Abstract
Ribonucleotides are frequently incorporated into DNA during eukaryotic replication. Here we map the genome-wide distribution of these ribonucleotides as markers of replication enzymology in budding yeast, using a new 5′-DNA end-mapping method, Hydrolytic End Sequencing. HydEn-Seq of DNA from ribonucleotide excision repair-deficient strains reveals replicase- and strand-specific patterns of ribonucleotides in the nuclear genome. These patterns support the role of DNA polymerases α and δ in lagging strand replication and of DNA polymerase ε in leading strand replication. They identify replication origins, termination zones and variations in ribonucleotide incorporation frequency across the genome that exceed three orders of magnitude. HydEn-Seq also reveals strand-specific 5′-DNA ends at mitochondrial replication origins, suggesting unidirectional replication of a circular genome. Given the conservation of enzymes that incorporate and process ribonucleotides in DNA, HydEn-Seq can be used to track replication enzymology in other organisms.
Keywords: ribonucleotide incorporation, replication origins, polymerase switching, mitochondrial replication, biomarkers
INTRODUCTION
Among the many eukaryotic DNA polymerases (Pols), e.g., 17 in humans and eight in budding yeast, three replicate the bulk of the nuclear genome 1,2. Synthesis at replication origins is initiated when an RNA primase synthesizes an RNA primer that is extended by limited DNA synthesis by Pol α 3. Pol ε is then proposed to catalyze the majority of leading strand replication 4–6 in a largely continuous manner. In contrast, the nascent lagging strand is synthesized as a series of ~180 nucleotide Okazaki fragments that are initiated by RNA primase followed by limited synthesis by Pol α. This is followed by extensive synthesis catalyzed by Pol δ 6–8 and subsequent maturation of Okazaki fragments into a continuous nascent lagging strand 9. The exact locations of polymerase switching during leading and lagging strand replication are under investigation 10,11 but remain uncertain. Equally uncertain is polymerase use after replication forks encounter difficult circumstances that may require switching to a different replicase or a more specialized DNA polymerase, e.g., to copy unusual DNA sequences or to bypass lesions 12,13. Replication enzymology differs for the mitochondrial genome, where both DNA strands are replicated by the same replicase, Pol γ, by mechanisms that also remain uncertain 14–16.
We have been using mutator alleles of yeast Pols ε, α and δ to infer their roles in nuclear DNA replication in vivo. These mutator alleles, pol2-M644G (Pol ε), pol1-L868M (Pol α) and pol3-L612M (Pol δ), generate single base replication errors at higher rates than their wild type parents. In the absence of mismatch repair (MMR), these errors remain in the genome and mark where each replicase synthesized DNA during replication. The results (see 6 and references therein) imply that in unstressed yeast cells, Pol ε is the primary leading strand replicase and Pols α and δ are primarily responsible for lagging strand replication. However, the resolution of this approach for tracking replication enzymology in vivo is limited by the high fidelity of replication. For example, the average genome-wide replication error rates of the mutator replicases are 1–2 × 10−7 6, such that single base replication errors are low-density markers of replication enzymology.
In the present study, we set out to track replication enzymology in vivo at much higher resolution using ribonucleotides rather than mutations. This approach takes advantage of several facts. The presence of an oxygen atom on the 2′-position of a ribose increases the sensitivity of the phosphodiester bond in nucleic acids to alkaline hydrolysis by five orders of magnitude. The active sites of Pols α, δ and ε can be engineered to increase the probability of ribonucleotide incorporation into DNA, to frequencies as high as 10−2 to 10−3 17. Disabling Ribonucleotide Excision Repair (RER) prevents removal of ribonucleotides from both the nascent leading strand and the nascent lagging strand 17–19. RER defective yeast cells are viable, including those encoding replicases that are promiscuous for ribonucleotide incorporation. These facts led us to propose 20 that ribonucleotides can be used as high-density markers of DNA polymerization reactions in vivo. Here we demonstrate that this is indeed the case, using a newly developed method to map ribonucleotides in the yeast genome at single nucleotide resolution. Initial results support the strand assignments for the nuclear replicases, confirm nuclear replication origins and identify new origins, reveal the locations of replication termination zones, quantify ribonucleotide incorporation for each of the four bases, establish that the distribution of ribonucleotides across the genome is non-uniform, and provide new information that is likely to be relevant to mitochondrial DNA replication.
RESULTS
Genome-wide mapping of ribonucleotides in DNA by HydEn-Seq
The new genome-wide mapping method, which we call HydEn-Seq (for Hydrolytic DNA End Sequencing (Fig. 1a, Supplementary Table 1), has been used to map ribonucleotides in five pairs of RER-deficient (rnh201Δ) versus RER-proficient (RNH201) yeast strains (Supplementary Table 2). One pair encodes wild type Pols α, δ and ε. A second pair encodes pol2-M644L, a Pol ε variant that incorporates fewer ribonucleotides than does a wild type strain 18. A third pair encodes pol2-M644G, a Pol ε variant that is promiscuous for ribonucleotide incorporation 18,21. A fourth pair encodes a pol3-L612G variant in which leucine 612 in the Pol δ active site is replaced with glycine, based on the prediction that like the analogous pol2-M644G (Pol ε) variant 18, the pol3-L612G variant would be even more promiscuous for ribonucleotide incorporation than our previously studied pol3-L612M allele 22. The fifth pair encodes a pol1-Y869A variant with alanine substituted for the “steric gate” tyrosine in the Pol α active site that normally prevents ribonucleotide incorporation 23. This allele is used to increase the frequency of ribonucleotide incorporation by Pol α over that observed in our previously studied pol1-L868M variant 17.
Figure 1. Mapping ribonucleotides by HydEn-Seq.
(a) HydEn-Seq protocol. The procedure was performed as described in Methods, using the oligonucleotides listed in Supplementary Table 1. (b) Alkaline agarose gel electrophoresis. The analysis was performed as previously described 18. Genomic DNA samples from the indicated yeast strains (lanes 1–10) were treated with alkali, separated by 1% alkaline agarose gel electrophoresis, and imaged after staining with SYBR Gold. Migration positions of two DNA size standards are indicated. (c) Densitometry scans of the gel image in (b). The Y-axis is scaled to maximum intensity for each pair of lanes. (d) Mean HydEn-seq end counts per haploid genome (see Nends calculation in Methods; error bars represent ranges of two to four independent measurements).
Alkaline hydrolysis of genomic DNA followed by electrophoresis in an alkaline agarose gel reveals that the genomes of all five rnh201Δ mutant strains contain more alkali-sensitive sites than their RNH201+ parents (Fig. 1b). Importantly, the genomes of the double mutant pol1-Y869A rnh201Δ, pol2-M644G rnh201Δ and pol3-L612G rnh201Δ strains contain many more alkali-sensitive sites than the strains with either single mutation alone (Fig. 1b,c,d), such that most of the 5′-DNA ends in these strains result from alkaline hydrolysis of ribonucleotides incorporated during replication by the variant derivatives of Pols α, δ or ε, respectively. This contrasts with the pol2-M644L rnh201Δ mutant strain, which harbors fewer ribonucleotides than the other rnh201Δ strains with variant replicases.
The locations of the 5′-DNA ends in the genomes of these strains were mapped by HydEn-Seq (Fig. 1a). Genomic DNA samples were hydrolyzed with 0.3M KOH 24, libraries were prepared from the resulting single-stranded DNA fragments, and 50 base, paired-end sequencing was performed on an Illumina HiSeq2500 instrument to identify the location of the 5′-DNA ends. Ribonucleotides are located immediately adjacent to the 5′-DNA ends (Fig. 1a). Two or more independent libraries were analyzed for each strain (Supplementary Table 3), with replicate libraries yielding similar results (Supplementary Table 4). Alignment of the fragments to a well-annotated reference genome 6 identified the DNA strand to which fragments align, and the location and identity of ribonucleotides in the genome. Read counts, scaled using the number of 5′-ends at the ends of chromosomes (Methods), confirm the relative ribonucleotide densities anticipated by agarose gel electrophoresis.
Strand specificity and origin identification
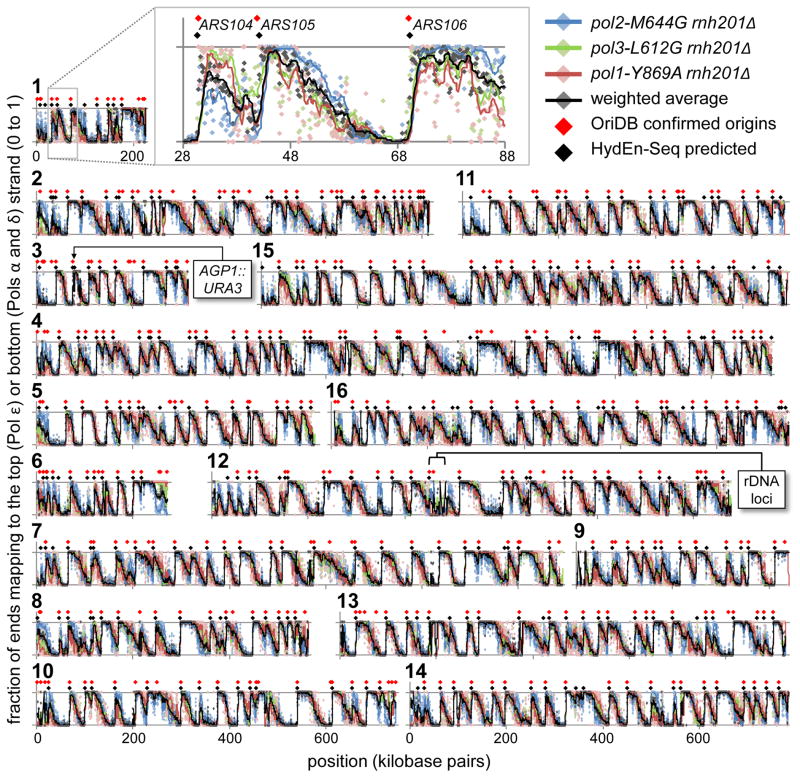
DNA fragments from the pol2-M644G rnh201Δ strain align with the two DNA strands in the nuclear genome in an alternating pattern complementary to alignments for the pol1-Y869A rnh201Δ and pol3-L612G rnh20Δ strains (Fig. 2, chromosome 10; Supplementary Fig. 1, 5′-DNA end read counts corresponding the Fig. 2, bottom; Fig. 3, all 16 chromosomes; Fig. 4a, heat maps). In contrast, a strand-specific pattern is not observed in the pol2-M644L rnh201Δ strain (Fig. 4a) or in RER-proficient strains (Supplementary Fig. 2). Thus the majority of 5′-DNA ends in the pol2-M644G rnh201Δ, pol1-Y869A rnh201Δ and pol3-L612G rnh20Δ strains are due to ribonucleotides incorporated during replication that are not removed because RER is defective. Comparing ribonucleotide maps in these three strains reveals numerous strand-specific transitions (diamonds in Figs. 2 and 3). Among these are 294 transitions that correspond to confirmed replication origins in the yeast origin database 25. Transitions are also observed at 72 locations (Fig. 3, listed in Supplementary Table 5) that have not yet been reported to be origins, but may be origins that are used in some cells in the population.
Figure 2. Strand-specific ribonucleotide mapping of chromosome 10.
(a) Top, map of chromosome 10 showing the fraction of end reads mapped to the top strand in bins of 200 base pairs, after background subtraction (see Methods). Origin prediction was complicated at chromosome ends (I) and in other highly repetitive regions (II). Middle, an expanded 200 kbp region of chromosome 10. Excursions (in purple) from the simplest polymerase division of labor (Pol α or δ lagging, Pol ε leading) fall into two classes: unexpected Pol α, δ or ε correspondence (III) and Pol α or δ divergence (IV). Bottom, 100 kbp region of chromosome 10. Inter-origin regions are more (e.g. ARS1012-ARS1014) or less (e.g. ARS1011-ARS1012) symmetrical, depending on fork progression rates and origins firing times. Some origins in the origin database have little effect on ribonucleotide strand bias (e.g. ARS1013), indicating either an incorrect call, minority participation in normally growing cells, unidirectional origin firing, or simply later firing, such that forks proceeding from adjacent origins approach to within current detection thresholds. (b) A stylized chromosome with two replication origins, showing the division of polymerase labor, as predicted from the direction of strand bias transitions at origins (compare with the ARS1012-ARS1014 region above). Roughly three quarters of previously confirmed replication origins (orange diamonds in panel (a); S. cerevisiae OriDB) align with abrupt transitions in strand preference (see Methods for quantitation). This allows algorithmic prediction of origins (black diamonds). ARS1013 was not detected via HydEn-seq. It is indicated (orange diamond) but not labeled.
Figure 3. Genome-wide replication origins located by HydEn-Seq.
(a) A 60 kbp segment of chromosome 1 showing the fraction of ends, after background subtraction, that mapped to the top strand from Pol ε data (pol2-M644G rnh201Δ; blue) and the fraction mapped to the bottom strand for Pol α and δ data (pol1-Y869A and pol3-L612G, in red and green, respectively). Grey points are the weighted average of the other three data sets in each bin (see Methods). All curves are trend lines smoothed over 10 bins. (b) As per (a), but for all 16 S. cerevisiae chromosomes. Shown for reference are the locations of the URA3 mutational reporter gene (near ARS306; used in our previous studies of leading and lagging strand replication fidelity 5, and the rDNA locus in chromosome 12 (not drawn to scale; the highly repetitive sequence precludes read mapping).
Figure 4. Distribution of ribonucleotides near origins in RER-deficient strains.
(a) Heat maps for the top and bottom strands of the nuclear genome in five different rnh201Δ strains, scaled per million reads and centered across a 4 kbp window of the 394 replication origins reported in the yeast origin database 25. (b) Meta-analysis of strand-specific ribonucleotides at 214 replication origins analyzed in a previous study 10, again scaled per million reads, in bins of 50 bp.
The ribonucleotide maps in the three rnh201Δ strains encoding the variant replicases strongly support earlier interpretations based on replication errors 4–7, that Pol ε synthesizes the majority of the nascent leading strand and Pols α and δ synthesize the majority of the nascent lagging strand of the budding yeast nuclear genome. Thus HydEn-Seq confirms a fundamental aspect of yeast replication enzymology. The evolutionary conservation among eukaryotic nuclear replicases and among type 2 RNases H in all three kingdoms of life (Supplementary Figure 3), suggests that the HydEn-Seq strategy used here may be applicable to tracking replication enzymology and identifying origins in other organisms. The ribonucleotide map in pol1-Y869A rnh201Δ strain further demonstrates that when RER is deficient, some DNA synthesized by Y869A Pol α survives Okazaki fragment maturation and resides in the mature lagging strand. This same conclusion was reached in earlier studies 6,7,26–28 that monitored replication errors rather than ribonucleotides using pol3-L612M strains that were either proficient or deficient in MMR but were RER proficient. It remains to be determined if DNA synthesized by Pol α survives Okazaki fragment maturation in wild type yeast.
Polymerase use at replication origins and termination zones
Heat maps (Fig. 4a) and meta-analyses of 5′-DNA ends in 50 base pair bins (Fig. 4b) reveal where strand switches at origins occur in all three replicase variant backgrounds. These transitions occur over several hundred base pairs centered on the Autonomously Replicating Sequence (ARS) Consensus Sequence (ACS, orange line in Fig. 4b). The results in the pol1-Y869A rnh201Δ strain are consistent with a role for Pol α in initiating synthesis on both strands at origins. The breadth of the strand transition at origins in this strain suggests that in a cell population, initiation occurs within a zone rather than at a single base pair. Deeper coverage of 5′-DNA ends in the future should allow higher resolution mapping, to investigate whether initiation occurs at a single base pair at some origins, as previously reported for one origin on chromosome 4 29. The strand transition at origins is much sharper in the pol2-M644G rnh201Δ strain than in the pol3-L612G rnh201Δ strain. Investigating this difference may eventually provide information that complements recent biochemical studies of initiation of leading and lagging strand replication (see 2 and references therein).
HydEn-Seq also reveals where mergers occur between forks arriving from adjacent origins and moving in opposite directions. The results suggest that termination occurs in zones that vary in location and breadth. In some cases (e.g., Fig. 2a, bottom right), the termination zone is broad and equidistant from adjacent origins, while in other cases (e.g., Fig. 2a, bottom left) the zone is narrower and or closer to one origin than the other. HydEn-Seq offers the opportunity to explore the mechanisms and genetic controls underlying these variations.
Ribonucleotide incorporation in wild type yeast
Studies of ribonucleotide incorporation in vitro by wild type Pols α, δ and ε predict that there should be 2.3 times more ribonucleotides incorporated into the nascent leading strand as compared to the nascent lagging strand 24. This prediction is supported by results in the RER-defective (rnh201Δ) strain encoding wild type replicases. In this strain, the strand-specific heat map (Fig. 4a) and the transition from one strand to the other as analyzed by meta-analysis (Fig. 4b) match those of the pol2-M644G rnh201Δ strain, and are opposite to those in the pol3-L612G 201Δ or pol1-Y869A rnh201Δ strains.
The observation that ribonucleotides are preferentially incorporated into the nascent leading strand in the strain encoding wild type replicases is relevant to the genome instability reported in the wild type replicase background when RER is defective. In this strain 30, the specificity of 2 to 5 base pair deletion mutations resulting from topoisomerase1 (Top1) cleavage at unrepaired ribonucleotides is indistinguishable from the 2 to 5 base pair deletion specificity in the pol2-M644G rnh201Δ strain 18 that primarily contains ribonucleotides in the nascent leading strand 5,21,22. The fact that ribonucleotides preferentially map to the nascent leading strand is also relevant to recent studies 22,31 suggesting that nicks generated by RNase H2 at ribonucleotides in the continuously replicated nascent leading strand may direct mismatch repair (MMR) to correct replication errors in that strand. This idea, when combined with the non-uniform distribution of ribonucleotides in the genome discussed below, implies that the potential contribution of this MMR signaling mechanism may vary across the genome. The preferential presence of ribonucleotides in the nascent leading strand may also be relevant to other suggested signaling functions for ribonucleotides in DNA 20,24.
Variations in ribonucleotide incorporation by base identity
Wild type Pols α, δ and ε have different preferences for incorporating each of the four different ribonucleotides in vitro 24. To determine if this is also true during replication in vivo, we analyzed fragments close to replication origins where (as explained previously 6) leading and lagging strand assignments can be made with the greatest confidence. Despite the fact that the 12 million base pair budding yeast genome is 62% A+T, the most abundant ribonucleotide present in the genome of the pol2-M644G rnh201Δ strain is rC, followed by rG, then rA and then rU (Fig. 5). These preferences re-capitulate the rank order for ribonucleotide incorporation by M644G Pol ε in vitro 24. The same rank order for ribonucleotide incorporation (rC > rG > rA > rU) is observed in the pol3-L612G rnh201Δ strain, but the proportions of the four rNTPs incorporated are different. For example, more rC and less rU are present in the pol3-L612G rnh201Δ genome as compared to the pol2-M644G rnh201Δ genome (Fig. 5). A different rank order is seen in the pol1-Y869A rnh201Δ strain, where after correcting for genome composition, the preference in the pol1-Y869A strains is rA ≈ rC ≈ rG > rU. In this strain, the non-rU rankings changed slightly among the three replicates examined.
Figure 5. Ribonucleotide base identity.
The proportion of each ribonucleotide base present in the nuclear genome of the three rnh201Δ strains encoding the indicated variant replicases. The base composition of the genome is shown on the left. Ribonucleotide proportions were calculated from the most highly strand-biased 10% of the genome (i.e. windows near replication origins; examples in Fig. 2a).
The low abundance of rU seen in all three genomes is consistent with the fact that, among the four dNTPs, dTTP is present at the highest concentration in strains encoding either wild type replicases 24 or the pol2-M644G variant 18, thereby reducing the probability of incorporating rU more than the other ribonucleotides. However, the dATP:rATP ratio is the lowest among the four ratios 24, yet rATP is only the most frequent ribonucleotide in one of the three strains. Thus, in addition to competition for incorporation within the polymerase active site based on mass action, other parameters may modulate ribonucleotide incorporation probability during replication in vivo. This includes the effect of DNA sequence context, as predicted by sequence context effects of ribonucleotide incorporation probability during DNA synthesis in vitro 24,32,33.
Non-uniform distribution of ribonucleotide in the genome
Several HydEn-Seq libraries contain an average of less than one 5′-DNA end read per base pair in the nuclear genome (Supplementary Table 3). It is therefore striking that end read counts vary from zero at many base pairs to more than 1,000 at others. This non-uniform distribution of ribonucleotides in the genome has implications for MMR signaling mentioned above, and for a second mechanism of genome instability wherein Top1 incises ribonucleotides in DNA to initiate the deletion of 2 to 5 base pairs within repetitive sequences 18,34. This instability is highly dependent on the DNA strand and sequence context in which the ribonucleotide resides 18. Variations in the location and density of ribonucleotides in DNA may also be relevant to recombination 35 and gross chromosomal rearrangements in yeast 36 and to chromosomal abnormalities in RNase H2-defective mouse cells 37,38.
In certain regions of the genome, strand-specific ribonucleotide density also deviates from the expectations of a simple division of labor among the three replicases. Initial analyses indicate that these “excursions” fall into at least two classes, those that show unexpected Pol α, δ or ε correspondence (e.g., purple bar designated III in Fig. 2a, middle) and those that show unexpected Pol α or δ divergence (designated IV). These excursions may result from ribonucleotides remaining in the genomes of these RER-deficient cells, which can lead to replicase pausing during DNA synthesis 18,39,40. Such pausing may elicit template switching or bypass synthesis (e.g., Pol ζ or Pol η) in a subsequent round of replication 41 or DNA synthesis associated with DNA repair or recombination after Top1 incision at ribonucleotides 21,34,35. An example is in Schizosaccharomyces pombe, where mating type switching occurs by recombination posited to be initiated by pausing of leading strand replication upon encountering a di-ribonucleotide imprint 42. Additional possibilities for some “excursions” detected by HydEn-Seq include events unrelated to ribonucleotides, such encounters of replication forks with transcription complexes, a bulky lesion, repetitive DNA or non-B form DNA or tightly bound proteins. There is no obvious limitation to monitoring the distance over which a newly recruited DNA polymerase may operate, e.g., within a short repair or lesion bypass patch or to the end of a chromosome during break-induced recombination 43.
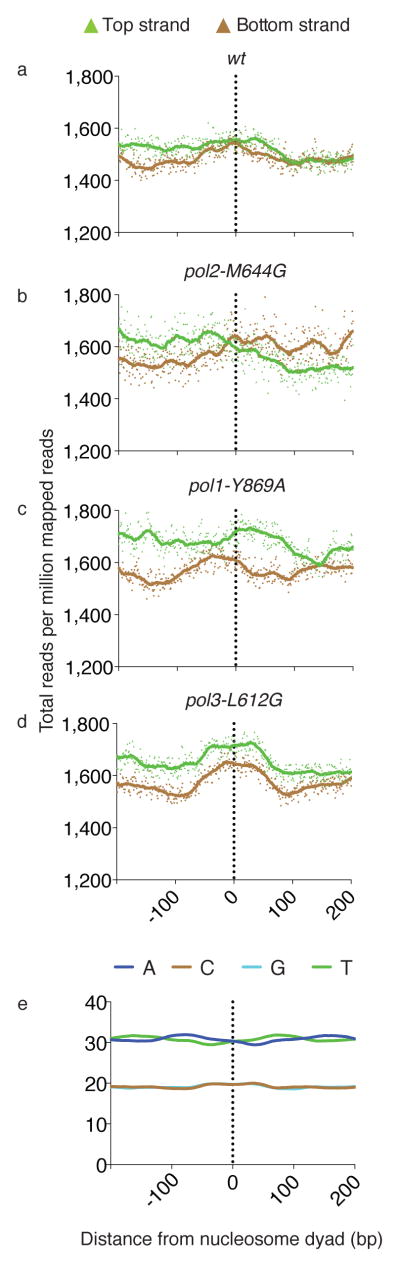
Ribonucleotide distribution relative to nucleosome dyads
Meta-analysis using nucleosome positioning data 6 reveals that ribonucleotide densities are elevated at positions corresponding to the nucleosome dyad (Fig. 6a–d). The elevations are subtle (note the scale on the Y axis). They may partly reflect a bias in sequence composition because nucleosome dyads are slightly enriched for G and C content (Fig. 6e), the preferred ribonucleotides incorporated during replication in the pol2-M644G rnh201Δ and pol3-L612G rnh201Δ strains. However, this may not be the sole explanation because (1) the peaks at the dyad are more prominent in the pol3-L612G rnh201Δ strain (Fig. 6d) as compared to the pol2-M644G rnh201Δ strain (Fig. 6b), yet these two strains have similar G+C versus A+T ribonucleotide incorporation preferences (Fig. 5), (2) the peaks are more prominent in the pol3-L612G rnh201Δ and pol1-Y869A rnh201Δ strains as compared to the pol2-M644G rnh201Δ strain, and (3) the peak in both strands in the pol3-L612G rnh201Δ strain are symmetrical around the dyad, whereas the peaks in the two strands in the pol1-Y869A rnh201Δ strain are offset and on opposite sides of the dyad. In the latter cases, lagging strand replicase features could be signatures of polymerization by Pol α and Pol δ during Okazaki fragment maturation, a process that is proposed to preferentially occur at the nucleosome dyad and to be phased according to the nucleosome repeat 10.
Figure 6. Meta-analysis of ribonucleotides at the nucleosome dyad.
(a) Meta-analysis of strand-specific ribonucleotide mapping at 37,888 nucleosome dyads 6 for the rnh201Δ strain, scaled per million reads and centered within a 400 bp window. Each dot indicates the number of 5′-DNA ends reads at one base pair. The vertical dotted line indicates the dyad. (b) As in (a) but for the pol2-M644G rnh201Δ strain. (c) As in (a) but for the pol1-Y869A rnh201Δ strain. (d) As in (a) but for the pol3-L612G rnh201Δ strain. The solid lines are the smoothed averages for a sliding window. (e) The base composition surrounding the nucleosome dyad.
Ribonucleotides at mitochondrial DNA replication origins
HydEn-Seq also reveals that the yeast mitochondrial genome contains large numbers of 5′-DNA ends generated by alkaline hydrolysis (Fig. 7). Most of these ends are in discrete, strand-specific (red and blue) peaks that span multiple base pairs. Eight of these peaks correspond to previously identified 44 mitochondrial replication origins (shaded green). Interestingly, the relative proportions of 5′-DNA ends in the major peaks are similar in all yeast strains examined. Thus the peaks are independent of the status of the nuclear replicases, which have no known role in mitochondrial replication, and they are also independent of the status of RNase H2, which has not been found in mitochondria 45.
Figure 7. HydEn-Seq maps of mitochondrial DNA.
Mitochondrial genomes for six strains are shown to indicate the base pair (bp) locations and proportions of strand-specific 5′-DNA ends detected by HydEn-Seq (blue for plus strand, red for minus). Previously assigned replication origins 44 are shaded in green, coding sequences in grey, tRNA genes in orange and genes for other non-coding RNAs in pink. Total mitochondrial end counts are shown for each strain with the number of replicate HydEn-Seq libraries for each in parentheses.
These observations are consistent with at least three hypotheses. The peaks may represent the ends of linear chromosomes, similar to the high density of 5′-ends observed at the ends of linear nuclear chromosomes. This possibility cannot yet be eliminated, but it seems unlikely because the most prominent peaks largely map to either the plus or minus strand, but not to both. Also, when the mitochondrial genome is rearranged in silico (Methods) to join the chromosome “ends” that were arbitrarily assigned and numbered when the genome was sequenced 44, no drop in the depth of coverage of fragments is observed at the junction relative to immediately adjacent regions. Thus, like mammalian mitochondrial genomes, the S. cerevisiae mitochondrial genome may largely, albeit not necessarily exclusively, be circular.
The second hypothesis is that some mtDNA fragments generated may be due to lesions other than ribonucleotides, such as strand-specific nicks or alkali-sensitive abasic sites resulting from oxidative stress. We cannot exclude this possibility, but it is currently disfavored by the fact that the 5′-DNA ends are distributed in a highly non-uniform and largely strand-specific manner.
The third hypothesis stems from previous studies showing that mammalian mtDNA contains ribonucleotides 46,47, and that human mitochondrial replicase (Pol γ) incorporates ribonucleotides during DNA synthesis in vitro 48. Moreover, eight of the most prominent, strand-biased 5′-DNA end peaks in the yeast mitochondrial genome correspond to previously identified 44 mitochondrial replication origins. These peaks, and perhaps similar strand-specific peaks detected within open reading frames and in sequences encoding RNAs, could reflect the presence of unrepaired residues of RNA primers made by mtRNA polymerase and used to initiate mtDNA replication, as has been reported in mammalian cells 16,49–52. The results suggest that the terminal ribonucleotides of RNA primers for mtDNA replication may not always be removed, either by mitochondrial RNase H1 53, which cannot incise at an RNA-DNA junction 45, or by strand displacement and flap cleavage, as for Okazaki fragment maturation during nuclear DNA replication 9. If this explanation holds, then the fact that 5′-DNA ends at the origins of mtDNA replication preferentially map to one strand or the other favors a unidirectional replication model for mtDNA in budding yeast.
DISCUSSION
This study demonstrates that ribonucleotides can be used to track replication enzymology at high resolution using a simple, 5-step library preparation procedure involving minimal use of enzymes and requiring less than two days to execute. While HydEn-seq is used here to map 5′-DNA ends primarily generated by alkaline hydrolysis at ribonucleotides, it can also be used to study other lesions in DNA, and it is not limited to spontaneous chemical hydrolysis but can be adapted to map 5′- and 3′-DNA ends generated by enzymatic hydrolysis. In addition to normal replication enzymology, HydEn-Seq should be useful to study polymerization changes in response to endogenous and exogenous environmental stress. The ability of HydEn-Seq to identify replication origins, termination zones and polymerase usage during replication should be applicable to other organisms in which replicases can be engineered to enhance ribonucleotide incorporation and RER can be inactivated. Polymerase structure-function studies have advanced to the point where it is now feasible to engineer replicases (e.g., see Supplementary Figure 3a), and more specialized polymerases in most polymerase families, to retain catalytic efficiency yet render them promiscuous for ribonucleotide incorporation. Theoretically, this may permit a variety of DNA synthesis reactions in cells to be studied by HydEn-Seq.
Following the idea that the high-density peaks in the mitochondrial genome may be due to unrepaired residues of RNA primers made by mtRNA polymerase, HydEn-Seq may also be useful to study RNA primers synthesized by RNA polymerases, RNA primases or Prim-Pols. The ability to map genomic locations that contain a high density of ribonucleotides can be used to explore the idea that ribonucleotides in DNA provide selective advantages to cells 24. Relevant here are ribonucleotides that may persist in certain locations even in RER-proficient cells, as exemplified by the di-ribonucleotide imprint used for mating type switching in Schizosaccharomyces pombe 54.
ONLINE METHODS
Materials
Oligonucleotides and yeast strains used in this study are listed in Supplementary Tables 1 and 2, respectively. The pol1-Y869A and pol3-L612G strains and their rnh201Δ derivatives were constructed as described earlier for pol1-L868M and pol3-L612M strains 7.
HydEn-Seq protocol
Yeast strains were grown to mid log phase (OD600=0.6) at 30°C in YPDA medium supplemented with 0.25 mg/ml adenine. DNA was isolated using the MasterPure™ Yeast DNA Purification Kit (Epicentre) without RNase A treatment. HydEn-Seq (Fig. 1) was performed by hydrolyzing one μg of genomic DNA with 0.3 M KOH for 2 hours at 55°C 24. Following ethanol-precipitation, the DNA fragments were treated for three minutes at 85°C, phosphorylated with 10 units of 3′-phosphatase-minus T4 polynucleotide kinase (New England Biolabs) for 30 minutes at 37°C, heat inactivated for 20 minutes at 65°C and purified using HighPrep™ PCR beads (MagBio). Phosphorylated products were treated for three minutes at 85°C, ligated to oligo ARC140 (Supplementary Table 1) overnight at room temperature using 10 units of T4 RNA ligase, 25% PEG8000 and 1 mM CoCl3(NH3)6, and purified using HighPrep™ PCR beads (MagBio). Ligated products were treated for 3 minutes at 85°C. The ARC76–ARC77 adapter was annealed to the second strand for five minutes at room temperature. The second strand was synthesized using four units of T7 DNA polymerase (New England Biolabs) and purified using HighPrep™ PCR beads (MagBio). Libraries were PCR amplified using primer ARC49 and primer ARC79 or ARC84 to ARC107, using KAPA HiFi Hotstart ReadyMix (KAPA Biosystems). Libraries were then purified using HighPrep™ PCR beads (MagBio) and pooled for sequence analysis. Paired-end sequencing was performed on a HiSeq2500 sequencer (Illumina) to identify the location of the 5′-DNA ends generated by alkaline hydrolysis.
HydEn-Seq trimming, filtering and alignment
All reads were trimmed for quality and adapter sequence using cutadapt 1.2.1 (-m 15 -q 10 -- match-read-wildcards)55. Pairs with one or both reads shorter than 15 nucleotides were discarded. Mate 1 of the remaining pairs was aligned to an index containing the sequence of all oligos utilized in the preparation of these libraries using bowtie 0.12.8 (-m1 -v2), and all pairs with successful alignments were discarded. Pairs passing this filter were subsequently aligned to the L03 S. cerevisiae reference genome 6 (-m1 -v2 -X10000 --best). Single-end alignments were then performed using mate 1 of all unaligned pairs (-m1 -v2). The count of 5′ ends of all unique paired-end and single-end alignments were determined for all samples, per-strand, across all chromosomes, combining all technical replicates, and shifted one base upstream to the location of the hydrolyzed ribonucleotide as summarized (Supplementary Table 2). These counts were converted to bigWig format for visualization on the UCSC browser. The distributions of counts per-nucleotide were determined using these values.
End count scaling and background subtraction
Two modes of end count scaling were used, depending on several factors. For analyses resulting in visual comparisons of individual libraries (i.e. heat maps and meta-analyses), end counts were normalized to counts per million uniquely mapped reads (divided by the values listed in Supplementary Table 2 under “Uniquely mapped ends” and then multiplied by 1,000,000). For analyses that required weighted averaging of multiple libraries and background subtraction (i.e. strand bias maps, origin predictions, and genomic ribonucleotide density estimates), end counts were scaled using n chromosomal 5′-end counts as internal standards (counts were divided by the values listed in Supplementary Table 2 under “Telomere End-Derived Scale Factor”; see below). To ensure that ends in these latter analyses originated only from replicase-inserted genomic ribonucleotides, scaled end counts from polymerase “L” variant strains (rnh201Δ, rnh201Δ, and pol2-M644L rnh201Δ) were subtracted from the scaled end counts of corresponding promiscuous-replicase strains (pol1-Y869A rnh201Δ, pol3-L612G rnh201Δ, and pol2-M644G rnh201Δ, respectively).
Calculating telomere end-derived scale factors and genomic ribonucleotide densities
The genomic ribonucleotide density (Rbulk) is
where Nbulk is the bulk end count, Lgenome is the length of the genome, and Ltelomere is the total length of all telomeric repeats in the reference genome:
where NC is the chromosome count (16 in S. cerevisiae). The mean number of 5′ chromosome ends per telomere ( ) is similar, but should be corrected for Rbulk in order to account for ribonucleotides found in telomeric repeats but not at chromosome ends (this correction never amounted to more than 2% of the final value):
where Ntelomere is the unadjusted total telomeric end count. serves as a scaling factor, allowing conversion of end counts in any bin into counts per position per genome (Supplementary Table 2). Where the bin is the whole genome, this results in an estimate of the mean fragment size ( ; always larger than the median fragment size as reported in 21,22:
and thence the mean number of ends per genome (Nends; always smaller than the median count per genome as reported in 21,22:
Predicting replication origins from HydEn-Seq maps
Replication origins were predicted from the weighted average fraction of scaled and background-subtracted ends (see above) mapping to the top strand in the Pol α, δ or ε variant strains. In each bin, the weighted average top-strand-fraction (f) was
where α, δ, and ε are the background-subtracted end counts from pol1-Y869A rnh201Δ, pol3-L612G rnh201Δ, and pol2-M644G rnh201Δ strains, respectively (each of which is itself the average of scaled counts from all replicate libraries). Parameters for origin calling were set based on results from a training set of OriDB confirmed replication origins on chromosome 11. Predicted origins were defined as regions where the bias changed abruptly over a defined distance in the weighted average curve in Fig. 4 (black; 200 bp bins; smoothed over 9 bins). In order for a position to be called an origin, either the average slope (the derivative) of the black curve had to exceed 0.00011 fractional units per bp in an 11-bin window (2.2 kbp) or 0.00016 fractional units per base pair in at least 3 of five surrounding bins (≥600 bp out of 1 kbp). These parameters attempt to define a sufficiently abrupt bias change over a region wide enough to exclude random noise.
Meta-analyses and preparation of heatmaps
Total counts of the per-strain 5′ ends intersecting same- and opposite-strand bins centered on genomic features of interest were determined using custom tools, excluding all mitochondrial annotations. Heatmaps, generated using the Partek Genomics Suite depict counts in all bins (normalized to ends per 1,000,000 uniquely mapped reads; see Supplementary Table 2), while meta-analyses depict the sum across all features.
Ribonucleotide frequencies
The composition of uniquely mapped ends was tallied in defined windows on each genomic strand. Windows were set in regions of high strand bias (≥99%) to ensure that nearly all ends represented ribonucleotides inserted by a particular replicase, depending on strand (leading versus lagging; e.g., grey or black bars, respectively in Fig. 3). The frequency of nucleotides occurring at 5′ ends intersecting these windows was determined using custom tools.
In silico mapping of the mitochondrial genome
The mitochondrial genome sequence was rearranged, such that the first 42,888 base pairs were removed from the start, and appended to the end. Read pairs from sample WT.1 (Supplementary Table 2) were aligned to this reordered mitochondrial genome, using bowtie 0.12.8 (-m1 -v2 -X10000). Based on start and end coordinates of the paired-end alignments, per-nucleotide coverage of HydEn-Seq fragments was determined using genomeCoverageBed (-d).
Analysis of polymerase and RNase H2 conservation
PSI-BLAST searches 56 (parameters in Supplementary Table 6) were conducted to find sequences homologous to S. cerevisiae replicases (catalytic subunit sequences from the Saccharomyces Genome Database) and the predicted RNH201 of Schizosaccharomyces pombe 972h- (gi accession number 19114596). Environmental sequences were excluded. For the replicases, PSI-BLAST was iterated until no new eukaryotic sequences were found. For RNH201, PSI-BLAST was iterates until >5,000 hits were acquired. The top hits were selected until the cumulative e-value exceeded 1.47. In all cases, partial sequences were culled and the remainder were aligned with CLUSTAL X (2.0) default parameters 57. Sequences with obvious deletions spanning active sites were culled, the remainder re-aligned, and trees built from the results (neighbor-joining, default parameters). The tree in Supplementary Figure 3 was constructed, in part, with the Interactive Tree of Life version 2.2.2 (http://itol.embl.de) 58.
Supplementary Material
Supplementary Table 1. Oligonucleotides. Listed are the oligonucleotides used for HydEn-Seq. Bold face indicates indexing. * indicates a phosphorothioate bond. ARC140 contains a 5′-amino group instead of a 5′-OH group, in combination with a C6 linker. This modification reduces formation of ARC140 concatemers during amplification. All oligos are from Integrated DNA technologies (Coralville, IA).
Supplementary Table 2. The 10 yeast strains used for HydEn-Seq as described in the text. The sources of several additional yeast strains subjected to HydEn-Seq but not analyzed in detail are given in Supplementary Table 3.
Supplementary Table 3. Yeast strains and library statistics. Listed are the genotypes and sources of the yeast strains used in this study, as well as relevant information on the HydEn-Seq libraries generated from these strains. a: orientation 1 of the URA3 gene located adjacent to ARS306. b: orientation 2 of the URA3 gene located adjacent to ARS306.
Supplementary Table 4. Correlation coefficients for comparisons among libraries. Library names are as given in Supplementary Table 3. For the purposes of calculating correlations, all data sets are binned in 200 base pair increments. Above the white diagonal, correlations are for raw end counts, on both strands. Below the white diagonal, correlations are for fractions of ends mapping to the top strand, as per Figs. 1 and S3. Solid black boxes outline collections of correlations between replicate libraries of the same genotype. Dashed black boxes indicate three emergent classes of libraries: (a) low-ribonucleotide data sets, including all RNH201+ samples and rnh201Δ samples with wild type, pol2-M644L, pol3-L612M, and pol1-L868M polymerase variants; (b) leading strand high-ribonucleotide samples, i.e. pol2-M644G rnh201Δ; and (c) lagging strand high-ribonucleotide samples, i.e. pol3-L612G, rnh201Δ and pol1-Y869A rnh201Δ.
Supplementary Table 5. List of confirmed and new origins observed by HydEn-Seq. Listed are 294 origins classified as Confirmed in the OriDB 25 that are also confirmed by HydEn-Seq, plus 72 positions identified by HydEn-Seq that we infer are previously unreported origins. The OriDB lists 98 more confirmed origins that were not observed in the present studies. An example is ARS1013, which is conspicuously absent from Figure 2a because it was not detected by HydEn-Seq.
Supplementary Table 6. RNase H2 subunit A BLAST parameters and hits.
Acknowledgments
We thank Matthew Young and Matthew Longley for helpful comments on the manuscript. This work was supported by the Division of Intramural Research of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (Project Z01 ES065070 to TAK), and by NIH grant 2R01GM052319-16A1 to PAM.
Footnotes
ACCESSION CODES
Sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE62181.
AUTHOR CONTRIBUTIONS
ARC, DJS and TAK designed the experiments, ARC, CDO, JSW, MFC and EPM performed the experiments, ARC, SAL, ABB, DCF, PAM and TAK analyzed the data, TAK wrote the manuscript, and all authors edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests. The authors declare no competing financial interests.
References
- 1.Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–70. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lujan SA, et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lujan SA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Research. 2014;24:1751–64. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larrea AA, et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107:17674–9. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–8. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuffee SR, Smith DJ, Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol Cell. 2013;50:123–35. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanrooij S, Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim Biophys Acta. 2010;1797:1378–88. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Gerhold JM, Aun A, Sedman T, Joers P, Sedman J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol Cell. 2010;39:851–61. doi: 10.1016/j.molcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Reyes A, et al. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013;41:5837–50. doi: 10.1093/nar/gkt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams JS, et al. RNA-DNA damage elicited by ribonucleotides incorporated during DNA replication is leading strand-specific. personal communication.
- 18.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–81. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–6. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–5. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–43. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siow CC, Nieduszynska SR, Muller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–6. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niimi A, et al. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–46. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282:2324–32. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–5. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielinsky AK, Gerbi SA. Chromosomal ARS1 has a single leading strand start site. Mol Cell. 1999;3:477–86. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 30.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–82. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–32. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–7. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase varepsilon. DNA Repair (Amst) 2012;11:649–56. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–4. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–4. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–34. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for Mammalian genome integrity and development. Cell. 2012;149:1008–22. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiller B, et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–26. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clausen AR, Murray MS, Passer AR, Pedersen LC, Kunkel TA. Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc Natl Acad Sci U S A. 2013;110:16802–7. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–7. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–31. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 45.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc Natl Acad Sci U S A. 1973;70:3339–43. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang MY, et al. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 48.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286:31490–500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–35. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 50.Bowmaker M, et al. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–9. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 51.Fuste JM, et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 52.Holt IJ, Reyes A. Human mitochondrial DNA replication. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerritelli SM, et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–15. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 54.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–2. [Google Scholar]
- 56.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Oligonucleotides. Listed are the oligonucleotides used for HydEn-Seq. Bold face indicates indexing. * indicates a phosphorothioate bond. ARC140 contains a 5′-amino group instead of a 5′-OH group, in combination with a C6 linker. This modification reduces formation of ARC140 concatemers during amplification. All oligos are from Integrated DNA technologies (Coralville, IA).
Supplementary Table 2. The 10 yeast strains used for HydEn-Seq as described in the text. The sources of several additional yeast strains subjected to HydEn-Seq but not analyzed in detail are given in Supplementary Table 3.
Supplementary Table 3. Yeast strains and library statistics. Listed are the genotypes and sources of the yeast strains used in this study, as well as relevant information on the HydEn-Seq libraries generated from these strains. a: orientation 1 of the URA3 gene located adjacent to ARS306. b: orientation 2 of the URA3 gene located adjacent to ARS306.
Supplementary Table 4. Correlation coefficients for comparisons among libraries. Library names are as given in Supplementary Table 3. For the purposes of calculating correlations, all data sets are binned in 200 base pair increments. Above the white diagonal, correlations are for raw end counts, on both strands. Below the white diagonal, correlations are for fractions of ends mapping to the top strand, as per Figs. 1 and S3. Solid black boxes outline collections of correlations between replicate libraries of the same genotype. Dashed black boxes indicate three emergent classes of libraries: (a) low-ribonucleotide data sets, including all RNH201+ samples and rnh201Δ samples with wild type, pol2-M644L, pol3-L612M, and pol1-L868M polymerase variants; (b) leading strand high-ribonucleotide samples, i.e. pol2-M644G rnh201Δ; and (c) lagging strand high-ribonucleotide samples, i.e. pol3-L612G, rnh201Δ and pol1-Y869A rnh201Δ.
Supplementary Table 5. List of confirmed and new origins observed by HydEn-Seq. Listed are 294 origins classified as Confirmed in the OriDB 25 that are also confirmed by HydEn-Seq, plus 72 positions identified by HydEn-Seq that we infer are previously unreported origins. The OriDB lists 98 more confirmed origins that were not observed in the present studies. An example is ARS1013, which is conspicuously absent from Figure 2a because it was not detected by HydEn-Seq.
Supplementary Table 6. RNase H2 subunit A BLAST parameters and hits.