Abstract
DNA interstrand cross-links (ICLs) prevent strand separation during DNA replication and transcription and are therefore extremely cytotoxic. In metazoans, a major pathway of ICL repair is coupled to DNA replication and requires the Fanconi anemia pathway. In most current models, collision of a single DNA replication fork with an ICL is sufficient to initiate repair. In contrast, we show here that in Xenopus egg extracts, two DNA replication forks must converge on an ICL to trigger repair. When only one fork reaches the ICL, the replicative CMG helicase fails to unload from the stalled fork, and repair is blocked. Arrival of a second fork, even when substantially delayed, rescues repair. We conclude that ICL repair requires a replication-induced X-shaped DNA structure surrounding the lesion, and we speculate how this requirement helps maintain genomic stability in S phase.
Introduction
DNA interstrand cross-links (ICLs) involve a covalent linkage between the Watson and Crick strands of DNA. Left unrepaired, a small number of ICLs can kill a mammalian cell1. This cytotoxicity is widely exploited for cancer chemotherapy, which employs bifunctional cross-linking agents such as the nitrogen mustards, platinum compounds, and mitomycin C1. It has been proposed that endogenous metabolites such as reactive aldehydes also cause ICLs in vivo2. Failure to repair ICLs and other lesions might be the underlying cause of Fanconi anemia, a rare bone marrow failure and cancer predisposition syndrome3.
In vertebrate cells, a major pathway of ICL repair occurs in the S phase of the cell cycle4. This mechanism requires the Fanconi anemia pathway, structure-specific endonucleases, translesion DNA polymerases, and recombinases2,5. We previously showed that in Xenopus egg extracts, a plasmid containing a site-specific cisplatin ICL (pICL) undergoes replication-coupled ICL repair (Supplementary Fig. 1a)6. In this system, replication initiates in a sequence non-specific manner and two replisomes converge on the ICL. Their leading strands stall ~20–40 nucleotides from the lesion (“–20” position) due to steric hindrance by the CMG (CDC45, MCM2-7, GINS) helicase, which translocates on the leading strand template ahead of DNA polymerase7. BRCA1-BARD1 complex then promotes the dissociation of CMG from the stalled forks8, followed by leading strand extension to within one nucleotide of the ICL (“approach” to “–1” position). Next, ubiquitylated FANCI-FANCD2 binds chromatin and helps recruit the XPF-ERCC1-SLX4 complex9,10. XPF-ERCC1 incises one parental strand (“unhooking”) and allows lesion bypass on the other parental strand by translesion DNA polymerases. Finally, the double-stranded DNA break generated by incisions is repaired by homologous recombination11. In this cell-free system, the cisplatin adduct remains attached to one parental strand.
The use of a small plasmid to model ICL repair in egg extracts inevitably leads to rapid convergence of two DNA replication forks on the lesion. In contrast, in vivo, where the average inter-origin distance is large (~100 kb)12, one replication fork should generally encounter an ICL well before a second fork arrives. Therefore, although convergent forks are generally viewed as being able to trigger ICL repair, it is widely assumed that a single fork is also sufficient2,4,5,13–18 (Supplementary Fig. 1b). In apparent agreement with single fork-induced repair, a psoralen-ICL flanked on one side by a replication roadblock (EBNA1 protein bound to FR repeats) can be repaired19. However, this result is ambiguous because fork arrest by EBNA1 is incomplete20. In addition, cell-free replication of a psoralen-ICL plasmid suggested that a single fork can trigger incisions, but these were not shown to require the Fanconi anemia pathway or to promote repair21.
Here, we set out to examine what happens when only a single fork strikes an ICL. We found that in Xenopus egg extracts, one DNA replication fork is completely inert for ICL repair. Specifically, the CMG complex is not unloaded from a single fork stalled at an ICL, the leading strand fails to approach to the lesion, and no downstream repair events are detected. Importantly, ICL repair is still productive when there is a major delay between the arrival of the first and second forks, as would occur in vivo. Finally, we showed that the order in which two forks strike the ICL does not affect the mechanism of repair. Together with previous results22, our data indicated that formation of an X-shaped structure surrounding an ICL is the essential trigger for ICL repair.
Results
A single fork stalled at an ICL fails to undergo approach
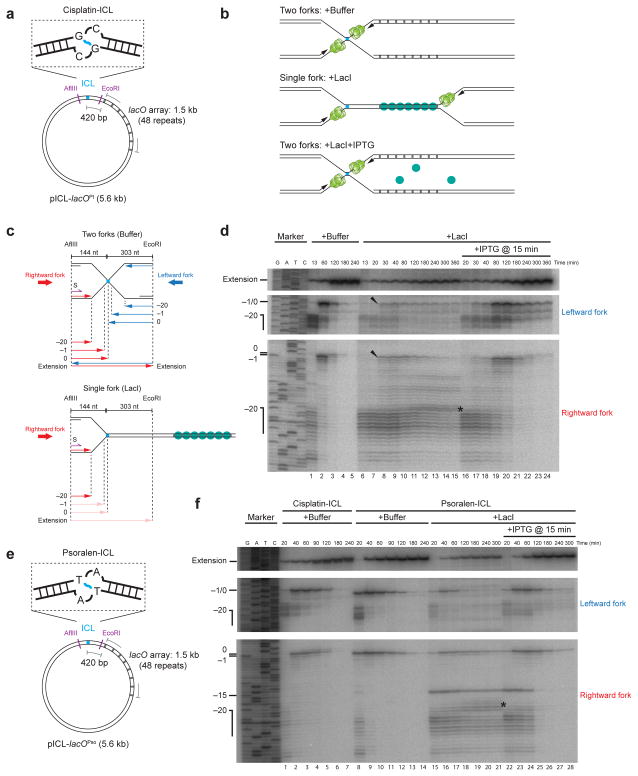
We wanted to directly compare what happens when one or two replication forks collide with an ICL. To this end, we constructed pICL-lacO, in which an array of 48 lac operator (lacO) sites was placed ~420 base pairs to the right of a cisplatin-ICL (Fig. 1a). Binding of LacI to the lacO sites should prevent the leftward replication fork from reaching the lesion23, allowing us to examine what happens when only the rightward fork encounters the ICL (Fig. 1b, middle cartoon). In the absence of an ICL, the LacI array inhibited replication fork progression for at least three hours (Supplementary Fig. 2a and Supplementary Fig. 2b, lanes 5–10)23. When IPTG was added 15 or 75 minutes after the initiation of DNA replication, the stalled replication forks restarted and completed DNA synthesis (Supplementary Fig. 2b, lanes 11–17 and lanes 18–24, respectively), although the rate of replication was reduced, likely due to residual binding of LacI to DNA in the presence of IPTG. Therefore, the LacI array can be used to control access of the leftward fork to the lesion in pICL-lacO.
Figure 1. A single fork stalled at an ICL fails to undergo approach.
(a) Cartoon of the cisplatin containing plasmid, pICL-lacOPt.
(b) Expected outcomes of pICL-lacO replication in the presence of buffer, LacI, and LacI + IPTG.
(c) Schematic illustration of nascent leading strands generated in the experiment shown in (d).
(d) Nascent strand analysis of pICL-lacO during replication-coupled repair, with or without LacI and IPTG, as indicated. Nascent strands, together with the sequencing ladder, were separated on a polyacrylamide gel and visualized by autoradiography. Top panel: extension products. Middle panel: nascent strands of the leftward fork. Bottom panel: nascent strands of the rightward fork. The ladder was generated from extension of primer S (shown in c, purple) on a control plasmid lacking the ICL. Black arrowheads: a small fraction of leading strands approaching to −1. Asterisk (*), background bands described in Supplementary Fig. 3d. Uncropped image of Fig. 1d is presented in Supplementary Data Set 1a.
(e) Cartoon of the psoralen-ICL containing plasmid, pICL-lacOPso.
(f) Nascent strand analysis of pICL-lacOPt (containing the cisplatin-ICL used throughout the paper) and pICL-lacOPso, with or without LacI and IPTG, as indicated. Asterisk (*), background bands described in Supplementary Fig. 3d. The −15 arrest of the leading strands in repair might come from specific stalling of the CMG helicase when it encounters a psoralen-ICL. Uncropped image of Fig. 1f is presented in Supplementary Data Set 1b.
We first investigated whether a single fork could undergo approach, an early event in ICL repair (Supplementary Fig. 1a). pICL-lacO was pre-incubated with buffer or LacI and then replicated in egg extract containing [α-32P]dATP to radiolabel nascent DNA strands. Replication intermediates were digested with AflIII and EcoRI and separated on a denaturing polyacrylamide gel to monitor nascent strand synthesis at nucleotide resolution (Fig. 1c). Without LacI, both the leftward and rightward leading strands reached the −20 position, after which they approached to the −1 position, before undergoing extension past the lesion (Fig. 1d, lanes 1–5)6. As expected, in the presence of LacI, arrival of the leftward leading strands was strongly reduced (~74% on average) (Fig. 1d, middle panel, compare lanes 1 and 6). Strikingly, although LacI did not affect arrival of rightward leading strands at the −20 position (Fig. 1d, bottom panel, compare lanes 1 and 6), their approach to −1 was dramatically inhibited (Fig. 1d, bottom panel, lanes 6–15). Specifically, ~70% of leftward strands failed to approach by 120 min, and more than 50% remained stalled at −20 for 6 hours (Supplementary Fig. 3a). Consistent with this inhibition of approach, there was on average a 70% reduction in extension products (Fig. 1d, lanes 6–15, top panel). While a fraction of leftward and rightward leading strands did approach to −1 (Fig. 1d, black arrowheads, Supplemental Fig 3a), this can be explained by the arrival of 26% of leftward forks at the lesion (Figure 1d, middle panel, lane 6), resulting in fork convergence. We speculate that residual arrival of leftward forks is due to occasional origin firing between the ICL and the LacI array, or incomplete inhibition of leftward fork progression by the LacI array, especially at late time points (Supplementary Fig. 2b, lanes 9, 10). Importantly, when we added LacI immediately after most forks had converged, leading strands underwent normal approach and extension (Supplementary Figs. 3b and 3c). Therefore, once forks have converged at the ICL, the LacI array does not inhibit repair. When a plasmid containing a lacO array (placO) was mixed with pICL in the presence of LacI, approach on pICL was unaffected, demonstrating that the LacI array does not inhibit approach in trans (Supplementary Fig. 3d). As shown in Fig. 1f, approach was also inhibited when a single fork encountered a psoralen-ICL. Our results show that in Xenopus egg extracts, a single fork stalled at an ICL is not able to undergo approach, the first event of ICL repair.
A single fork stalled at an ICL remains competent for repair
In vivo, one fork will usually strike an ICL well before a second fork arrives. To mimic this situation in vitro, we allowed one fork to strike an ICL in the presence of LacI. We then disassembled the LacI barrier at different times using IPTG, and measured approach. As shown in Figs. 1d, 1f and Supplementary Fig. 4, when IPTG was added 15 or 75 minutes after replication initiation, approach and extension were restored. Thus, a single, stalled fork remains competent for ICL repair for extended periods of time. We conclude that ICL repair is productive even when there is an extensive delay between the arrival of the first and second forks, as would normally occur in vivo.
CMG is not evicted from a single fork stalled at an ICL
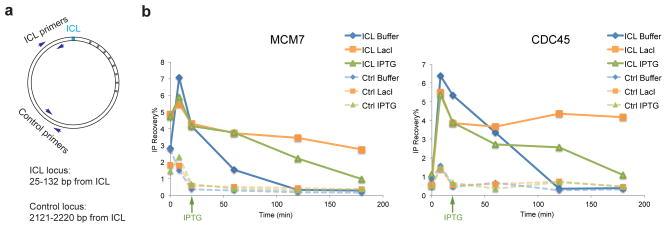
We previously showed that leading strand approach requires dissociation of CMG from the stalled replisomes7,8. We therefore postulated that the failure of a single fork to undergo approach might be caused by defective CMG dissociation. To test this idea, we examined CMG localization at the ICL locus and a control locus distal to the ICL using chromatin immunoprecipitation (ChIP) (Fig. 2a). In the presence of buffer, the MCM7 and CDC45 subunits of the CMG helicase accumulated at the ICL and then dissociated (Fig. 2b and Supplementary Fig. 5a, solid blue lines)7. In the presence of LacI, only ~30% of MCM7 and CDC45 dissociated from the ICL (consistent with residual fork convergence), while the majority persisted (Fig. 2b and Supplementary Fig. 5a, solid orange lines), and only dissociated upon IPTG addition (Fig. 2b and Supplementary Fig. 5a, solid green lines). Dissociation of CMG from the control locus was unaffected by LacI (Fig. 2b and Supplementary Fig. 5a, dashed lines). Our data indicate that CMG does not dissociate from a single fork that has stalled at an ICL, accounting for the failure in approach.
Figure 2. CMG is not evicted from a single fork stalled at an ICL.
(a) Schematic of primers used in chromatin immunoprecipitation (ChIP).
(b) MCM7 and CDC45 ChIP anaylsis at different time points during repair. pICL-lacO was replicated with or without LacI, and IPTG was added immediately before the 20 minute time point, as indicated (green arrow). A repetition of this experiment is shown in Supplementary Fig. 5a.
A single, stalled fork is not incised
We next addressed whether a single fork could trigger ICL unhooking, the signature event of ICL repair. To address this question, pICL-lacO was replicated with or without LacI in extract containing [α-32P]dATP, and replication intermediates were separated on a native agarose gel and visualized by autoradiography. Without LacI, replication fork convergence gave rise to a discrete “Figure 8” structure (Fig. 3a and Fig. 3b, blue arrowhead) that later disappeared as a result of FANCI-FANCD2-dependent incisions, and ultimately accumulated as supercoiled plasmid (Fig. 3b, lanes 1–6)6,9. In the presence of LacI, we observed the expected “theta” intermediate due to fork stalling (Fig. 3a and Fig. 3b, green arrowhead). However, the theta structure persisted for at least three hours (Fig. 3b, lanes 7–12). Replication of pQuant, a plasmid lacking an ICL or lacO sites was not affected by LacI (Fig. 3b). These results indicate that a single fork is not subject to incisions.
Figure 3. A single, stalled fork does not undergo incisions.
(a) Schematic of repair intermediates expected in panel (b).
(b) Replication intermediates of pICL-lacO separated on a native agarose gel. pICL-lacO and an internal control plasmid lacking the lacO array (pQuant) were pre-incubated with buffer or LacI and replicated in the presence of [α-32P]dATP. IPTG was added as indicated. Sc: supercoiled. Oc: open circular. Blue arrowhead: “Figure 8” DNA structure. Green arrowhead: “Theta” DNA structure.
(c) Schematic of incision assay. Before dual incisions, single cutting with BlpI yields X-shaped products (blue strands, upper cartoon), whereas after dual incisions, BlpI digestion yields linear molecules (blue strands, lower cartoon).
(d) Incision assay. pICL-lacO was replicated with or without LacI and IPTG, as indicated. The repair intermediates were digested with BlpI, separated on an alkaline (denaturing) gel, and visualized by Southern blotting to detect parental strands. Unreplicated pICL-lacO was used to generate size markers for the X-shaped structure and linear structure, which came from a small fraction of uncrosslinked background plasmids in the pICL-lacO preparations. Uncropped image of Fig. 3d is presented in Supplementary Data Set 1c.
(e) XPF and SLX4 ChIP analysis. pICL-lacO and pQuant were replicated with or without LacI, and IPTG was added immediately before the 20 minute time point where indicated (green arrow). At different times, samples were withdrawn for XPF and SLX4 ChIP using primer pairs for the ICL locus (Fig. 2a) or pQuant (Ctrl). A repetition of this experiment is shown in Supplementary Fig. 5c.
To measure more directly whether parental strands were incised, replication intermediates of pICL-lacO were linearized with BlpI, separated on an alkaline gel, and probed by Southern blotting to visualize parental strands. In the absence of incisions, a large, X-shaped molecule should be visible (Fig. 3c, upper cartoon, blue strands), whereas after dual incisions in one parental strand, the X-shaped species should be converted to linear forms (Fig. 3c, lower cartoon, blue strand). Without LacI, the X-shaped structure declined and linear molecules appeared (Fig. 3d, lanes 2–5), as expected9. In contrast, in the presence of LacI, the reduction of X-shaped molecules was greatly inhibited (~70%), and the accumulation of linear molecules was attenuated (Fig. 3d, lanes 6–12; Supplementary Fig. 5d). Incisions were restored by the addition of IPTG (Fig. 3d, lanes 13–19). We conclude that a single fork stalled at an ICL is inefficiently incised.
Next, we addressed what leads to the observed incision defect. Ubiquitylated FANCI-FANCD2 binds to chromatin and promotes recruitment of the XPF-ERCC1-SLX4 complex, which unhooks the ICL9,10. LacI moderately reduced FANCD2 binding to the ICL as measured by ChIP (Supplementary Fig. 5b). In contrast, LacI caused a more substantial reduction in the recruitment of XPF and SLX4 to the ICL, and this effect was reversed by IPTG (Fig. 3e, Supplementary Fig. 5c). Some of the SLX4 and XPF recruitment in the presence of LacI might be due to residual fork convergence in this condition. These results suggest that although a single stalled fork containing CMG can recruit FANCI-FANCD2, fork convergence is necessary for efficient recruitment of XPF-ERCC1-SLX4, either because binding requires CMG dissociation or the presence of an X-shaped structure.
The order of replisome arrival does not affect repair
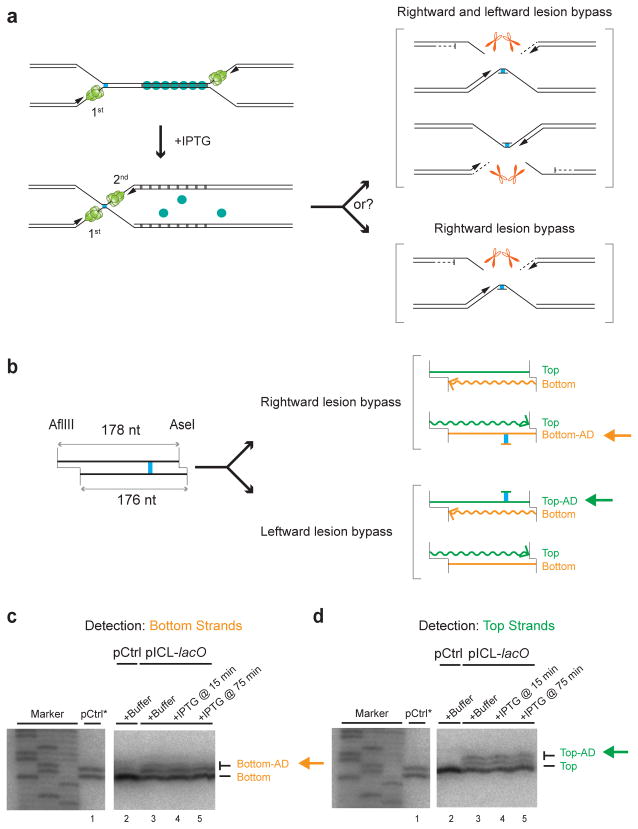
The convergence of two forks on an ICL creates an apparently symmetrical structure (Supplementary Fig. 1a). However, one parental strand subsequently undergoes incision while the other acts as the template for lesion bypass (Supplementary Fig. 1a)6. We asked whether the order in which two forks arrive at an ICL dictates which parental strand is incised and which is used for lesion bypass. To address this question, the leftward fork was stalled with LacI for 15 or 75 minutes, and then released with IPTG, so that the rightward fork reached the ICL first (Fig. 4a). If the rightward leading strand is now used exclusively for lesion bypass, the cisplatin adduct that persists after ICL repair6 should be attached primarily to the bottom strand (Fig. 4a, bottom bracket); if the two leading strands are still used for bypass with equal probability, the adduct should be detected equally on both parental strands (Fig. 4a, top bracket). We digested the final repair products with AflIII and AseI, such that the top and bottom parental strands differed in size by 2 nt (Fig. 4b). The DNA was then separated on a sequencing gel, and the top or bottom parental strands were visualized by strand-specific Southern blotting. Importantly, when arrival of the leftward fork was delayed with LacI, both the bottom (Fig. 4c, lanes 4, 5) and top (Fig. 4d, lanes 4, 5) parental strands retained adducts. Furthermore, the ratio between adducted and un-adducted strands was unaffected by LacI (Figs. 4c and 4d, compare lane 3 with lanes 4 and 5). We conclude that the order in which the two replisomes arrive at the ICL does not affect which parental strand is incised and which is used as the template for lesion bypass. This result suggests that a single fork remains wholly uncommitted to ICL repair until a second fork arrives. How the strand used for lesion bypass is ultimately chosen remains unclear.
Figure 4. The order of replisome arrival at an ICL does not determine which leading strand undergoes lesion bypass.
(a) Scheme to determine whether the order in which the two forks arrive at an ICL dictates which parental strand is used as the lesion bypass template.
(b) Schematic depiction of final repair products after AflIII and AseI digestion, depending on which leading strand undergoes lesion bypass. Note that AflIII and AseI generate different sized overhangs, allowing us to differentiate top (178 nt) and bottom (176 nt) strands. Top-AD or Bottom-AD: top or bottom strand containing an adduct.
(c, d) Strand-specific Southern blotting to detect the adducts. pCtrl or pICL-lacO was replicated in the presence of buffer or LacI, and IPTG was added at the indicated times. After 6 hours, repair products were digested with AflIII and AseI, separated on a sequencing gel, and analyzed with strand-specific Southern blotting to visualize the bottom (c) or top (d) strands. To generate size markers for the top (178 nt) and bottom (176 nt) strands (lane 1), pCtrl was replicated in the presence of [α-32P]dATP (pCtrl*), and was analyzed on the same sequencing gel as the strand-specific Southern after AflIII and AseI digestion. The fact that no top (c, lane 2) or bottom strand (d, lane 2) was detected in Southern blotting of pCtrl established the strand-specificity of the blotting protocol. Uncropped images of Figs. 4c and 4d are presented in Supplementary Data Set 1d.
Discussion
In this paper, we addressed whether one or two DNA replication forks are necessary to trigger ICL repair. Due to the challenge of engineering site-specific ICLs on mammalian chromosomes and the difficulty of manipulating the abundance of DNA replication forks in cells, this question has not been addressed in vivo. Instead, we used a replication fork barrier to control the access of DNA replication forks to an ICL in Xenopus egg extracts, which recapitulate physiological ICL repair2,5,6,9–11,24. In contrast to most current models, we found that two DNA replication forks must converge on an ICL to trigger repair. Strikingly, a single fork does not support even the first step in repair, CMG dissociation, which is presumably critical to initiate lesion bypass and to expose the ICL to the incision machinery.
Implications for ICL repair in cells
Our results raise the question of how two forks arrive at an ICL in vivo. Given an average inter-origin distance of 100 kb12, coordinated firing of adjacent origins25, and an average fork-rate of 1.5 kb/minute26, the maximum time delay between the arrival of the first and second forks at most ICLs should be ~60 minutes in vivo. We have shown that a single fork stalled at an ICL does not collapse and remains competent for repair for at least 60 minutes. Therefore, in vivo, ICL repair could rely on the convergence of forks from adjacent origins. In some cases, a second fork might be delivered more rapidly due to firing of a nearby dormant origin27. Interestingly, cisplatin treatment causes selective loss of telomeres28. This observation is consistent with our model because fork convergence cannot occur when an ICL is located beyond the last origin of replication at the chromosome end.
Recently, the Seidman group reported that a single fork can bypass a psoralen-ICL without repairing it, dependent on the DNA translocase FANCM (“traverse”)22. This observation implies that during traverse, CMG or another DNA helicase bypasses ICLs to allow fork progression. We have not observed traverse in Xenopus egg extracts, even on a psoralen-ICL (see discussion in Supplementary Fig. 6, legend). This failure is not due to a lack of FANCM, which is present in egg extracts29 (R. Amunugama and J.C.W., unpublished results), but perhaps because some other activity is absent in early embryos. Importantly, the lagging strand of a traversed fork is equivalent to the leading strand of a converging fork (Supplementary Fig. 6, compare purple and green strands). Thus, both traverse and fork convergence generate an X-shaped DNA structure surrounding the ICL that we propose is the essential trigger for ICL repair. The requirement for this structure helps explain why both 5′ and 3′ directed flap endonucleases have been implicated in ICL repair4,24.
Relationship of FANCI-FANCD2 recruitment and CMG unloading
An important question concerns the interdependence of CMG unloading with other, early events in ICL repair. We recently showed that BRCA1-BARD1 is required for CMG unloading and FANCD2 recruitment at ICLs8, raising the possibility that CMG eviction might be needed to make room for FANCI-FANCD2 near the lesion. Disfavoring this idea, we also showed that inhibition of approach with aphidicolin impairs CMG unloading while having little or no effect on BRCA1-BARD1 or FANCD2 recruitment8, suggesting that CMG removal is not required for efficient FANCD2 loading. This conclusion is further supported by our present finding that FANCD2 is recruited to single forks, which retain CMG. Although we cannot rule out that CMG obstructs FANCI-FANCD2 binding to the ICL itself, our results argue that it does not prevent FANCI-FANCD2 recruitment in the general vicinity of the lesion.
Implications for genome stability
What is the advantage of coupling ICL repair to fork convergence? During an unperturbed S phase, DNA replication forks are expected to stall transiently at DNA sequences and chromatin structures that are difficult to replicate30,31. If CMG unloading were possible from single forks, the helicase might sometimes be unloaded from transiently stalled replisomes. Given that there is no known pathway to reload the CMG complex in S phase of metazoans, the inadvertent dissociation of CMG is predicted to cause fork collapse, incomplete DNA synthesis, and genome instability. Making CMG unloading absolutely dependent on fork convergence avoids this problem since the helicase will only be lost when replication is locally completed. Fork traverse past an ICL also avoids this problem because CMG or another DNA helicase will continue unwinding on the other side of the ICL to allow completion of DNA synthesis. We recently showed that a single fork is sufficient to trigger repair of a DNA protein cross-link (DPC), indicating that some helicase-blocking lesions can be repaired in the absence of fork convergence and CMG unloading23. In this case, the DPC is highly exposed and thus amenable to destruction even in the presence of CMG, allowing fork bypass. In conclusion, our work strongly suggests that the formation of an X-shaped structure surrounding an ICL, either by fork convergence or fork traverse, is essential to initiate ICL repair. The unexpected failure of single, stalled forks to trigger ICL repair is probably essential to avoid inadvertent fork collapse in S phase.
Online Methods
LacI protein purification
The LacI-Biotin protein was purified according to a protocol from Kenneth Marians’ laboratory (personal communication). Briefly, NEB T7 express cells were co-transformed with pBirAcm, which contains an IPTG inducible birA gene for overexpression of the biotin ligase (Avidity, Denver, CO), and pET11a[lacI::avi] (a gift from Kenneth Marians’ laboratory), containing the lacI gene encoding an AviTag at the C terminus. Transformed cells were grown on LB plates containing ampicillin and chloramphenicol, and single colonies were picked and amplified in LB containing the same antibiotics. Cells were lysed in lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 5 mM EDTA, 10% sucrose, 0.2 mg/ml Lysozyme, 0.1% Brij 58, cOmplete protease inhibitor (Roche, Nutley, NJ)), and chromatin-bound LacI was extracted from the pellets by sonication. Nucleic acids were removed from the extracted fraction by adding polymin P (final concentration 0.05%), and LacI-Biotin was precipitated by adding ammonium sulphate to a final concentration of 37%. The LacI-Biotin pellet was then resuspended in 50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, cOmplete protease inhibitor (Roche, Nutley, NJ), applied to a soft-link avidin column and eluted with biotin-containing buffer (50 mM Tris pH 7.5, 1 mM EDTA, 100 mM NaCl, 1 mM DTT, 5 mM biotin). LacI-Biotin was dialyzed against 50 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl, 1 mM DTT, 38% glycerol overnight, frozen in liquid nitrogen, and stored at −80 °C. A more detailed LacI-Biotin purification protocol is available upon request.
Preparation of pICL-lacO
To make the backbone of pICL-lacO, we first engineered an EcoRI site 295 nt downstream of the second BbsI site in the parental plasmid of pICL32. We then cloned the lacO array (48 lacO repeats) between the EcoRI and SacI sites. The parental plasmid was then amplified and digested with BbsI. To make the cisplatin pICL-lacO, a 20 nucleotide cisplatin-ICL duplex was prepared and ligated into the tandem BbsI sites of the backbone plasmid32. To make the trioxsalen (“psoralen” in the main text) pICL-lacO, complementary primers containing only one thymidine were annealed in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH pH 7.4, and 2 mM magnesium acetate) at a concentration of 50 μM each. DNA-trioxsalen crosslinking was carried out using 2.6 μM annealed DNA in crosslinking buffer (10 mM Tris pH 7.5, 1 mM EDTA, 50 mM NaCl) and 87.6 μM trioxsalen. The reaction was exposed to 365 nm UVA light for six periods of 15 minutes each, at a power of 4 mW/cm2. After every cycle fresh trioxsalen was added to 87.6 μM. Crosslinked DNA was purified from a 20% polyacrylamide 8 M urea gel. The purified DNA was end-labeled with [γ-32P]ATP and run on a gel, which revealed that > 99.9% of the DNA contained a DNA interstrand cross-link. The purified crosslinked duplex was then ligated into the tandem BbsI sites of its corresponding backbone plasmid.
Cisplatin crosslinked duplex (with the cross-link between the two highlighted Gs):
5′-CCCTCTTCCGCTCTTCTTTC-3′
5′-GCACGAAAGAAGAGCGGAAG-3′
Psoralen crosslinked duplex (with the cross-link between the two highlighted Ts):
5′-CCCCGGGGCTAGCC-3′
5′-GCACGGCTAGCCCC-3′
The generation of the lacO array (48 lacO repeats) will be presented in detail elsewhere (J.M.D. and J.C.W. in preparation). The sequences of the plasmids and primers used for mutagenesis are available upon request.
Xenopus egg extracts and DNA replication
Xenopus egg extracts were prepared as described33. For DNA replication, plasmids were first incubated in a high-speed supernatant (HSS) of egg cytoplasm (final concentration of 7.5 ng DNA/μL extract) for 20 minutes at room temperature to license the DNA, followed by the addition of two volumes of nucleoplasmic egg extract (NPE) to initiate replication. For replication with LacI, plasmid (75 ng/μL) was incubated with an equal volume of 40 μM LacI for 30 minutes before HSS addition. IPTG was added at a final concentration of 10 mM in egg extracts. In Supplementary Fig. 3c, pICL-lacO was incubated in HSS without pre-binding of LacI, and LacI was added at the indicated times relative to NPE addition. In all figures, the 0 minute time point refers to the time of NPE addition. For DNA labeling, reactions were supplemented with [α-32P]dATP, which is incorporated into nascent strands during replication. Where indicated, pQuant, an undamaged plasmid lacking the lacO array was included (0.375 ng/μL final concentration in HSS) and served as an internal replication standard. For Fig. 3a, replication was stopped by adding 0.5 μl of each reaction to 10 μl of replication stop solution A (5% SDS, 80 mM Tris pH 8.0, 0.13% phosphoric acid, 10% Ficoll) supplemented with 1 μl Proteinase K (20 mg/ml) (Roche, Nutley, NJ). Samples were incubated for 1 hour at 37 °C prior to separation by 0.8% native agarose gel electrophoresis. DNA samples were then detected using a phosphorimager33. For all other applications except ChIP, replication reactions were stopped in 10 volumes of replication stop solution B (50 mM Tris pH 7.5, 0.5% SDS, 25 mM EDTA), and replication intermediates were purified as previously described6. All experiments were performed at least twice, and a representative result is shown.
ChIP and quantitative real-time PCR
ChIP was performed essentially as described11. Briefly, 3 μL reaction samples were crosslinked in 47 μL of 1% formaldehyde in ELB (10 mM HEPES-KOH pH 7.7, 2.5 mM MgCl2, 50 mM KCl, 250 mM sucrose) for 10 minutes at room temperature. Crosslinking was stopped by the addition of 5 μL 1.25 M glycine followed by passage through a Micro Bio-Spin 6 Chromatography column (Bio-Rad, Hercules, CA, USA) to remove excess formaldehyde. The flow-through was diluted to 500 μL with sonication buffer (20 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 5 μg/mL Aprotinin+Leupeptin, 2 mM PMSF) and subjected to sonication, yielding DNA fragments ~300–500 bp in size.
Following immunoprecipitation (IP), formaldehyde cross-links were reversed and DNA was purified for analysis by quantitative real-time PCR. The recovery rate was determined by the amount of IP samples relative to the input samples. For quantitative PCR, where three technical replicates were performed for each sample, replicates that deviated from the average value by greater than 0.3 standard deviations were discarded. FANCD26[Rabbit 20019], CDC4534[Rabbit 534], MCM735[Rabbit 456], XPF10[Rabbit 20682; Rabbit 20683], and SLX410[Rabbit 24153; Rabbit 24256] polyclonal antibodies were previously described and validated. For MCM7, SLX4, and FANCD2 IP, antibodies were purified from serum using Protein A Sepharose beads (GE Healthcare, Piscataway, NJ, USA) and 5 μg of IgG was used for IP per sample. For CDC45 and XPF, 1 μL of serum was used directly for immunoprecipitation.
PCR primer pairs used in ChIP:
ICL primer pair:
5′-AGCCAGATTTTTCCTCCTCTC-3′
5′-CATGCATTGGTTCTGCACTT-3′;
Control primer pair on pICL-lacO:
5′-AACGCCAATAGGGACTTTCC-3′
5′-GGGCGTACTTGGCATATGAT-3′;
Control primer pair on pQuant:
5′-TACAAATGTACGGCCAGCAA-3′
5′-GAGTATGAGGGAAGCGGTGA-3′.
Nascent strand analysis
Nascent strand analysis was performed as described6. Briefly, pICL-lacO was replicated in the presence of [α-32P]dATP and purified repair intermediates were digested with AflIII and EcoRI, followed by addition of 0.5 volumes Gel loading Buffer II (Denaturing PAGE) (Life Technologies, Grand Island, NY, USA). For the mixing experiment of pICL and placO in Supplementary Fig. 3d, the repair intermediates were digested with only AflIII, which cuts on both sides of the ICL on pICL. Radiolabeled nascent strands were then separated on a 7% denaturing polyacrylamide gel, transferred to filter paper, dried, and visualized using a phosphorimager. Sequencing gel markers were generated with primer S (5′-CATGTTTTACTAGCCAGATTTTTCCTCCTCTCCTG-3′) using the Cycle Sequencing kit (USB Corporation, Cleveland, OH, USA).
Incision assay
The incision assay was performed as described before36. Briefly, pICL-lacO was replicated and digested with BlpI, which cuts the plasmid once. In parallel, unreplicated pICL-lacO was digested with BlpI to serve as size markers for the X-shaped structure and linear DNA, which came from a small fraction of uncrosslinked plasmids present in our pICL-lacO preparations. After digestion, the DNA was seperated on a 1% agarose gel under denaturing conditions (50 mM NaOH, 1 mM EDTA) for 18 hours at 0.85 Volts/cm. Subsequently, Southern blotting was performed by capillary transfer in transfer buffer (1.5 M NaCl, 0.4 M NaOH) onto a nylon membrane (Hybond-N+, Amersham). After transfer, the membrane was washed in 4X SSC for 5 minutes, and UV irradiated to crosslink the DNA to the membrane. Prehybridization was performed with 25 ml of hybridization buffer (4X SSC, 2% SDS, 1X Blocking reagent (Roche, Nutley, NJ), 0.1 mg/ml Salmon sperm DNA (Life Technologies, Grand Island, NY, USA)) for 30 minutes at 45 °C. Hybridization was carried out overnight with 25 μl of probe prepared by Roche random labeling kit (Roche, Nutley, NJ). After overnight hybridization, the membrane was washed 4 times with 0.5X SSC, 0.25% SDS for 15 minutes at 45 °C. The dried membrane was exposed to a phosphorimager screen.
Strand-specific Southern blot
Strand-specific Southern blot was performed as described6. Briefly, AflIII and AseI digested samples were separated on a 7% polyacrylamide gel and transferred to a nylone membrane (Hybond-N+, Amersham). After transfer, the membrane was rinsed in 4X SSC for 5 minutes, and UV irradiated to crosslink the DNA to the membrane. The membrane was then pre-hybridized with 25 ml hybridization buffer (ULTRAhyb from Ambion) for at least 3 hours at 42 °C. Strand-specific probes generated by a PCR based primer extension reaction6, were added to the hybridization buffer and incubated with the membrane at 42 °C overnight. After overnight hybridization, the membrane was washed 2 times with 2X SSC, 0.1% SDS for 5 minutes at 42 °C. The dried membrane was exposed to a phosphorimager screen.
Uncropped images of gels and autoradiographs used in this study can be found in Supplementary Data Set 1.
Supplementary Material
Acknowledgments
We thank S. Elledge, L. Zou, A. Smogorzewska, P. Knipscheer, and the members of the Walter laboratory for feedback on the manuscript. M.B. was supported by Human Frontiers Science Program long-term fellowship LT000773/2010-L and European Molecular Biology Organization long-term fellowship (ALTF 742-2009). A.M. was supported by Natural Sciences and Engineering Research Council of Canada scholarship. M.A.C. was supported by UK Royal Society grant UF100717 and the Fell Fund grant 103/789. J.C.W. was supported by US National Institutes of Health grants GM62267 and HL098316. J.C.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
J.M.D. generated the lacO array (48 lacO repeats) and validated its use as a replication fork barrier; M.B. and J.Z. prepared pICL-lacOPt; A.M. and M.A.C. prepared psoralen crosslinked oligonucleotides; J.Z. and J.C.W. designed and analyzed the experiments; J.Z. performed all the experiments; J.Z. and J.C.W. prepared the manuscript.
References
- 1.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 2.Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Med. 2013;3:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Räschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 Promotes Unloading of the CMG Helicase from a Stalled DNA Replication Fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein Douwel D, et al. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long DT, Räschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 13.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 15.Williams HL, Gottesman ME, Gautier J. The differences between ICL repair during and outside of S phase. Trends Biochem Sci. 2013;38:386–393. doi: 10.1016/j.tibs.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunting SF, Nussenzweig A. Dangerous liaisons: Fanconi anemia and toxic nonhomologous end joining in DNA crosslink repair. Mol Cell. 2010;39:164–166. doi: 10.1016/j.molcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legerski RJ. Repair of DNA interstrand cross-links during S phase of the mammalian cell cycle. Environ Mol Mutagen. 2010;51:540–551. doi: 10.1002/em.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanishi K, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis NA, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014 doi: 10.1038/nature13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Breton C, Hennion M, Arimondo PB, Hyrien O. Replication-fork stalling and processing at a single psoralen interstrand crosslink in Xenopus egg extracts. PLoS ONE. 2011;6:e18554. doi: 10.1371/journal.pone.0018554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-Protein Crosslink by Replication-Coupled Proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 25.Leonard AC, Méchali M. DNA replication origins. Cold Spring Harb Perspect Biol. 2013;5:a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duderstadt KE, Reyes-Lamothe R, van Oijen AM, Sherratt DJ. Replication-fork dynamics. Cold Spring Harb Perspect Biol. 2014;6:a010157–a010157. doi: 10.1101/cshperspect.a010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishibashi T, Lippard SJ. Telomere loss in cells treated with cisplatin. Proc Natl Acad Sci USA. 1998;95:4219–4223. doi: 10.1073/pnas.95.8.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobeck A, Stone S, Landais I, de Graaf B, Hoatlin ME. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem. 2009;284:25560–25568. doi: 10.1074/jbc.M109.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 31.León-Ortiz AM, Svendsen J, Boulton SJ. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair (Amst) 2014;19:152–162. doi: 10.1016/j.dnarep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Enoiu M, Ho TV, Long DT, Walter JC, Schärer OD. Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol Biol. 2012;920:203–219. doi: 10.1007/978-1-61779-998-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- 34.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 35.Fang F, Newport JW. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J Cell Sci. 1993;106( Pt 3):983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- 36.Knipscheer P, Räschle M, Schärer OD, Walter JC. Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol Biol. 2012;920:221–243. doi: 10.1007/978-1-61779-998-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.