Abstract
The proteasome controls the concentrations of most proteins in eukaryotic cells. It recognizes its protein substrates through ubiquitin tags and initiates degradation at disordered regions within the substrate. Here we find that the proteasome has pronounced preferences for the amino acid sequence composition of the regions at which it initiates degradation. Specifically, proteins where the initiation regions have biased amino acid compositions show longer half-lives in yeast. The relationship is also observed on a genomic scale in mouse cells. These preferences affect the degradation rates of proteins in vitro, can explain the unexpected stability of natural proteins in yeast, and may affect the accumulation of toxic proteins in disease. We propose that the proteasome’s sequence preferences provide a second component to the degradation code and may fine-tune protein half-life in cells.
The ubiquitin proteasome system (UPS) adjusts cellular protein concentrations by selecting specific proteins for destruction and hydrolyzing them into small peptides. The proteasome is the protease at the center of this system and is a 2.5 MDa multi-subunit particle. Proteins are targeted to the proteasome through a two-part degradation signal or degron. The degron consists of a proteasome-binding tag, typically a polyubiquitin chain, and a disordered region at which the proteasome engages its substrate and initiates degradation1–3. We will refer to this disordered region as the initiation site or region. Once the substrate is properly engaged, the proteasome degrades it sequentially4. Many proteins are ubiquitinated but not degraded by the proteasome in cells and ubiquitin tags serve as signals in processes such as membrane trafficking and chromatin rearrangements2. In some cases the proteasome likely does not recognize the ubiquitinated protein, perhaps because the ubiquitin tag is first recognized by receptors associated with the competing process5. In other cases, the proteasome may recognize a protein but fail to initiate degradation, for example if the protein lacks a disordered region of sufficient length at the appropriate location6,7.
In this study we investigate why some proteins resist proteasomal degradation despite containing both proteasome binding tag and disordered regions and to discover the missing component of the degron. We begin by analyzing three specific proteins, the diffusible proteasome substrate receptor Rad23, the ubiquitin-conjugating enzyme Cdc34 and a fragment of Huntingtin protein that accumulates in Huntington’s Disease (Htt exon 1). We propose that the proteasome is unable to initiate degradation of these proteins because it does not recognize the amino acid sequences of their disordered regions. We go on to investigate the proteasome’s amino acid sequence preferences by comparing the degradation of proteins that differ only in their initiation regions. We find that degradation rates correlate with the amino acid composition of the initiation regions: the more diverse an amino acid sequence, the better it is recognized by the proteasome. The same correlation holds on a genome scale between the half-lives of approximately 4000 mouse proteins and the sequence of their disordered regions. We propose that the amino acid sequence composition of disordered regions fine-tunes protein half-life and that genetic mechanisms that generate diversity in sequence composition may represent a source of phenotypic variation.
Results
Rad23 escapes proteasomal degradation
Rad23 and the related Dsk2 and Ddi1 proteins contain a ubiquitin-like (UbL) domain, which binds to the proteasome, and one or more ubiquitin associated (UBA) domains, which bind to poly-ubiquitin chains in proteasome substrates. The substrates are degraded while the UbL-UBA proteins escape intact8–10. UbL-UBA proteins contain disordered linker regions but the linkers are flanked by folded domains, which impede degradation11 (Fig. 1a). We noticed that the amino acid sequences of the linkers in the yeast UbL-UBA proteins are strongly biased in that some amino acids occur more frequently than expected, though different amino acids dominate in different linkers6,12 (see also their sequence annotation in PFAM6,12). Therefore, we asked how well the proteasome would initiate degradation at the linkers if its access were not constrained by flanking domains at both ends.
Figure 1. Rad23 linker regions do not act as efficient initiation sites.
(a) Sketch of S. cerevisiae Rad23 protein. UbL: ubiquitin-like domain; UBA: ubiquitin associated domain; RBD Rad4 binding domain; L1, L2, L3: linkers and their lengths in number of amino acids. (b) In vitro degradation kinetics for model proteins by purified S. cerevisiae proteasome. The proteins consist of four ubiquitin domains fused in series followed by an E. coli dihydrofolate reductase (DHFR) domain and different disordered tails at the C terminus. 15 and 102: 15 or 105 amino acid long tails derived from S. cerevisiae cytochrome b2. The graph plots the amount of protein estimated by electronic autoradiography in SDS PAGE gels bands shown over time as a percentage of the initial protein as described in the Methods section. Data points represent mean values determined from three repeat experiments; error bars show standard errors of the mean.
The three linkers in Rad23 range from 45 to 75 amino acids in size (Fig. 1a). Polypeptide tails of this length at the C termini of model substrate proteins support efficient proteasomal initiation and degradation6. We tested whether the Rad23 linkers would also support degradation of these same model substrates. The proteins consisted of the compact 17 kDa protein E. coli dihydrofolate reductase (DHFR) and we targeted them to the proteasome by fusing four ubiquitin moieties in series to their N termini3,13. The resulting ubiquitin DHFR proteins were not degraded by the proteasome unless a disordered tail was also attached to the C terminus of DHFR to allow the proteasome to engage its substrates and initiate degradation3,6,7. For this study, we constructed five ubiquitin - DHFR test substrates: in three of them we attached each of the Rad23 linkers to the C terminus of DHFR (Ub4-DHFR-L1, Ub4-DHFR-L2, Ub4-DHFR-L3), in one we attached a 102 amino acid long tail derived from S. cerevisiae cytochrome b2 that we knew would allow the proteasome to initiate degradation6 (Ub4-DHFR-102), and in one we attached a tail, also derived from cytochrome b2, that is too short to function as a good initiation region6 (Ub4-DHFR-15) (Fig. 1b; the amino acid sequences of the initiation regions are shown in the Supplementary Table 1). We synthesized the proteins by in vitro transcription and translation in E. coli extract, and presented them to purified yeast proteasome in the presence of ATP and an ATP-regenerating system. The proteasome degraded the proteins with the established initiation site (Ub4-DHFR-102), but not any of the proteins with Rad23 linkers (Ub4-DHFR-L1, Ub4-DHFR-L2, Ub4-DHFR-L3) or the short tail (Ub4-DHFR-15) as initiations regions (Fig. 1b). Thus, we concluded that some property of the Rad23 linkers prevents the proteasome from engaging the substrates and initiating degradation. This property serves as an additional mechanism protecting Rad23 from degradation.
Cdc34 lacks a good proteasomal initiation site
The amino acid compositions of the Rad23 linkers are biased in the sense that some amino acids are noticeably overrepresented while others are lacking6. We wondered whether biased amino acid sequences occurred in other natural proteins that unexpectedly escape degradation by the proteasome. Cdc34 is a ubiquitin activating enzyme (E2) and it becomes ubiquitinated like many other proteins that are part of the UPS14,15. The ubiquitin moieties attached to Cdc34 are linked through Lys48 of ubiquitin as found in bona fide proteasome degrons16 and Cdc34 contains a C-terminal disordered region of 130 amino acids, yet Cdc34 is not degraded. The amino acid composition of Cdc34’s disordered tail is strongly biased with 50 of the 130 amino acids in the tail being aspartate or glutamate residues. These residues are concentrated in a 99 amino acids long acidic region (Fig. 2a). We asked whether the proteasome would be able to engage Cdc34 at its acidic region and, if not, whether the acidic region would act in a dominant manner and prevent the proteasome from initiating degradation at other regions within the protein.
Figure 2. The acidic region of Cdc34 does not support proteasomal degradation.
(a) Schematic representation of a ubiquitinated Cdc34 protein. The amino acid sequence of the acidic region is shown with aspartates and glutamates in green. (b) In vitro degradation kinetics for model proteins by purified S. cerevisiae proteasome. The proteins consist of four ubiquitin domains fused in series followed by an E. coli dihydrofolate reductase (DHFR) domain and different disordered tails at the C terminus. Acidic: C-terminal 86 amino acids of Cdc34; 102: 102 amino acid long tails derived from S. cerevisiae cytochrome b2; where no tail is indicated the protein ended with the C terminus of DHFR. The graph plots the amount of protein estimated by electronic autoradiography in SDS PAGE gels bands shown over time as a percentage of the initial protein as described in the Methods section. Data points represent mean values determined from three repeat experiments; error bars show standard errors of the mean.
We first tested whether the isolated C-terminal tail of Cdc34 would allow the proteasome to initiate degradation on a model substrate in vitro. We constructed three test substrates as described above, ubiquitin DHFR without a tail (Ub4-DHFR), ubiquitin DHFR with a good initiation region (Ub4-DHFR-102), and ubiquitin DHFR with the C-terminal 86 amino acids of Cdc34 (Ub4-DHFR-acidic tail) and presented them to yeast proteasome. As expected, the Ub4-DHFR protein without a tail remained stable and Ub4-DHFR-102 was degraded effectively3,6,7 (Fig. 2b). Ub4-DHFR-acidic tail also remained stable (Fig. 2b), suggesting that the Cdc34 acidic region does not allow the proteasome to initiate degradation.
Next we asked whether the acidic region has a dominant effect on the stability of Cdc34 and protects Cdc34 from degradation even in the presence of other sequences at which the proteasome would otherwise be able to initiate degradation. To test this possibility, we attached two tails of different lengths and amino acid sequences to the C terminus of Cdc34 and followed their degradation by yeast proteasome (Fig. 3a). One tail was a 95 amino acid sequence derived from S. cerevisiae cytochrome b2, the other tail consisted of 39 amino acids and was derived from an internal region of the E. coli lac repressor17. We synthesized Cdc34 with and without the tails by in vitro transcription and translation in E. coli extract, purified the proteins, and induced them to autoubiquitinate by incubating them with ubiquitin, ATP, and the E1 enzyme Ube1 (Fig. 3b). Wildtype Cdc34, despite its extensive ubiquitination, remained stable as expected, but Cdc34 with either of the two tails was degraded effectively (Fig. 3a). Degradation was by the proteasome because it was inhibited by the proteasome inhibitor MG132 and by depletion of ATP in the reaction (Supplementary Fig. 1). Thus, the acidic region does not protect Cdc34 from proteasomal degradation dominantly. Instead, it appears that Cdc34 escapes degradation simply because it lacks an effective initiation region.
Figure 3. The acidic region does not stabilize Cdc34 dominantly against proteasomal degradation.
(a) In vitro degradation kinetics of ubiquitinated Cdc34 proteins with different tails fused to their C termini as initiation regions by purified S. cerevisiae proteasome. NRR: asparagine-rich region composed of four copies of residues 373-386 from transcription factor Spt23; SRR: a serine-rich region derived from four copies of residues 178-196 of transcription factor ICP4, 95: 95 amino acid long tail derived from S. cerevisiae cytochrome b2, 39: 39 amino acid long tail derived from E. coli lac repressor; where no tail is indicated the protein ended with the C terminus of Cdc34. The graph plots the amount of protein estimated by electronic autoradiography in the SDS PAGE gels bands (indicated by brackets) over time as a percentage of the initial protein as described in the Methods section. Data points represent mean values determined from three repeat experiments; error bars show standard errors of the mean. (b) In vitro ubiquitination kinetics for the Cdc34 proteins shown in (a) as estimated by electronic autoradiography of SDS PAGE gels (indicated by brackets; (Ub)n: poly-ubiquitinated species). The bar graph plots of the percent of protein ubiquitinated relative to the amount of starting protein. The experiment was repeated three times and one of these is shown.
The acidic region in Cdc34 is characterized by a large number of aspartate and glutamate residues and it is possible that its high negative charge density prevents proteasome binding. Thus, we tested whether sequences with differently biased amino acid compositions and without the high charge density support degradation. We constructed Cdc34 variants by attaching tails rich in asparagine residues (Cdc34-NRR) or in serine residues (Cdc34-SRR) to Cdc34’s C terminus and investigated proteasomal degradation (Fig. 3a). Both Cdc34 variants were ubiquitinated effectively (Fig. 3b) but escaped degradation, just as wildtype Cdc34 (Fig. 3a). Thus, Cdc34 resists degradation because the proteasome is unable to engage it effectively at its disordered region. However, it is not the acidity of the region by itself that prevents degradation but some other property of the amino acid sequence (see also below).
Cdc34 is stable in vivo due to poor proteasomal initiation
The experiments described above suggest that the amino acid sequence of the initiation region can affect proteasomal degradation in vitro. In vivo, other factors may affect Cdc34 degradation and therefore we followed the stability of Cdc34 with and without tails in yeast. We added an HA tag to the N terminus of the Cdc34 proteins described above and expressed them from a centromeric plasmid under the control of a constitutive promoter. We then monitored the accumulation of the different Cdc34 fusion proteins at steady state in mid log phase by anti-HA Western blotting (Fig. 4).
Figure 4. The lack of an efficient initiation site also affects the stability of Cdc34 in vivo.

Steady state accumulation of N-terminally HA tagged Cdc34 and Cdc34 with C-terminal tails in S. cerevisiae. Protein levels were determined by Western blotting for the HA tag in SDS PAGE gels of S. cerevisiae protein extracts. Proteasome degradation was tested by the addition of the proteasome inhibitor MG-132 as indicated. Protein loading levels in each lane were estimated in all lysates by Western blotting for the integral ER membrane protein Scs2.
The Cdc34 fusions accumulated at the levels expected from the in vitro experiments. HA-tagged wildtype Cdc34 (Cdc34 in Fig. 4) was easily detected but the Cdc34 proteins with tails that served as proteasome initiation sites in vitro (Cdc34-95 and Cdc34-39 in Fig. 4) could barely be detected unless the proteasome was inhibited by MG132. In contrast, Cdc34 with the asparagine-rich tail (Cdc34-NRR in Fig. 4) accumulated just as wildtype Cdc34. The 95 amino acid tail did not increase the extent of detectable ubiquitination of Cdc34-95 compared to Cdc34 (Supplementary Fig. 2). Thus, in vivo, just as in vitro, the amino acid sequence of the region in Cdc34 at which the proteasome initiates degradation affects the stability of the protein.
Proteasomal initiation of degradation on model substrates
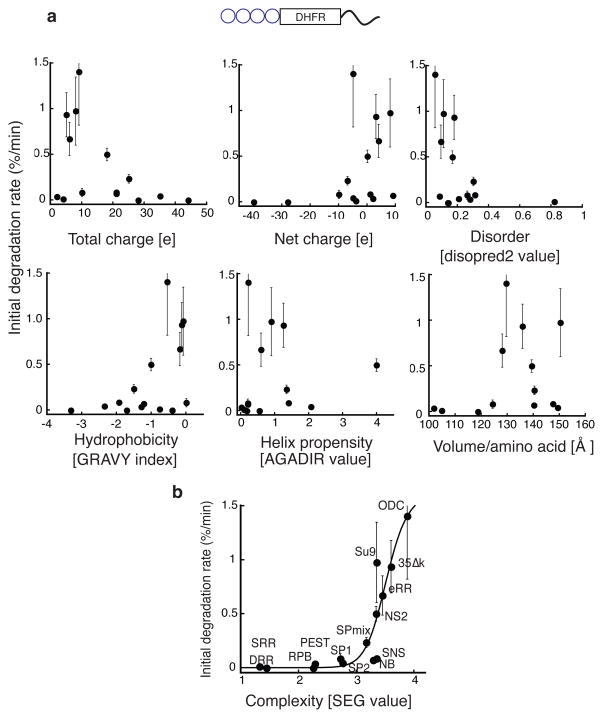
To test whether the amino acid sequence effects are specific to Cdc34 or reflect general preferences by the proteasome for its initiation region, we again monitored degradation of the Ub4-DHFR model substrates but now with different C-terminal imitation regions or tails (Fig. 5a). We tested 15 sequences derived from different proteins, most of which are not known to be involved in protein degradation by the UPS (Supplementary Table 2). We synthesized radiolabeled substrates by coupled transcription and translation in E. coli extract and presented them to purified yeast proteasome. The proteins were degraded with rates that ranged over at least one order of magnitude with some proteins remaining stable over the entire reaction and others degrading with halftimes of minutes (Supplementary Fig. 3). All the tails appeared to be largely disordered as tested by their sensitivity to a non-specific protease and as judged by circular dichroism spectrophotometry (Supplementary Fig. 4). The degradation rates did not correlate with chemical properties of the tails such as total charge, net charge, hydrophobicity, helical propensity, disorder prediction score and side chain volume. Instead, the degradation rates appeared to correlate well with the bias of the amino acid composition of the tails (Fig. 5b). We quantified the sequence bias with the SEG program18,19. Tails with biased amino acid compositions, in which fewer amino acids were represented, received a low score whereas tails with diverse, or complex, amino acid sequences received a higher score. We found that tails with biased amino acid compositions supported degradation poorly whereas tails with diverse amino acid sequences supported degradation well (Fig. 5b).
Figure 5. Proteasome’s preferences for the amino acid composition of initiation regions.
Plots of in vitro initial degradation rates of model proteins with different C-terminal tails by purified S. cerevisiae proteasome as a function of different properties of the tails. The proteins consisted of four ubiquitin moieties fused in frame followed by an E. coli dihydrofolate reductase (DHFR) domain and a disordered tail at the C terminus. (a) Initial rates of degradation (Supplementary Figure 3) were plotted against total charge (linear fit R=0.46), net charge (linear fit R=0.37), hydrophobicity (GRAVY scale, ProtParam in ExPASy (http://www.expasy.org); linear fit R=0.59), helix propensity (calculated by Agadir (http://agadir.crg.es)63; linear fit R=0.10), volume per amino acid (linear fit R=0.27), disorder (calculated by DISOPRED2 (http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1)64; linear fit R=0.43). (b) Initial rates degradation were plotted against the amino acid sequence complexity calculated by SEG algorithm18,19 (sigmoidal fit R=0.90; linear fit R=0.70). The solid line is a fit of the sigmoid curve to the data. Data points represent means of n measurements, and error bars show standard errors (NB: n=4; NS2: n=4; SNS: n=4; SPmix: n=4; SP1: n=3; SP2: n=3; GRR: n=3; SRR: n=3; DRR: n=3; PEST: n=3; RPB: n=3; eRR: n=3; ODC: n=5; 35DK: n=6; Su9: n=3; the amino acid sequence for the various peptides is given in the supplementary information).
The different amino acid sequences of the initiation sites could affect degradation either at the initiation step or at the proteolysis step6,20–24. The proteasome contains three pairs of proteolytic sites, each pair with different preferences for the amino acids preceding the cleaved peptide bond, which allows the proteasome core to hydrolyze most amino acid sequences25–27. To test how well the proteolytic core is able to digest the different tails we analyzed here, we activated purified yeast proteasome with 0.01% SDS to allow it to degrade peptides without ATP hydrolysis. We synthesized peptides corresponding to the simplest regions in ten of the tails, presented them to the activated proteasome and followed degradation with the amine-reactive dye fluorescamine, which detects the α-amino groups produced during hydrolysis28,29 (Supplementary Fig. 5). The proteasome degraded all tails well and the proteolysis rates did not correlate with the complexity of their amino acid composition (Supplementary Fig. 5). Some of the biased sequences such as those of the SRR and SP2 peptides did not allow the proteasome to initiate degradation (Fig. 5, Supplementary Fig. 3) but were digested efficiently by the proteasome in the peptide proteolysis assays (Supplementary Fig. 5). Thus, we propose that the proteasome has pronounced preferences for the amino acid sequence at which it initiates degradation. This interpretation is in agreement with the observation that Cdc34 is degraded when an initiation region is provided after the acidic region (Fig. 2).
Amino acid sequence bias affects protein abundance globally
Protein regions with biased amino acid sequences are often disordered but disordered regions do not always show biased amino acid composition30,31. Therefore, we asked whether amino acid sequence bias correlates with cellular protein stability globally on a genomic scale.
We found previously that mouse proteins with disordered regions longer than 30 amino acids at either the N or C terminus have shorter lifetimes than proteins without disordered tails32 by comparing experimentally determined half-lives of 4502 mouse proteins33 with predicted disorder at their N- and C- termini. The reason for these relationships is presumably that the proteasome requires the disordered regions to engage these proteins and initiate degradation. We now find that the correlation depends on the amino acid composition of the disordered regions. The half-lives of proteins whose disordered regions have biased amino acid compositions are comparable to the half-lives of proteins without disordered regions. Among proteins with N-terminal disordered regions, proteins with biased tail sequences have significantly longer half-lives than proteins with unbiased tail sequences (Wilcoxon rank sum P = 2.3 x 10−2; median half-life difference (ΔH) = 8.1 hours, Fig. 6a and Supplementary Table 3). This trend also holds for proteins with C-terminal disordered regions (Wilcoxon rank sum P = 1.1 x 10−3; ΔH = 13.4 hours, Fig. 6b and Supplementary Table 3). The differences do not seem to be due to variable numbers of ubiquitination sites (Supplementary Fig. 6). Thus, the proteasome’s amino acid sequence preferences may be part of the code that influences protein half-lives on a genomic scale.
Figure 6. Amino acid bias in N- and C-terminal disorder stabilizes proteins in mouse.
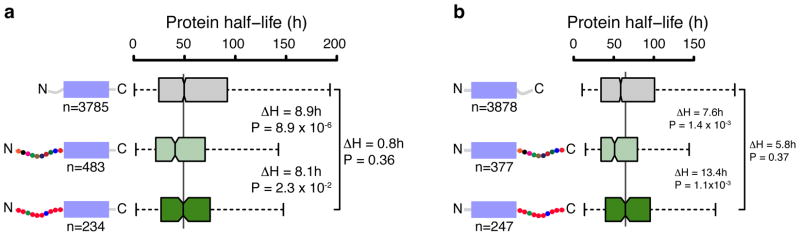
Boxplots showing the distribution of protein half-lives in mouse cells33 grouped by the length and amino acid composition of intrinsically disordered segments at (a) the N terminus and (b) C terminus. Proteins were classified into short disordered (<30 amino acids are disordered; gray box), long (>30 amino acids) disordered without amino acid bias (light green box) and long disordered with amino acid bias (dark green box). The black line within the box represents the median and the colored boxes represent the first and third quartiles. The notches correspond to ~95% confidence interval for the median. Whiskers connected to the boxes by the dashed lines show the data points up to 1.5 times the interquartile range from the box. Points beyond the whiskers were considered outliers and not shown. The number of data points for each group (n), differences between the half-life medians (in hours) of two compared groups (ΔH) and P-values indicating the significance in differences in half-life distributions are shown. Half-lives were modeled from relative abundance measurements over time33 but estimated stability differences between proteins are robust because the same model is applied to all proteins. Statistical significance of differences was assessed using the Wilcoxon rank sum test, which is non-parametric and hence does not assume any particular property of the distribution of the data.
Biased amino acid sequences in disease
Huntingtin (Htt), the protein associated with Huntington’s disease (HD), has a strikingly biased amino acid composition and this bias is associated with the etiology of HD. The intensity of the HD phenotype increases with the accumulation of a protein fragment that corresponds to exon 1 of a mutated Huntingtin (Htt) gene in nuclear inclusions and most evidence suggests that HD is caused by a gain of toxic function in Htt mutants34. This raises the question why Htt accumulates in the first place and is not cleared from the cell. Htt protein and ubiquitin are co-localized in HD brain inclusions34,35 and there is good evidence that the Htt protein itself is ubiquitinated36–39. The aggregates that accumulate in HD also contain proteasome subunits, suggesting that the proteasome attempts to degrade them40. Exon 1 of the Huntingtin gene encodes a short (17 amino acid long) protein interaction domain41 followed by a stretch of at least 23 glutamines (PolyQ) and then a proline-rich region (PRR) of 50 amino acids (Fig. 7a). We asked whether its biased amino acid composition impedes initiation of proteasomal degradation just as observed for Cdc34 and Rad23. To test this hypothesis, we attached four ubiquitin moieties linearly to the N terminus of Htt exon1 as a proteasome binding tag and then presented it to yeast proteasome. Ubiquitin-tagged Htt exon1 protein containing 34 or 52 glutamines was degraded slowly if at all, but attaching a 95 amino acid disordered tail derived from S. cerevisiae cytochrome b2 to its C terminus accelerated its degradation greatly (Fig. 7b). Htt exon 1 protein tends to aggregate but the differences in degradation were not caused by different aggregation states because the proteins were largely if not completely monomeric under the assay conditions (Supplementary Fig. 7).
Figure 7. Degradation of Htt is dependent on initiation region complexity.
(a) Schematic representation of protein fragment encoded by Huntingtin (Htt) exon1. (b–c) In vitro degradation of model proteins by purified S. cerevisiae proteasome. The proteins were targeted to the proteasome by four ubiquitin moieties fused in series to the N termini of the different constructs. The extent of degradation was plotted as the percentage of protein remaining at different times. (b) Degradation of Htt exon1. The Ubiquitin tag was followed by the protein sequence corresponding to Htt exon 1 with 34 or 52 residues in the glutamine repeat regions. Where indicated, a disordered tail of 95 amino acids derived from S. cerevisiae cytochrome b2 was placed at the C terminus of Htt exon1. (c) Degradation of the glutamine repeat region of Htt exon1. The ubiquitin tag was followed by an E. coli dihydrofolate reductase (DHFR) domain and one of three different tails. 52Q: 52 glutamine residues; 52Q—95: 52 glutamine residues followed by a 95 amino acid long C-terminal tail derived from S. cerevisiae cytochrome b2. Proteins with the glutamine repeat regions in panels b and c remained soluble under the experimental conditions as judged by size exclusion chromatography (see Supplementary Fig. 7). (d) Degradation of the proline repeat region of Htt exon1. The ubiquitin tag was followed by a DHFR domain and Htt’s polyproline region with or without a 95 amino acid tail. Data points represent the mean of five repeat experiments.
We then tested how well the glutamine and proline regions of exon 1 can support proteasomal initiation individually using the ubiquitin-DHFR model proteasome substrates described above. Neither the 52 amino acid long polyQ region (Fig. 7c) nor the PRR (Fig. 7d) region allowed the proteasome to initiate degradation. In contrast, the constructs were rapidly degraded when we attached a 95 amino acid long disordered tail after the polyQ or PRR regions (Fig. 7c, 7d). The constructs were also degraded when we replaced the polyQ or PRR regions with a complex (i.e., not biased) sequence of similar length (a 50 amino acid sequence derived from S. cerevisiae cytochrome b2) showing that 50 amino acid tails are in principle able to support degradation. Again, the glutamine containing substrates remained soluble in these assays and did not aggregate (Supplementary Fig. 7). Thus it appears that the amino acid sequence of Htt exon1 is a poor initiation site for the proteasome.
Discussion
Most proteins are targeted to the proteasome by a ubiquitin tag and their degradation also requires an unstructured or disordered region in the substrate, which we call the initiation site or region, at which the proteasome engages its substrate and initiates degradation1. The disordered region has to be located near the ubiquitin tag on the substrate7 and it has to be long enough to allow the substrate to access the proteasome’s degradation machinery6. Here we propose that the amino acid sequence of the disordered region also affects degradation and that the proteasome has distinct preferences for the binding site at which it initiates degradation. In particular, it seems that the proteasome can engage proteins inefficiently at polypeptide sequences with biased amino acid compositions and very biased sequences can escape proteasomal initiation entirely.
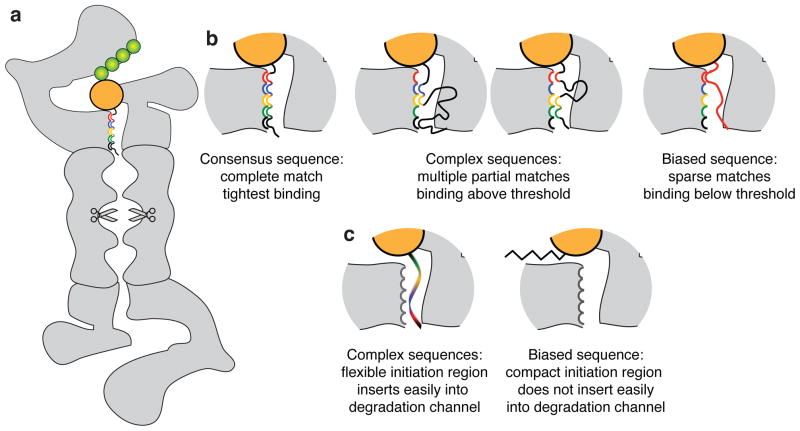
Amino acid repeat sequences were first discovered to be associated with unexpected stability in the Epstein Barr virus protein EBNA1 (ref. 22). Epstein Barr virus infects B lymphocytes and forms stable episomes whose maintenance requires the virally encoded protein EBNA1 (ref. 42). EBNA1 is protected from proteasomal degradation by glycine-alanine (GA) repeats20,22,23,43. Biased amino acid sequences also reduce the processivity of the proteasome and cause the production of partially degraded protein fragments by stalling the progression of the proteasome along its substrate21,24,44,45 but the molecular mechanism of these effects is not known. How can biased amino acid sequences impede substrate engagement by the proteasome? One possibility is that amino acid composition is directly related to the binding affinity to the proteasome. Since the proteasome must bind and degrade many different substrates, it is likely that the proteasome’s receptor for the initiation regions recognizes several patterns of chemical features (hydrophobic, positive or negative charge, etc.) that are complementary to the receptor surface rather than one strict consensus sequence (Fig. 8a). A diverse set of sequences would then be able to bind the receptor well enough to serve as initiation sites for the proteasome. In other words, the threshold for serving as a good initiation sequence would be achieved by partial matches between the receptor surface and the initiation site sequence (Fig. 8b). Under these circumstances, the likelihood of an amino acid sequence binding to the proteasome above any given threshold would correlate with the complexity of the sequence: biased sequences would be less likely to satisfy the required interactions with the receptor surface than complex sequences. Thus, the correlation between initiation of degradation and sequence complexity could reflect specific sequence binding preferences by the proteasome.
Figure 8. Amino acid sequence composition bias may affect recognition of initiation sites by the proteasome directly.
Schematic representation of initiation sites of different amino acid compositions being recognized by the 26S proteasome to illustrate two different models for the relationship between sequence complexity and proteasome recognition. The 26S proteasome is represented by grey shapes, ubiquitin by green spheres, the folded domain of the substrate by the large yellow sphere, and the initiation region by the black–red–blue–yellow–green tails (a). Biased sequences may bind less tightly to their receptor if they satisfy fewer open interactions than complex sequences (b). Alternatively, biased sequences may form compact rigid structures and access their binding site in the degradation channel less easily than flexible disordered regions (c).
A second possibility is that amino acid composition affects the structure and compactness of the initiation site and thus its ability to reach its receptor on the proteasome. Disordered polypeptide sequences adopt a diverse set of conformations that are in equilibrium with each other. Amino acid composition affects the conformational ensemble46–49 and may thus affect the ability of an initiation region to reach its receptor on the proteasome (Fig. 8c).
Our biochemical experiments tested only 15 different sequences and so the relationship between amino acid sequence composition and degradation cannot be extrapolated to make general predictions. Nevertheless, the relationship is reflected in the three different natural proteins investigated here. In all three proteins, the disordered regions that are available as proteasome initiation sites have biased amino acid sequences. Interestingly, inspection of the linker regions in the UbL-UBA proteins suggests that the property of bias is conserved between UbL-UBA proteins but that the nature of the bias is not: different amino acids are overrepresented in different linkers and in different UbL-UBA proteins50. Even more strikingly, the stability of soluble proteins in mouse cells correlates with the amino acid sequence complexity in the disordered regions in these proteins. Thus, the proteasome’s amino acid sequence preferences seem to fine-tune protein turnover globally. Disordered regions in proteins typically evolve more rapidly and with fewer restraints than the regions that form folded domains51, presumably because changes in disordered regions are less likely to affect the structure of a folded region and hence the molecular function mediated by the folded domain. Therefore evolving the sequence composition of the disordered region of the protein provides a simple genetic mechanism to change protein levels without directly affecting the molecular function of a protein.
For any one specific protein, its stability will depend on the relationship between the ubiquitin modifications and initiation region. For example, in the UbL-UBA protein Rad23, the three linkers connecting UbL and UBA domains represent the only possible initiation regions for the proteasome. The amino acid compositions of the linkers are clearly biased so that it is straightforward to relate sequence composition to degradation by the proteasome. In other proteins, only one particular stretch of their disordered regions may have a biased amino acid composition so that the proteasome can initiate degradation elsewhere within the protein. Thus, to be able to predict the effect of biased sequences on protein degradation more broadly, we need to understand where proteins are ubiquitinated and where exactly the proteasome initiates degradation relative to the ubiquitination site. It is also possible that mechanisms exist to create disordered regions throughout proteins, for example when ubiquitination of a folded domain leads to its unfolding52. Finally, it is quite possible that accessory factors such as Cdc48–p97 make the proteasome less dependent on the presence of disordered regions for degradation53.
The proteasome’s struggle to initiate degradation at biased amino acid sequences may contribute to the accumulation of some proteins that form aggregates associated with neurodegenerative diseases. Here, we find that, at least in vitro, the proteasome struggles to initiate degradation of the Htt fragment that accumulates in HD. Proteins related to other neurodegenerative diseases also carry abnormal glutamine repeats in their disease-associated forms54. Examples are atrophin-1, which is associated with dentatorubropallidoluysian atrophy, androgen receptor, which is associated with spinobulbar muscular atrophy, and ataxin proteins, which are associated with spino-cerebellar ataxia. The neuronal aggregates of these proteins contain ubiquitin yet the proteasome apparently fails to degrade them54. Similarly, tau protein forms aggregates in neurons of Alzheimer’s Disease patients called neurofibrillary tangles and the protein in the tangles is ubiquitinated55,56. Tau protein contains long stretches largely made up of five amino acids and thus it is possible that tau accumulates because the proteasome struggles to initiate degradation at the repeat sequences.
The Htt exon 1 fragment is made up almost entirely of glutamine and proline repeats so that no other sequences are available for proteasomal initiation but this is not the case for the other proteins listed. To know if and how the initiation site sequence contributes to their stability we need to map the ubiquitination sites and determine how the proteasome selects its initiation sites. Other mechanisms likely contribute to failure of proteasome degradation45,57–60 and different mechanisms of Htt accumulation can demand opposing therapeutic strategies. If Htt protein is not recognized by the proteasome, interventions that enhance the interaction between Htt protein and the proteasome are needed. Indeed, enhancing proteasome activity can reduce the accumulation of disease associated protein aggregates61,62. On the other hand, if Htt protein clogs up the proteasome, it may be beneficial to decrease Htt protein interaction with the proteasome. Thus, it will be important to determine which mechanism is relevant physiologically.
In summary, we find that the proteasome has pronounced preferences for the amino acid sequence of the substrate at the site at which it initiates degradation. These preferences affect substrate selection and may represent a second component of the proteasome targeting code superimposed on the ubiquitination code. Amino acid sequence variation within disordered regions can affect cellular protein half-life without directly affecting molecular function and may be a general genetic mechanism that has important implications in linking genotype to phenotype.
Online Methods
Substrate proteins
Protein substrates were derived from S. cerevisiae cytochrome b2, Cdc34 or Rad23, E. coli DHFR, and H. sapiens Huntingtin. Their coding sequences were cloned either into the plasmid pGEM-3Zf (+) (Promega) for in vitro expression or into the yeast CEN plasmid p416 GPD for in vivo experiments as described previously6.
N-terminal ubiquitin tags were composed of four copies of the coding region for ubiquitin, each containing the mutation Gly76 to Val and connected to the next ubiquitin by the linker sequence Gly-Ser-Gly-Gly-Gly as described previously13. C-terminal tails derived from cytochrome b2 sequences were attached to DHFR and the other proteins through a short linker and lysine residues in the tails were replaced by arginine. The tail sequences are given in Supplementary Tables 1 and 2. Ub4-DHFR-15, Ub4-DHFR-64, Ub4-DHFR-102, Ub4-DHFR-Q34-102 and Ub4-DHFR-Q52-102 all contained hexahistidine tag on the C-terminus; Ub4-DHFR-50 did not. In Ub4-DHFR-PRR, the tail consisted of the proline-rich region of Huntingtin exon1 (residues 41-90 in the Htt exon1 variant with a 34 residue glutamine repeat), followed by residues 1-95 from cytochrome b2 where indicated. The tail of Ub4-DHFR-acidic tail contained the last 86 amino acids of Cdc34. In Ub4-DHFR-L1, Ub4-DHFR-L2, and Ub4-DHFR-L3, the tails consisted of the Rad23 linker regions as follows: L1 connects the UbL and the N-terminal UBA domains and corresponds to amino acids 77-144 of Rad23, L2 connects the N-terminal UBA and the Rad4-binding domain and corresponds to amino acids 186-250 of Rad23, and L3 connects the Rad4-binding domain and the C-terminal UBA domains and consists of 296-355 of Rad23.
The constructs containing Huntingtin exon 1 consisted of the N terminal four ubiquitin tag described in the preceding paragraph followed by the sequence of huntingtin exon 1 with either 34 or 52 residue glutamine repeats, i.e., amino acids 1-90 (34 Q) or 1-109 (52Q) of huntingtin. The huntingtin sequence was followed by amino acids 1-50 or 1-95 of cytochrome b2 as indicated.
The Cdc34 constructs in Figure 2 consisted of the entire CDC34 coding sequence followed by the tails described below and the constructs expressed in yeast included an N-terminal HA tag. The C-terminal tails consisted of residues 321–354 of the E. coli lac repressor but with lysine residues replaced by arginines (Cdc34-39), four copies of residues 373-386 from transcription factor Spt23 (Cdc34-NRR), four copies of residues 178-196 of transcription factor ICP4 (Cdc34-SRR), or residues 1-95 of S. cerevisiae cytochrome b2. Tail sequences are given in Supplementary Tables 1 and 2.
In vitro autoubiquitination
In vitro translated radiolabeled substrates were ubiquitinated at 30 °C for 16 hours in a reaction mixture containing 25 mM Tris pH 7.5, 50 mM NaCl, 4 mM MgCl2, 4 mg/ml ubiquitin, 10 mM ATP, 1 μM DTT, and 200 nM human recombinant Ube1.
Protein expression and purification
Yeast proteasome was purified from S. cerevisiae strain YYS40 (MATa rpn11::RPN11 3×FLAG-HIS3 leu2 his3 trp1 ade2 can1 ssd1) by immunoaffinity chromatography using FLAG antibodies (M2 agarose affinity beads, Sigma) as described previously with modifications65. Proteasome preparations were analyzed by SDS PAGE and compared to published compositions66. A typical gel with assigned bands is shown below (Supplementary Fig. 8). Each proteasome preparation was checked for activity by testing degradation of the proteasome substrate Ub4-DHFR-95 and for contamination by proteases by testing for stability of proteins that lack a proteasome binding tag (DHFR-95).
For in vitro degradation experiments, radioactive substrates were expressed from a T7 promoter by a coupled in vitro transcription–translation reaction using E. coli T7 S30 Extract System for Circular DNA (Promega) containing [35S] methionine following the manufacturers protocol. Htt substrates were expressed from a T7 promoter by in vitro transcription–translation using the RTS 100 E. coli HY Kit (Roche) containing [35S] methionine according to the manufacturer’s instruction in 25 μL reactions. After synthesis, the substrates were either partially purified by high-speed centrifugation followed by precipitation in two volumes of saturated (NH4)2SO4 or affinity purified using Talon magnetic beads (Clontech) as described previously45.
Proteasomal degradation assays
Degradation assays were performed as described previously6. Briefly, assays were carried out at 30 °C by adding radiolabeled substrates to 50 nM of purified yeast proteasome in a reaction buffer containing a creatine-phosphate creatine kinase ATP regenerating system. Samples were removed at designated times, added to SDS PAGE sample buffer to stop the reaction and analyzed by SDS-PAGE. Protein amounts were determined by electronic autoradiography (Instant Imager; Packard). Each assay was repeated at least three times. Initial degradation rates are given by the slope of the decay curves at time zero in Supplementary Fig. 3 and are calculated as the product of the amplitude and the rate constant of the decay curve determined by non-linear fitting to a single exponential decay in the software package Kaleidagraph (version 4.1, Synergy Software). Original images of autoradiographs can be found in Supplementary dataset 1.
In vivo protein abundance determination
Cdc34 fusion proteins were expressed under the control of the constitutive glyceraldehyde 3-phosphate dehydrogenase, GPD, promoter from a CEN plasmid (p416 GPD) with a URA3 selection marker in S. cerevisiae strain BY4741 pdr5Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pdr5::kanMX4). Cells were grown to mid-log phase and lysed by vortexing with glass beads (BioSpec Products). Protein extracts were prepared and analyzed by Western blotting using standard protocols as described6. Cdc34 protein was detected with a monoclonal anti-HA antibody (1:5,000; Sigma, #H9658) and an Alexa-800-labelled goat anti-mouse secondary antibody (1:20,000; Rockland Immunochemicals, #610-132-121), and Scs2p was detected by an anti-SCS2 polyclonal antibody (1:1,000; gift from J. Brickner, Northwestern University) and an Alexa-680 goat anti-rabbit secondary antibody (1:20,000; Invitrogen, #A21109). Protein amounts were estimated by direct infrared fluorescence imaging (Odyssey LICOR Biosciences). Original images of Western blots can be found in Supplementary dataset 1.
Peptide proteolysis assay
Peptide proteolysis assays were carried out according to the method of Evans and Ridella 67. Proteolysis reactions were performed with 200 nM purified yeast proteasome at 30 °C in 5% (v/v) glycerol, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.4), 0.01% SDS, 1 mM DTT, 1 mM ATP, 10 mM creatine phosphate, 0.1 mg/ml creatine phosphokinase. 250 μM (final concentration) peptides were added to purified proteasome in reaction buffer to start the proteolysis. Samples were withdrawn at the indicated times, added to an equal volume of 10% TCA and incubated for 5 min at 65 °C. The mixtures were neutralized by 25 volumes of 0.2 M Na2HPO4 on ice. Finally, 1/40 volume of 25 mg/mL fluorescamine in DMSO was added and mixed vigorously. Fluorescence was measured using an excitation wavelength of 390 nm and an emission of 475 nm. Peptides were custom-synthesized (Genscript Corp. NJ) apart from peptides NB, NS2, SP1, SP2, and SP231, which were gifts from R. A. Lamb (Northwestern University).
Determination of the global effect of amino acid bias on protein stability
Mouse protein half-life data was obtained from Schwanhäusser et al.33. Intrinsic disorder was predicted for all studied protein sequences (downloaded from UniProtKB/Swiss-Prot, http://www.uniprot.org/) using three complementary methods: DISOPRED264, IUPRED long68, and PONDR VLS169. Based on the length of the disordered segments, proteins were first classified as those that had short (stretches of ≤30 disordered residues) and long disordered termini (stretches of >30 disordered residues; for N- and C-termini separately). Amino acid bias within N- and C- terminal long disordered segments was identified using LPS-annotate70,71 employing default parameters. Using a stringent detection p-value cut-off of <1x10−10, the proteins with long disordered termini were classified into those with and without amino acid bias. Statistical significance of the difference in the distributions of half-life values among different classes of proteins was estimated using the non-parametric Wilcoxon rank sum test.
Supplementary Material
Acknowledgments
We thank J. Brickner and R. A. Lamb (Northwestern University), M. Glickman (Technion), and Y. Saeki (Tokyo Metropolitan Institute of Medical Science) for advice and reagents, as well as A. Zokarkar for early experiments on this project. We are also grateful to K. Brown, A. Gnanam, and E. J. Cho (The University of Texas at Austin) for help with the CD experiments. This work was supported by the NIH (R01GM63004, U54GM105816, U54CA143869, AM), the Welch Foundation (F-1817, AM), the Program to Disseminate Tenure Track System from the Ministry of Education, Culture, Sports, Science and Technology, Japan (TI), the UK Medical Research Council (MC_U105185859, SC and MMB) and the European Molecular Biology Organization (Long Term Fellowship, SC; Young Investigator Program, MMB).
Footnotes
Author contributions
SF, TI, EI, HY and GK performed experiments, analyzed data and co-wrote the paper. MMB and AM directed experiments, analyzed data and co-wrote the paper.
References
- 1.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 4.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 5.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schauber C, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 9.Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Heinen C, Acs K, Hoogstraten D, Dantuma NP. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun. 2011;2:191. doi: 10.1038/ncomms1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stack JH, Whitney M, Rodems SM, Pollok BA. A ubiquitin-based tagging system for controlled modulation of protein stability. Nat Biotechnol. 2000;18:1298–1302. doi: 10.1038/82422. [DOI] [PubMed] [Google Scholar]
- 14.Goebl MG, Goetsch L, Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee A, Gregori L, Xu Y, Chau V. The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J Biol Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- 16.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 17.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 18.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 19.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Meth Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 20.Daskalogianni C, et al. Gly-Ala repeats induce position- and substrate-specific regulation of 26 S proteasome-dependent partial processing. J Biol Chem. 2008;283:30090–30100. doi: 10.1074/jbc.M803290200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoyt MA, et al. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharipo A, Imreh M, Leonchiks A, Imreh S, Masucci MG. A minimal glycine-alanine repeat prevents the interaction of ubiquitinated I kappaB alpha with the proteasome: a new mechanism for selective inhibition of proteolysis. Nat Med. 1998;4:939–944. doi: 10.1038/nm0898-939. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Coffino P. Repeat sequence of Epstein-Barr virus-encoded nuclear antigen 1 protein interrupts proteasome substrate processing. J Biol Chem. 2004;279:8635–8641. doi: 10.1074/jbc.M310449200. [DOI] [PubMed] [Google Scholar]
- 24.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 25.Pratt G, Rechsteiner M. Proteasomes cleave at multiple sites within polyglutamine tracts: activation by PA28gamma(K188E) J Biol Chem. 2008;283:12919–12925. doi: 10.1074/jbc.M709347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juenemann K, et al. Expanded Polyglutamine-containing N-terminal Huntingtin Fragments Are Entirely Degraded by Mammalian Proteasomes. J Biol Chem. 2013;288:27068–27084. doi: 10.1074/jbc.M113.486076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 28.Kisselev AF. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- 29.Udenfriend S, et al. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science. 1972;178:871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 30.Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P, Fersht AR. Structure and Function of Intrinsically Disordered Proteins. Chapman & Hall/CRC Press; 2010. [Google Scholar]
- 32.van der Lee R, et al. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Rep. 2014;8:1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 34.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 35.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 36.Kalchman MA, et al. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–94. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 37.Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jana NR, Zemskov EA, Gh W, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 39.Waelter S, et al. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyttenbach A, et al. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and polyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 43.Levitskaya J, et al. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 44.Kraut DA, Matouschek A. Proteasomal degradation from internal sites favors partial proteolysis via remote domain stabilization. ACS Chem Biol. 2011;6:1087–1095. doi: 10.1021/cb2002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraut DA, et al. Sequence- and species-dependence of proteasomal processivity. ACS Chem Biol. 2012;7:1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 47.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller-Spath S, et al. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moesa HA, Wakabayashi S, Nakai K, Patil A. Chemical composition is maintained in poorly conserved intrinsically disordered regions and suggests a means for their classification. Mol Biosyst. 2012;8:3262–3273. doi: 10.1039/c2mb25202c. [DOI] [PubMed] [Google Scholar]
- 51.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagai T, Levy Y. Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc Natl Acad Sci USA. 2010;107:2001–2006. doi: 10.1073/pnas.0912335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beskow A, et al. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 54.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 55.Bancher C, et al. An antigenic profile of Lewy bodies: immunocytochemical indication for protein phosphorylation and ubiquitination. J Neuropathol Exp Neurol. 1989;48:81–93. doi: 10.1097/00005072-198901000-00007. [DOI] [PubMed] [Google Scholar]
- 56.Iqbal K, Grundke-Iqbal I. Ubiquitination and abnormal phosphorylation of paired helical filaments in Alzheimer’s disease. Mol Neurobiol. 1991;5:399–410. doi: 10.1007/BF02935561. [DOI] [PubMed] [Google Scholar]
- 57.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 58.Hipp MS, et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 61.Kruegel U, et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muñoz V, Serrano L. Development of the multiple sequence approximation within the AGADIR model of α-helix formation: Comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers. 1997;41:495–509. doi: 10.1002/(SICI)1097-0282(19970415)41:5<495::AID-BIP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 64.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Saeki Y, Isono E, Toh-e A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Meth Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 66.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans CH, Ridella JD. An evaluation of fluorometric proteinase assays which employ fluorescamine. Anal Biochem. 1984;142:411–420. doi: 10.1016/0003-2697(84)90485-8. [DOI] [PubMed] [Google Scholar]
- 68.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 69.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61 (Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 70.Harbi D, Kumar M, Harrison PM. LPS-annotate: complete annotation of compositionally biased regions in the protein knowledgebase. Database (Oxford) 2011;2011:baq031. doi: 10.1093/database/baq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.