It has been applied in quite a few researches involving diverse sample matrices including TCMs, plant materials, dietary supplements, and fruits [10-13]. The method in combination with chemometric methods was demonstrated to be successful in assessing the similarities and differences in chemical profiles from different samples. Furthermore, when combined with ultra high-performance liquid chromatography high-resolution MS method (UHPLC/HRMS), putative identification of the chemical compounds responsible for the differentiation of the samples could be obtained [14].
The present study aimed to differentiate AFI from its counterfeit PTFI using FIMS method combined with chemometrics. UHPLC/HRMS was used to provide complementary data for identification of the characteristic chemical compounds.
2. Experimental
2.1. Materials
Nine samples of AFICA (AFICA-01 ~ AFICA-09) and twelve samples of PTFI (PTFI-01 ~ PTFI-12) were gifted and authenticated by Professor Yuan-Shiun Chang from the College of Pharmacy at China Medical University. The authentication of the herbs was confirmed using DNA-sequence-based methods (Authen Technologies, Richmond, CA). Acetonitrile and methanol were Optima* grade (Fisher Scientific, Pittsburgh, PA, USA). Formic acid is MS grade (Sigma/Aldrich, St. Louis, MO, USA). Purified water was produced by Thermo Barnstead Nanopure Life Science UV/UF Water Purification System. Reference compounds including naringin, hesperidin and neohesperidin were from Chengdu Must Bio-technology Co., LTD (Chengdu, China, purity >95%).
2.2. Sample preparation
Five-hundred milligrams of each dried ground sample was mixed with 10.0 mL of methanol:water (5:5, v/v) in a 15-mL centrifuge tube. All samples were sonicated for 60 min at room temperature. The extracts were centrifuged at 4,000 rpm for 15 min (IEC Clinical Centrifuge, Damon/IEC Division, Needham Heights, MA, USA). The supernatant of each sample was diluted 100 times and then filtered through a 17-mm (0.45 μm) PVDF syringe filter (VWR Scientific, Seattle, WA, USA) for analysis. To avoid errors arising from unexpected degradation of the chemical compounds, the sample analyses were completed within 24 h after the extraction. The injection volume for each sample was 5 μL. Each sample was analyzed five times for the FIMS experiment, three times for the quantification experiment and one time for the UHPLC/HRMS experiment.
2.3. Ultra high-performance liquid chromatography high-resolution mass spectrometry system
The UHPLC-HRMS system consisted of a Thermo LTQ Orbitrap XL mass spectrometer with an Accela 1250 binary pump, a PAL-HTC-Accela autosampler, an Accela 1250 PDA detector, and an Agilent column compartment (G1316A).
For FIMS, a guard column was used to minimize potential contamination for MS system, but it did not provide meaningful separation. Mobile phases consisted of 0.1% formic acid in H2O (A) and 0.1% formic acid in acetonitrile (B) with isocratic elution at 50:50 (v:v) and a flow rate at 0.5 mL/min for 2.0 min. Electrospray ionization (ESI) was performed in the negative ion mode from m/z 100 to 1000 to obtain the FIMS fingerprints. The parameters of the mass spectrometer were optimized with hesperidin by auto-tune using the Xcalibur software through infusion of the reference compound. The following conditions were used: sheath gas flow rate, 80 (arbitrary units); aux gas flow rate, 15 (arbitrary units); spray voltage, -4.5 kV; heated capillary temperature, 300 °C; capillary voltage, −40.0 V; tube lens offset, -150 V. Spectra were averaged over retention time between 0.2 and 1.2 min. Five repeat analyses of the 21 different samples provided 105 spectra.
The chromatographic separation was carried on a Thermo Hypersil GOLDTM aQ analytical HPLC column (200 × 2.1 mm, 1.9 μm) with a flow rate of 0.30 mL/min. The column heater was kept at 60 °C. Mobile phase A consisted of 0.1% formic acid in water and B consisted of 0.1% formic acid in acetonitrile. The elution gradient was 10% B (v/v) over 0–2 min, 10% to 95% B over 2–25 min. Quantification of the three analytes was performed using a detection wavelength at 280 nm. ESI was performed in the negative ion mode to obtain the HRMS data using fourier transform MS (FTMS). The conditions for FTMS were as follows: sheath gas flow rate, 80 AU; aux and sweep gas, 15 AU; spray voltage, −4.5 kV; capillary temperature, 300 °C; capillary voltage, −40 V; and tube lens offset, −150 V. The mass range was from m/z 100 to 1,000 m/z with a resolution of 30,000, isolation width of 1.5 amu, and maximum ion injection time of 500 ms. The most intense ions were selected for the data-dependent scan with collision energy at 30−35%.
2.4. Data processing for FIMS fingerprints
The mass spectrum for each sample consisted of a vector (counts versus mass for m/z 100–1000). The spectra were exported to Excel (Microsoft, Inc., Belleview, WA, USA) for data pre-processing and then to Solo (Eigenvector Research, Inc. Wenatchee, WA, USA) for principal component analysis (PCA) and soft independent modeling of class analogy (SIMCA). The preprocessing in Microsoft Excel involved combining the 105 spectra, sorting the data according to sample names, and filling the mass matrix (each spectrum had a different number of masses since any count of the ion below the detection threshold would not be exported into the mass list from the spectrum). A zero was inserted for each missing m/z in a mass list so that the number of data points of the mass list of each sample was 901. The resulting two-dimensional matrix (105 samples versus 901 masses) was then exported to Solo for PCA and SIMCA. Preprocessing in Solo, prior to PCA and SIMCA, consisted of normalization (normalized to unit vector) and mean centering.
3. Results and discussion
3.1. PCA of the FIMS fingerprints of AFICA and PTFI samples
PCA mathematically transforms a number of possibly correlated variables into a smaller number of uncorrelated variables called principal components (PCs). PCA score plots obtained from the generated PCs provides visual patterns that can be easily understood and avoids subjective decisions.
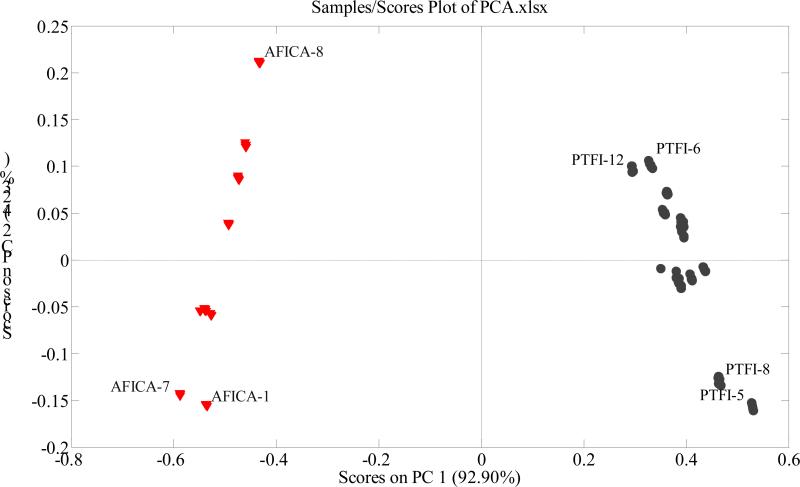
The variables used in the present study were the intensity values of the ions between 100 and 1000 (901 variables), and the observations were the samples (21 × 5 = 105). The dataset was exported to Solo for PCA. According to the PCA scores plot (Fig. 1), the two groups of samples were clearly separated from each other with the first two principal components cumulatively accounting for 97.13% of the total variance. All the AFICA samples are clustered on the left of the plot, with PC1 scores below zero. All the PTFI samples are clustered on the right, with PC1 scores above zero.
Figure 1.
PCA scores plot of AFICA and PTFI using the intensities of the 901 ions as variables.
Generally, the loadings plots clarify not only how much a variable contributes to the PC but also how well that PC takes that variable into account over the data points. Moreover, loadings plots indicate the relationship between variables. The PC1 loadings plot (Supplementary Fig. A) indicates that the ions at m/z 191, 285, 301, 341, 579, 593, and 609 are responsible for the separation of samples on PC1. The samples with positive intensity values for ions at m/z 191, 285, 341 and 593 get positive PC1 scores, whereas, the samples with positive intensity values for ions at m/z 301, 579, and 609 get negative PC1 scores. The position of each sample in PCA scores plot depends on the combined effects of intensity values of these ions. All the PTFI samples have positive intensity values for ions at m/z 285, 341 and 593, leading to their right positions on the scores plot; and all the AFICA samples have positive intensity values for ions at m/z 301, 579, and 609, leading to their positions to the left in the scores plot.
According to PC2 loadings plot (Supplementary Fig. B), the ion at m/z 579 contributes to PC2 scores of the samples positively, whereas, the ions at m/z 191, 301, 593 and 609 contribute to PC2 scores of the samples negatively. In the present study, PC2 didn't play critical role in classifying two groups of samples.
In summary, the seven ions were found to be the characteristic markers which played the most important role in differentiation between AFICA and PTFI samples in the present study.
3.3. Soft independent modeling of class analogy (SIMCA)
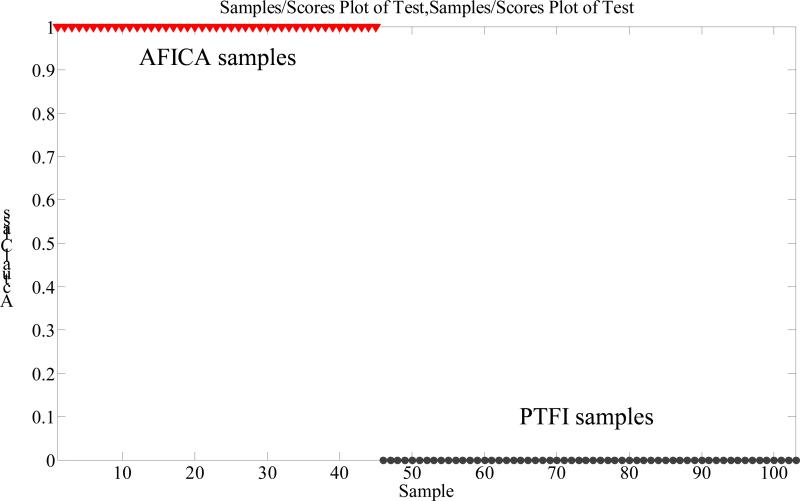
SIMCA is one of the supervised pattern recognition techniques using training set to conduct a model in order to predict unknown samples [15]. In order to confirm the statistical significance of the PCA results, SIMCA was performed in the present study. The recognition ability (Fig. 2) proved to be highly satisfactory and suggested a 100% correct classification for samples from each group. The results further confirmed the PCA grouping results.
Figure 2.
SIMCA modeling and prediction results.
3.4. Identification of the seven characteristic ions
The retention time (tR, min), UV λmax (nm), [M-H]− wt, [M-H]− formula, error (ppm) between theoretical and measured values, MS2 and MS3 ions of each characteristic peak as well as its assignment are summarized in Table 1. The ESI-MSn spectra of these peaks are shown in supplementary document (Supplemental Fig. C to Fig. I).
Table 1.
Information of retention time (tR, min), UV λmax (nm), [M-H]− wt, [M-H]− formula, error (ppm) between theoretical and measured values, MS2 and MS3 ions of the characteristic peaks.
| Peak No. | tR (min) | UV λmax (nm) | [M-H]−wt | [M-H]− formula | error (ppm) | MS2 ions | MS3 ions | Identification |
|---|---|---|---|---|---|---|---|---|
| 1a | 1.62 | / | 191.0555 | C7H11O6 | 0.485 | 173 [M-H-H2O]− 147 [M-H-CO2]− |
Quinic acid | |
| 1b | 1.62 | / | 341.1073 | C12H21O11 | −1.557 | 179 [M-H-hexosyl]− 161 [M-H-hexosyl-H2O]− |
Dihexose | |
| 2 | 8.65 | 222, 283 | 579.1707 | C27H31O14 | −0.228 | 271 [M-H-rutinosyl]− | 227 [M-H-rutinosyl-CO2]−, 177, 165, 151 [M-H-rutinosyl-120]−, 107 [M-H-rutinosyl-120-CO2]− | Narirutin |
| 3 | 9.01 | 230, 282, 327 sh | 579.1703 | C27H31O14 | −0.918 | 561 [M-H-H2O]− 459 [M-H-120]− 313 [M-H-120-rhamnosyl]− 295 [M-H-120-rhamnosyl-H2O]− 271 [M-H-neohesperidosyl]− 235 [M-H-neohesperidosyl-2H2O]− |
441 [M-H-120-H2O]−, 357 [M-H-120-C7H2O]−, 339 [M-H-240]−, 313 [M-H-120-rhamnosyl]−, 271 [M-H-neohesperidosyl]−, 235 [M-H-neohesperidosyl-2H2O]−, 151 [M-H-120-neohesperidosyl]− 177 [M-H-neohesperidosyl-94]−, 151 [M-H-neohesperidosyl-120]− |
Naringin |
| 4 | 9.31 | 284, 328 sh | 609.1808 | C28H33O15 | −0.547 | 301 [M-H-rutinosyl]− | Hesperidin | |
| 5 | 9.65 | 284, 328 sh | 609.1810 | C28H33O15 | −0.397 | 489 [M-H-120]− 343 [M-H-120-rhamnosyl]− 325 [M-H-120-rhamnosyl-H2O]− 301 [M-H-neohesperidosyl]− |
Neohesperidin | |
| 6 | 11.35 | 225, 283 | 593.1855 | C28H33O14 | −1.655 | 285 [M-H-rutinosyl]− | 270 [M-H-rutinosyl-CH3]−, 243 [M-H-rutinosyl-C2H2O]−, 164, 151 | Neoponcirin |
| 7 | 11.63 | 226, 282 | 593.1845 | C28H33O14 | −3.341 | 575 [M-H-H2O]− 473 [M-H-120]− 431 [M-H-162]− 327 [M-H-120-rhamnosyl]− 309 [M-H-120-rhamnosyl-H2O]− 285 [M-H-neohesperidosyl]− |
270 [M-H-neohesperidosyl-CH3]−, 243 [M-H-neohesperidosyl-C2H2O]−, 164, 151 | Poncirin |
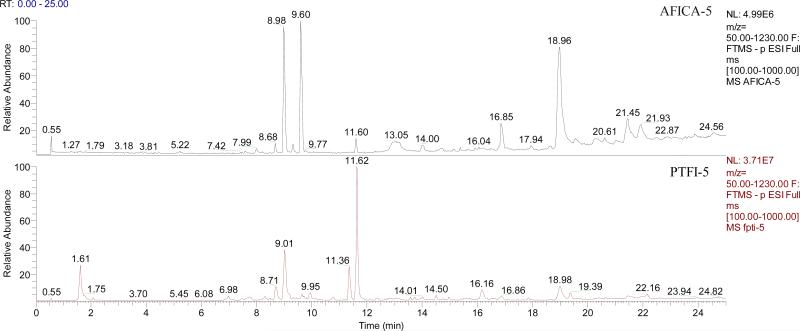
Representative UHPLC/HRMS total ion chromatograms (TICs) of AFICA-5 and PTFI-5 are shown in Fig. 3. Discrepancies in chemical profiles between AFICA and PTFI samples suggest that the two groups of samples probably have different pharmacological effects.
Figure 3.
UHPLC/HRMS total ion chromatograms of AFICA-5 and PTFI-5.
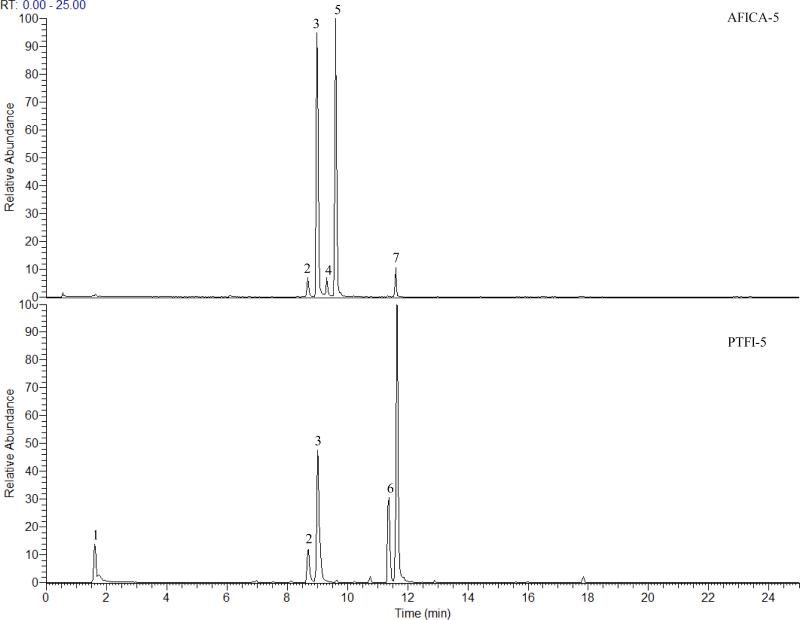
Combined extracted ion chromatograms (EICs) for AFICA-5 and PTFI-5 for the seven characteristic ions are displayed in Fig. 4. The ions at m/z 191 and 341 were from peak 1 (1a and 1b, Table 1) with retention time at 1.62 min, of which the HRMS measurements were 191.0555 (C7H11O6, 0.485 ppm) and 341.1073 (C12H21O11, -1.577 ppm), respectively. The ion at m/z 683 was also found in MS spectrum which was tentatively identified as the [2M-H]− ion of the ion at m/z 341. In MS2 spectrum, the deprotonated ion at m/z 191 yielded characteristic ions at m/z 173 and 147, corresponding to losses of neutral fragments of H2O and CO2, respectively. This compound was tentatively identified as quinic acid according to the literature [16,17]. The ion at m/z 341 produced a base peak at m/z 179 [MH-hexosyl]− as well as the ion at m/z 161 [M-H-hexosyl-H2O]− in MS2 spectrum, which was tentatively identified as dihexose [16].
Figure 4.
Extracted ion chromatogram at m/z 191, 285, 301, 341, 579, 593 and 609 of AFICA-5 and PTFI-5.
Two peaks (peak 2 and peak 3), retention times at 8.65 (UV λmax: 222, 283 nm) and 9.01 (UV λmax: 230, 282, 327 nm) min, had ions at m/z 579. HRMS measurement of the deprotonated ion [M-H]− of peak 2 was 579.1707, suggesting the chemical composition of C27H31O14 (-0.228 ppm). The ion at m/z 271 [M-H-308]− was observed as the predominant product ion in the MS2 spectra, suggesting the neutral loss of a rutinosyl. In the MS3 experiment, the ion at m/z 151 [M-H-rutinosyl-120]− was found to be the base peak, resulting from the Retro-Diels-Alder (RDA) reactions, which further lost a neutral CO2, leading to a fragment ion at m/z 107 [M-H-rutinosyl-120-CO2]−. Other fragment ions at m/z 165, 177 and 227 [M-H-rutinosyl-CO2]− were also observed. This peak was tentatively identified as narirutin according to the MS behavior reported in literature [18-21]. HRMS measurement of the deprotonated ion [M-H]− of peak 3 was 579.1703, also suggesting the chemical composition of C27H31O14 (-0.918 ppm). Its major fragment ions in MS2 spectrum were 561 [M-H-H2O]−, 459 [M-H-120]−, 313 [M-H-120-rhamnosyl]−, 295 [M-H-120-rhamnosyl-H2O]−, 271 [M-H-neohesperidosyl]−, and 235 [M-H-neohesperidosyl-2H2O]−. It is worth noting here the loss of 120Da may be caused by either the glycan [22] or ring C [23]. In the MS3 experiment, the ion at m/z 459 [M-H-120]− yielded the base peak at m/z 357 [M-H-120-C7H2O]− as well as a series of fragment ions at m/z 441 [M-H-120-H2O]−, 339 [M-H-240]−, 313 [M-H-120-rhamnosyl]−, 271 [M-H-neohesperidosyl]−, 235 [M-H-neohesperidosyl-2H2O]− and 151 [M-H-120-neohesperidosyl]−. The ion at m/z 271 [M-H-neohesperidosyl]− yielded the base peak at m/z 151 [M-H-neohesperidosyl-120]− and another predominant ion at m/z 177 in MS3 experiment. Finally, the peak was identified as naringin and confirmed by the reference compound.
The ion at m/z 609 was from peak 4 and peak 5 with retention times at 9.31 (UV λmax: 284, 328 nm) and 9.65 (UV λmax: 284, 328 nm) min, respectively. They are identified as hesperidin and neohesperidin, respectively. HRMS measurement of peak 4 gave a [M-H]− ion at m/z 609.1808 (C28H33O15, -0.547 ppm) as well as the ion at m/z 301, resulting from a cleavage of rutinosyl. In the MS2 spectrum, the deprotonated ion produced a predominant base peak at m/z 301 [M-H-rutinosyl]−. Peak 5 showed the HRMS measurement of [M-H]− at m/z 609.1810 also with the formula of C28H33O15 (-0.397 ppm). Interestingly, due to the difference of the chemical structures between neohesperidin and hesperidin, a series of fragment ions at m/z 489 [M-H-120]−, 343 [M-H-120-rhamnosyl]−, 325 [MH-120-rhamnosyl-H2O]− and 301 [M-H-neohesperidosyl]− were observed in MS2 spectrum of neohesperidin. The MS behavior was consistent with that reported in literatures [19-21].
The ion at m/z 593 was from peak 6 and peak 7 with retention times at 11.35 (UV λmax: 225, 283 nm) and 11.63 (UV λmax: 226, 282 nm) min, respectively. HRMS measurement of peak 6 gave a [M-H]− ion at m/z 593.1855 (C28H33O14, -1.655 ppm) as well as the ion at m/z 285, resulting from a cleavage of rutinosyl. In the MS2 experiment, the deprotonated ion produced a predominant base peak at m/z 285 [M-H-rutinosyl]−, which further yielded a series of fragment ions at m/z 270 [M-H-rutinosyl-CH3]−, 243 [M-H-rutinosyl-C2H2O]−, 164, and 151 in MS3 experiment.
According to the literature [20, 24], it was tentatively identified as neoponcirin (isosakuranetin-7-rutinoside). Peak 7 is an isomer of neoponcirin. The structural difference between the two compounds was the position of the linkage to rhamnosyl and glucosyl, which leads to different MS behaviors. Peak 7 showed the HRMS measurement of [M-H]− at m/z 593.1845 (C28H33O14, -3.341 ppm) along with its formic acid adduct ion at m/z 639 [M+HCOOH-H]−. In the MS2 experiment, the deprotonated ion yielded a series of fragmentation ions at m/z 575 [M-H-H2O]−, 473 [M-H-120]−, 431 [M-H-162]−, 327 [M-H-120-rhamnosyl]− and 309 [M-H-120-rhamnosyl-H2O]− and a base peak at 285 [M-H-neohesperidosyl]−. It can be seen that this glycoside with neohesperidose favored eliminating of rhamnosyl and a fragment with molecular weight of 120 Da. In MS3 experiment, the ion at m/z 285 produced fragmentation ions at m/z 270 [M-H-neohesperidosyl-CH3]−, 243 [M-H-neohesperidosyl-C2H2O]−, 164, and 151. This peak was tentatively identified as poncirin (isosakuranetin-7-neohesperidose) according to literature [20, 21, 24].
3.5. Contents of naringin, hesperidin and neohesperidin in all the tested samples
As described above, the seven characteristic ions were found in eight peaks. Naringin, hesperidin and neohesperidin were then quantified using reference compounds. The results are shown in Table 2. First, naringin was found to be the main chemical compound in all the samples. The concentrations ranged from 64.63 ± 1.87 to 189.12 ± 6.82 mg/g in AFICA samples, which were close to the values reported in literatures [25-27]. However, the values were between 10.36 ± 0.47 to 22.03 ± 1.40 mg/g in PTFI samples, which were much lower than that in AFICA samples. Second, hesperidin and neohesperidin were not detected in any of the PTFI samples, but they were detected in all the AFICA samples (1.95 ± 0.13 to 5.16 ± 0.18 mg/g for hesperidin and 56.85 ± 0.82 to 105.99 ± 1.25 mg/g for neohesperidin, respectively). Finally but most importantly, synephrine is the chemical marker for quality control of AFI samples specified in the Chinese Pharmacopoeia (edition 2010), the concentration of which should not be less than 3 mg/g. However, due to the high polarity of synephrine, sodium dodecyl sulfate is needed for good retention on the chromatographic column. This is not compatible with MS detection. The results obtained in the present study show that AFICA have high concentrations of naringin, hesperidin and neohesperidin, all of which can be quantified using both UV detector and mass spectrometer. Therefore, these three compounds can be considered as alternatives for assessment of the herb besides synephrine.
Table 2.
Contents of naringin, hesperidin and neohesperidin in AFICA and PTFI samples (mg/g, expressed as mean ± SD).
| Mean quantification value (mg/g) | |||
|---|---|---|---|
| Sample ID | Naringin | Hesperidin | Neohesperidin |
| AFICA-1 | 64.63 ± 1.87 | 5.16 ± 0.18 | 56.85 ± 0.82 |
| AFICA-2 | 117.84 ± 6.20 | 1.57 ± 0.15 | 62.44 ± 0.77 |
| AFICA-3 | 139.12 ± 6.59 | 1.51 ± 0.07 | 58.50 ± 0.93 |
| AFICA-4 | 129.50 ± 7.01 | 1.35 ± 0.03 | 60.76 ± 0.55 |
| AFICA-5 | 129.19 ± 6.28 | 2.63 ± 0.00 | 85.61 ± 0.75 |
| AFICA-6 | 126.05 ± 6.35 | 2.59 ± 0.06 | 85.21 ± 0.76 |
| AFICA-7 | 93.44 ± 3.65 | 4.22 ± 0.18 | 78.63 ± 1.25 |
| AFICA-8 | 189.12 ± 6.82 | 1.95 ± 0.13 | 57.96 ± 0.80 |
| AFICA-9 | 161.68 ± 4.91 | 3.88 ± 0.35 | 105.99 ± 1.25 |
| PTFI-1 | 14.30 ± 0.97 | ND | ND |
| PTFI-2 | 22.03 ± 1.40 | ND | ND |
| PTFI-3 | 20.94 ± 1.42 | ND | ND |
| PTFI-4 | 20.02 ± 0.49 | ND | ND |
| PTFI-5 | 12.43 ± 0.95 | ND | ND |
| PTFI-6 | 19.69 ± 1.08 | ND | ND |
| PTFI-7 | 13.09 ± 0.88 | ND | ND |
| PTFI-8 | 10.36 ± 0.47 | ND | ND |
| PTFI-9 | 18.50 ± 0.93 | ND | ND |
| PTFI-10 | 18.96 ± 1.39 | ND | ND |
| PTFI-11 | 16.40 ± 0.56 | ND | ND |
| PTFI-12 | 20.82 ± 1.04 | ND | ND |
4. Conclusion
Two previous reports on the identification of AFI are of interests. Both of them were HPLC based. One used multivariate analysis just as this study, however, it did not use loading plot nor the SIMCA model [28]. The other used a targeted approach, not a comprehensive approach as described in this study [29]. Compared with the FIMS metabolic fingerprinting method, both methods were time-consuming (HPLC analysis were both well over 60 minutes per sample) and labor-intensive. The FIMS metabloic fingerprinting method was demonstrated to be a simple, fast, and reliable for differentiating AFICA from its counterfeit PTFI. Furthermore, the characteristic ions observed from the PCA loadings plots were identified with the aid of UHPLC-HRMS. Naringin, hesperidin and neohesperidin were quantified using UV detector at 280 nm. The results confirmed the authenticity of AFICA samples and also indicated that naringin and neohesperidin were the main chemical compounds in AFICA, both with concentrations greater than 50 mg/g. It's worth noting that the two compounds might be more acceptable for quality assessment on AFICA compared with synephrine.
Supplementary Material
Highlights.
Aurantii Fructus Immaturus and Fructus Poniciri Trifoliatae Immaturus were differentiated.
Seven characteristic ions from eight compounds were screened out as chemical markers.
The eight compounds were identified.
Naringin, hesperidin and neohesperidin were quantified.
The results help using Aurantii Fructus Immaturus correctly in clinic.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.State Pharmacopoeia Commission, Pharmacopoeia of the People's Republic of China, vol. 1, 2010. Chinese Medical Science and Technology Press; Beijing: 2010. pp. 230–231. [Google Scholar]
- 2.Li S, Pan PM, Lai CS, Lo CY, Dushenkov S, Ho CT. Isolation and syntheses of polymethoxy flavones and hydroxylated polymethoxy flavones as inhibitors of HL-60 cell lines. Bioorg. Med. Chem. 2007;15:3381–3389. doi: 10.1016/j.bmc.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Manthey JA, Guthrie N. Antiproliferative activities of citrus flavonoids against six human cancer cell lines. J. Agric. Food Chem. 2002;50:5837–5843. doi: 10.1021/jf020121d. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka T, Kawabata K, Kakumoto M, Makita H, Hara A, Mori H, Satoh K, Hara A, Murakami A, Kuki W, Takahashi Y, Yonei H, Koshimizu K, Ohigashi H. Citrus auraptene inhibits chemically induced colonic aberrant crypt foci in male F344 rats. Carcinogenesis. 1997;18:2155–2161. doi: 10.1093/carcin/18.11.2155. [DOI] [PubMed] [Google Scholar]
- 5.Mokbel MS, Watanabe Y, Hashinaga F, Suganuma T. Purification of the antioxidant and antimicrobial substance of ethyl acetate extracts from Buntan (Citrus grandis Osbeck) fruit peel. Pak. J. Biol. Sci. 2006;9:145–150. [Google Scholar]
- 6.Takase H, Yamamoto K, Hirano H, Saito Y, Yamashita A. Pharmacological profile of gastric mucosal protection by marmin and nobiletin from a traditional herbal medicine, Aurantii Fructus Immaturus. Jpn. J. Pharmacol. 1994;66:139–147. doi: 10.1254/jjp.66.139. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima A, Yamakuni T, Haraguchi M, Omae N, Song SY, Kato C, Nakagawasai O, Tadano T, Yokosuka A, Mimaki Y, Sashida Y, Ohizumi Y. Nobiletin, a citrus flavonoid that improves memory impairment, rescues bulbectomy-induced cholinergic neurodegeneration in mice. J. Pharmacol. Sci. 2007;105:122–126. doi: 10.1254/jphs.sc0070155. [DOI] [PubMed] [Google Scholar]
- 8.Xie ZW. Research on duration and changes of Zhi Shi and Zhi Qiao as ancient and present drugs. Res. Tradit. Chin. Med. 1991;1:19–22. [Google Scholar]
- 9.Hall RD. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol. 2006;169:453–468. doi: 10.1111/j.1469-8137.2005.01632.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Lin LZ, Harnly JM. Mass spectroscopic fingerprinting method for differentiation between Scutellaria lateriflora and the Germander (Teucrium canadense and T. chamaedrys) Species. J. AOAC Int. 2010;93:1148–1154. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Niu YG, Xie ZH, Shi HM, Chen P, Yu LL. Differentiating leaf and whole-plant samples of di- and tetraploid Gynostemma pentaphyllum (Thunb.) Makino using flow-injection mass spectrometric fingerprinting method. J. Funct. Foods. 2013;5:1288–1297. [Google Scholar]
- 12.Zhao Y, Sun JH, Yu LL, Chen P. Chromatographic and mass spectrometric fingerprinting analyses of Angelica sinensis (Oliv.) Diels-derived dietary supplements. Anal. Bioanal. Chem. 2013;405:4477–4485. doi: 10.1007/s00216-012-6668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P, Harnly JM, Lester GE. Flow injection mass spectral fingerprints demonstrate chemical differences in Rio Red grapefruit with respect to year, harvest time, and conventional versus organic farming. J. Agric. Food Chem. 2010;58:4545–4553. doi: 10.1021/jf904324c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun JH, Chen P. A flow-injection mass spectrometry fingerprinting method for authentication and quality assessment of Scutellaria lateriflora-based dietary supplements. Anal. Bioanal. Chem. 2011;401:1577–1584. doi: 10.1007/s00216-011-5246-2. [DOI] [PubMed] [Google Scholar]
- 15.Berrueta LA, Alonso-Salces RM, Heberger K. Supervised pattern recognition in food analysis. J. Chromatogr. A. 2007;1158:196–214. doi: 10.1016/j.chroma.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Harnly JM, Harrington Pde B. Flow injection mass spectroscopic fingerprinting and multivariate analysis for differentiation of three Panax species. J. AOAC Int. 2011;94:90–99. [PMC free article] [PubMed] [Google Scholar]
- 17.Fang N, Yu S, Prior RL. LC/MS/MS characterization of phenolic constituents in dried plums. J. Agric. Food Chem. 2002;50:3579–3585. doi: 10.1021/jf0201327. [DOI] [PubMed] [Google Scholar]
- 18.Xu F, Liu Y, Zhang Z, Yang C, Tian Y. Quasi-MSn identification of flavanone 7-glycoside isomers in Da Chengqi Tang by high performance liquid chromatography-tandem mass spectrometry. Chin. Med. 2009;4:15. doi: 10.1186/1749-8546-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao HY, Fan MX, Wu X, Wang HJ, Yang J, Si N, Bian BL. Chemical profiling of the Chinese herb formula Xiao-Cheng-Qi decoction using liquid chromatography coupled with electrospray ionization mass spectrometry. J. Chromatogr. Sci. 2013;51:273–285. doi: 10.1093/chromsci/bms138. [DOI] [PubMed] [Google Scholar]
- 20.Cho HE, Ahn SY, Kim SC, Woo MH, Hong JT, Moon DC. Determination of flavonoid glycosides, polymethoxyflavones, and coumarins in herbal drugs of Citrus and Poncirus fruits by high performance liquid chromatography-electrospray ionization/tandem mass spectrometry. Anal. Lett. 2014;47:1299–1323. [Google Scholar]
- 21.Liu WY, Zhou C, Yan CM, Xie SL, Feng F, Wu CY, Xie N. Characterization and simultaneous quantification of multiple constituents in Aurantii Fructus Immaturus extracts by HPLC-DAD-ESI-MS/MS. Chin. J. Nat. Med. 2012;10:456–463. [Google Scholar]
- 22.Cuyckens F, Rozenberg R, de Hoffmann E, Claeys M. Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 2001;36:1203–1210. doi: 10.1002/jms.224. [DOI] [PubMed] [Google Scholar]
- 23.Shi P, He Q, Song Y, Qu H, Cheng Y. Characterization and identification of isomeric flavonoid O-diglycosides from genus Citrus in negative electrospray ionization by ion trap mass spectrometry and time-of-flight mass spectrometry. Anal. Chim. Acta. 2007;598:110–118. doi: 10.1016/j.aca.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Feng F. Identification of components in Zhi-Zi-Da-Huang decoction by HPLC coupled with electrospray ionization tandem mass spectrometry, photodiode array and fluorescence detectors. J. Pharm. Biomed. Anal. 2009;49:1157–1165. doi: 10.1016/j.jpba.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZX, Zheng YZ, Liang LJ, Dong TX, Zhan HQ, Zhao KJ. Analysis of HPLC fingerprints of Citrus aurantium and Citrus sinensis and the contents of naringin and synephrine. Zhongguo Yao Fang. 2011;22:3711–3714. [Google Scholar]
- 26.Jiang H, Li J, Shi RB. Determination of three flavanones in the effective fraction of flavones in Fructus Aurantii Immaturus by HPLC. Zhongguo Yao Fang. 2008;19:2127–2128. [Google Scholar]
- 27.Wang Q, Yuan D. Determination of flavonoids in Fructus Aurantii Immaturus and Fructus Aurantii from different habitat by HPLC. Heilongjiang Yi Yao. 2008;21:1–3. [Google Scholar]
- 28.Chuang CC, Wen WC, Sheu SJ. Classification of Aurantii Fructus samples by multivariate analysis. J. Sep. Sci. 2007;30:1827–1832. doi: 10.1002/jssc.200700016. [DOI] [PubMed] [Google Scholar]
- 29.Chuang CC, Wen WC, Sheu SJ. Original identification on the commercial samples of Aurantii Fructus. J. Sep. Sci. 2007;30:1235–1241. doi: 10.1002/jssc.200600522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.