Abstract
Microhomology-mediated end-joining (MMEJ) is an error-prone alternative double-strand break repair pathway that utilizes sequence microhomology to recombine broken DNA. Although MMEJ is implicated in cancer development, the mechanism of this pathway is unknown. We demonstrate that purified human DNA polymerase θ (Polθ) performs MMEJ of DNA containing 3’ single-strand DNA overhangs with two or more base-pairs of homology, including DNA modeled after telomeres, and show that MMEJ is dependent on Polθ in human cells. Our data support a mechanism whereby Polθ facilitates end-joining and microhomology annealing then utilizes the opposing overhang as a template in trans which stabilizes the DNA synapse. Polθ exhibits a preference for DNA containing a 5’-terminal phosphate, similar to polymerases involved in non-homologous end-joining. Lastly, we identify a conserved loop domain that is essential for MMEJ and higher-order structures of Polθ which likely promote DNA synapse formation.
Keywords: Microhomology-mediated end-joining, alternative end-joining, double-strand break, double-strand break repair, DNA polymerase
Genome instability in the form of chromosome breaks, deletions and rearrangements is a hallmark of cancer cells and a driver of tumorigenesis. Mounting evidence indicates that an error-prone alternative form of double-strand break (DSB) repair called microhomology-mediated end-joining (MMEJ) promotes inter- and intra-chromosome rearrangements associated with DNA deletions by utilizing sequence microhomology to recombine broken DNA ends1–6. MMEJ has been documented in various eukaryotic organisms, and genetic data demonstrate that this process is distinct from the classical non-homologous end-joining (NHEJ) pathway1,2,4. For example, MMEJ functions in a Ku and Ligase IV independent manner and is therefore referred to as an alternative end-joining (alt-EJ) pathway2,4. Considering that the majority of Ku and Ligase IV independent end-joining occurs via a microhomology-mediated mechanism, MMEJ appears to be the major form of alt-EJ2,7,8. Although the genetic requirements for MMEJ have not fully been identified, studies in mammalian cells demonstrate that MMEJ is promoted by PARP-1, Ligase III, CtIP and Mre111,3,4,9–12.
Recent evidence indicates that MMEJ is induced during S-phase and G2 and functions in response to replicative stress, albeit at substantially lower levels than homology directed repair (HDR) which is highly accurate and therefore important for preserving genome integrity3. Intriguingly, MMEJ and HDR utilize the same initial DNA resection machinery which includes Mre11 and CtIP and facilitates the formation of 3’ single-strand DNA (ssDNA) overhangs at DSBs3. MMEJ, however, only requires limited resection, whereas HDR depends on extensive resection which is performed by additional factors (i.e. Exonuclease I, Bloom’s helicase, Dna2)3. Importantly, MMEJ utilizes sequence microhomology exposed by limited DNA resection to join DNA ends or stabilize spontaneously annealed end joining intermediates1,2. This mechanism is in contrast to NHEJ which does not require DNA resection due to its ability to join blunt ended DNA and DNA containing short overhangs with little or no homology4. Recent comprehensive studies in human cells demonstrate that the majority of chromosome rearrangements formed by NHEJ and MMEJ contain 0–2 base-pairs (bp) and 2–6 bp of microhomology, respectively, at their junctions7,8. These and other studies demonstrate that MMEJ typically results in relatively large DNA deletions (~30–200 bp), whereas deletions due to NHEJ are substantially shorter in length (<10 bp)7,8,13.
Although MMEJ occurs infrequently in mammalian cells, it is implicated in multiple processes involving chromosome rearrangements. For example, seminal studies showed that MMEJ promotes class switch recombination in NHEJ deficient B cells6. More recent studies demonstrate that MMEJ promotes VDJ recombination in NHEJ proficient B cells harboring mutations in RAG recombination genes5. Lastly, studies show that MMEJ promotes error-prone replication repair and telomere fusions1,3,8.
Although the mechanism of MMEJ is unknown, genetic studies in invertebrates suggest a central role for the atypical A-family DNA polymerase theta (Polθ)13–15. For example, studies in Drosophila indicate that Polθ promotes MMEJ of DSBs induced by a sequence specific endonuclease, whereas in C. elegans polq-1 was shown to be required for MMEJ in response to replication fork collapse at G-quadruplexes13–15. Recent genetic studies in mice also indicate the involvement of Polθ in MMEJ. For example, Polq was shown to promote class-switch recombination and confer resistance to DSB inducing agents16.
The Polθ gene is highly unusual in that it encodes for a N-terminal helicase like domain, a large central domain, and a C-terminal polymerase domain17,18. Although the functions of the helicase and central domains are unknown, the polymerase domain encoded by human POLQ—referred to herein as Polθ—has been characterized as a highly promiscuous enzyme18–20. For example, unlike most polymerases, Polθ promotes extension of ssDNA and partial ssDNA (pssDNA) substrates19. Polθ also exhibits low-fidelity DNA synthesis, translesion synthesis, lyase activity, and is implicated in interstrand crosslink repair, base excision repair, and DNA end-joining13,14,16–18,20–22.Thus, although Polθ is among the A-family of polymerases (i.e. Klenow fragment, Thermus aquaticus (Taq) Pol, bacteriophage T7 Pol), which normally perform high-fidelity DNA synthesis, it exhibits similar characteristics as Y- and X-family polymerases whose activities include translesion synthesis and NHEJ, respectively. Here, we set out to examine the ability of Polθ to promote MMEJ in vitro and investigate whether the unusual characteristics of the polymerase contribute to this activity.
RESULTS
Human Polθ promotes MMEJ in vitro and in vivo
We examined whether Polθ promotes end-joining of substrates modeled after partially resected DSBs (pssDNA) containing variable microhomology lengths (Fig. 1a,b). MMEJ was performed by incubating Polθ with radio-labeled pssDNA in the presence of deoxy-ribonucleotides (dNTPs) for 30 min, then terminating the reaction by the addition of EDTA and proteinase K which degrades the polymerase. MMEJ of pssDNA is indicated by a product approximately twice the size of the substrate in a non-denaturing gel which is indicated by a black asterisk. Polθ converted pssDNA-6 into a double-size product (Fig. 1c), indicating MMEJ. Polθ similarly promoted MMEJ of pssDNA-4 (Fig. 1c), but was less efficient in joining pssDNA-2 (Fig. 1c), and failed to join pssDNA-0 which lacks microhomology (Fig. 1c). Polθ completed MMEJ of pssDNA-4 in less than 20 min, but only joined a fraction of pssDNA-2 substrates after 60 min (Supplementary Fig. 1a).
Figure 1. Polθ promotes microhomology-mediated end-joining in vitro and in vivo.
a, Model of Polθ involvement in MMEJ. Polθ is proposed to extend paired 3’ overhangs at DNA synapses. b, pssDNA and ssDNA substrates. * = 32P. (c,d) Non-denaturing gels showing MMEJ reactions with pssDNA (c) and ssDNA (d). * = MMEJ products. e, Schematic of solid-phase MMEJ assay (left). (Left panel) Denaturing gel showing DNA purified from pellet and supernatant fractions following MMEJ in the presence (lanes 2 and 4) and absence (lanes 1 and 3) of Polθ. (Right panel) Denaturing gel showing MMEJ reactions with pssDNA-4 (lanes 1 and 2) and ssDNA-4 (lanes 3 and 4). * = MMEJ products. f, Non-denaturing gel showing MMEJ reactions with the indicated Pols. * = MMEJ products. g, MMEJ GFP reporter assay. Schematic of GFP reporter with microhomology, I-SceI site, stop codon and GFP gene indicated (top). Plot of % GFP cells following transient expression of I-SceI and transfection with Polθ siRNA and scrambled siRNA. Error bars, s.d. (n = 9 independent experiments)(left). Western blots of Polθ and β-actin following transfection with Polθ siRNA (lane 2) and scrambled siRNA (lane 1)(right).
We next examined the ability of Polθ to perform MMEJ of ssDNA versions of the substrates used in Figure 1c (see Fig. 1b). Remarkably, we found that ssDNA was a poor substrate for Polθ MMEJ compared to pssDNA. For example, Polθ failed to join ssDNA containing <6 bp of microhomology and performed very limited MMEJ of ssDNA-6 (Fig. 1d). Thus, the polymerase almost exclusively joins substrates modeled after partially resected DNA when homology is limited (i.e. microhomology ≤6 bp). We note that Polθ produced mostly lower molecular weight byproducts on substrates that do not support efficient MMEJ, such as ssDNA substrates and pssDNA containing <4 bp microhomology (Fig. 1c and 1d). Previous studies suggest that Polθ may extend ssDNA and pssDNA lacking microhomology via terminal transferase activity which could account for these small byproducts19. However, we demonstrate that Polθ lacks appreciable terminal transferase activity, but instead extends substrates that do not support efficient MMEJ by performing ‘snap-back’ replication (Supplementary Fig. 2).
We devised a solid-phase experiment as a control to confirm MMEJ. Here, Polθ joining of radio-labeled to biotinylated pssDNA-4 substrates is detected by tethering of radio-labeled MMEJ products to streptavidin beads (Fig. 1e). Only the double-size product representative of MMEJ was retained on the beads in a Polθ dependent manner (Fig. 1e, left panel); this product is due to MMEJ since it was exclusively generated from pssDNA-4 (Fig. 1e, right panel). We note that Polθ performed strand displacement synthesis during MMEJ (Fig. 2c), which explains the ~48 nt length of MMEJ products in the denaturing gel. MMEJ products also appeared in the supernatant due to radio-labeled substrates joined to one another (Fig. 1e, left panel, lane 4). Smaller byproducts were exclusively in the supernatant, thus they are not MMEJ products, but are due to ‘snap-back’ replication activity (Fig. 1e, left panel, lane 4; Supplementary Fig. 2). These data along with restriction and length analysis of end-joining products unequivocally demonstrate that Polθ promotes MMEJ (Supplementary Fig. 1b and 1c).
Figure 2. Polθ uses the opposing overhang as a template in trans which stabilizes the DNA synapse.
a, Model of Polθ overhang extension of pssDNA-4. Microhomology is outlined. b, Non-denaturing (left) and denaturing (center, right) gels showing MMEJ reactions in the presence of indicated dNTPs. Lane 3 in right panel represents a 48 nt marker based on model in panel a. * = MMEJ products. c, Model of Polθ strand displacement synthesis during MMEJ of pssDNA-4 conjugated with Cy3 and black-hole quencher (BHQ). Plot of fluorescence intensity following MMEJ in the presence (grey) and absence (black) of dNTPs. RU = relative units. Error bars, s.d. (n = 3 independent experiments).
We next investigated whether MMEJ is specific to Polθ by performing end-joining with polymerases from various families: Y-family (Polη, Polκ); X-family (Polµ); B-family (Polδ), and; A-family (Klenow fragment)(Fig. 1f). Remarkably, only Polθ promoted the MMEJ product (lane 2). Klenow fragment produced a smaller product with low efficiency, suggesting this related enzyme might exhibit a limited form of end-joining (lane 7). Polδ degraded the DNA due to exonuclease activity (lane 6). Since all the polymerases were active on a primer-template (Supplementary Fig. 3), these data indicate that MMEJ is specific to Polθ.
We next examined whether MMEJ is dependent on Polθ in human cells. Using a previously characterized green fluorescence protein (GFP) MMEJ reporter system stably incorporated into U20S cells23,24, we demonstrated that downregulation of Polθ expression via siRNA resulted in suppression of MMEJ of an I-SceI induced DSB as indicated by a reduction in GFP expressing cells (Fig. 1g)(Supplementary Data Set 1). Together, these data demonstrate that human Polθ promotes MMEJ in vitro and in vivo.
Polθ utilizes the opposing overhang as a template in trans
Since the melting temperature of the 4 bp microhomology sequence (GGCC, <10° C) is substantially lower than the reaction temperature (37° C), the DNA synapse is likely stabilized by overhang extension (Fig. 2a). To test this, we performed MMEJ with limiting mixtures of dNTPs (Fig. 2b, left and middle panels). Failure to produce a stable MMEJ product in reactions lacking necessary complementary dNTPs will indicate the dependence of using the opposing overhang as a template in trans. Consistent with this model, Polθ failed to promote MMEJ with only one or two dNTPs present in the reaction (compare lanes 3–5 in left and middle panels). Instead, multiple products appeared as a smear, suggesting repetitive cycles of abortive ‘snap-back’ replication (Supplementary Fig. 2). Polθ promoted MMEJ when only dGTP was omitted which is expected to allow extension by 12 nucleotides (nt)(compare lanes 3 and 6, left and middle panels). The polymerase, however, extended the overhang in a similar manner to when all four dNTPs are present (compare lanes 3 and 6, middle panel), which we attribute to the high frequency of nucleotide misincorporation and mismatch extension activity exhibited by Polθ20,25. Using a denaturing sequencing gel, we show that the MMEJ product generated in the presence of all four dNTPs is approximately one nucleotide longer than the expected 48 nt product (Fig. 2b, right panel). This is likely due to the ability of Polθ to incorporate dAMP at the end of the template like Klenow fragment as shown previously16.
We next examined whether Polθ performs strand displacement activity during MMEJ. To test this, we performed the MMEJ reaction using pssDNA-4 modified with Cy3 and a blackhole quencher (BHQ)(Fig. 2c). In this scenario, strand displacement is indicated by an increase in Cy3 fluorescence due to dissociation of the short strand conjugated with BHQ. We observed a significant increase in Cy3 fluorescence only in the presence of dNTPs, demonstrating that Polθ performs strand displacement synthesis during MMEJ. Together, the data in Figure 2 demonstrate that Polθ facilitates MMEJ by utilizing the opposing overhang as a template in trans, which stabilizes the DNA synapse.
Template preferences for Polθ MMEJ
We next examined the template preferences for Polθ MMEJ. Cellular studies indicate that MMEJ requires <20 nt of resection which is performed by resection initiation factors, Mre11 and CtIP3. Strikingly, we find that Polθ MMEJ requires a similar overhang length (<18 nt)(Fig. 3a), suggesting that the polymerase exclusively acts on DNA after initial resection by Mre11, which was recently shown to be 15–20 nt in yeast26. Considering that a 5’-terminal phosphate stimulates replication across gaps by X-family Pols involved in NHEJ, we tested whether this is also the case for Polθ during MMEJ27–29. Remarkably, a 5’-terminal phosphate increased the rate of MMEJ products generated by Polθ (Fig. 3b), indicating that Polθ exhibits an affinity for the 5’-terminal phosphate like NHEJ Pols. Since microhomology annealing likely stabilizes the DNA synapse, we examined whether MMEJ is more proficient in the presence of GC-rich microhomology. Indeed, we find that MMEJ was facilitated by GC-rich microhomology (Fig. 3c). This demonstrates that MMEJ is promoted by hydrogen bonds between overhangs which appear to stabilize the end-joining intermediate.
Figure 3. Template preferences for Polθ MMEJ.
a, Schematic of pssDNA-4 substrates with variable length overhangs (left). (Middle panels) Non-denaturing gels showing MMEJ reactions with the indicated pssDNA. (Right) Plot of % MMEJ products. b, Plot of % MMEJ products generated from pssDNA-4 with (black) and without (grey) a 5’-terminal phosphate for the indicated times. c, Non-denaturing gels showing a time course of MMEJ reactions with pssDNA-2 containing CG (left) and AT (right) 3’ terminal microhomology. * = MMEJ products.
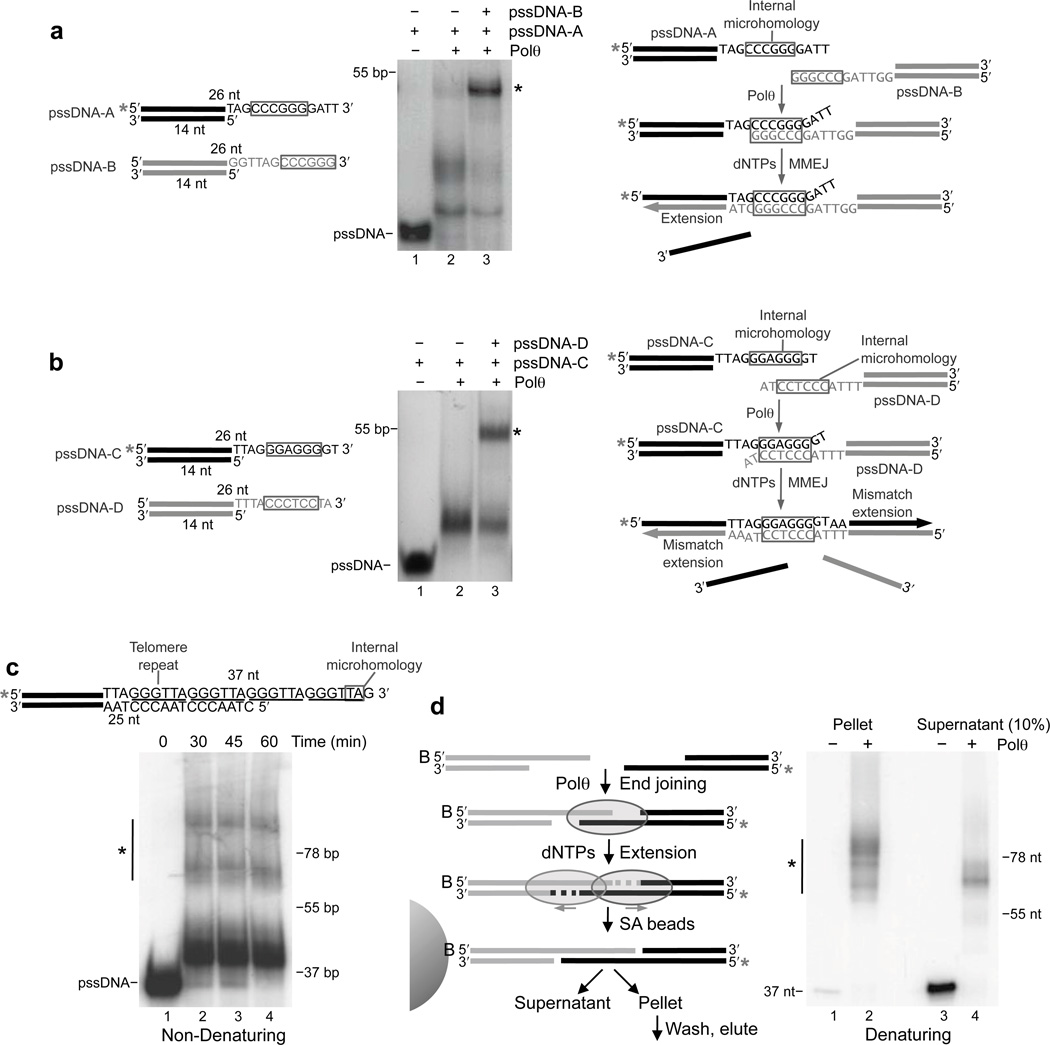
Although previous studies indicate that MMEJ requires limited resection promoted by Mre11 and CtIP3, it remains unknown whether microhomology exposed by this process lies internally or at the 3’ terminus of the resulting overhangs. For example, the experiments presented above exclusively used pssDNA substrates that contain terminal microhomology. However, microhomology may also be positioned internally relative to the 3’ termini of overhangs. We therefore investigated whether Polθ can perform MMEJ of pssDNA containing internal microhomology. Indeed, Polθ promoted efficient MMEJ when the microhomology region is located 5 nt away from the 3’ terminus on one substrate (Fig. 4a). In this scenario, the polymerase probably only extends the overhang containing terminal microhomology (Fig. 4a, right schematic). Next, we tested whether Polθ is capable of performing MMEJ when microhomology is located internally on both substrates. Indeed, the polymerase promoted efficient MMEJ when microhomology was located 3 nt away from the 3’ terminus on both substrates (Fig. 4b). The ability of Polθ to efficiently extend mismatched primers in previous studies suggests that the polymerase performs mismatch extension of both overhangs in this scenario (Fig. 4b, right schematic)25. Polθ also performed MMEJ when microhomology was located 4 nt away from both 3’ termini, albeit with lower efficiency (Supplementary Fig. 4).
Figure 4. Polθ promotes MMEJ of DNA containing internal microhomology.
(a,b) Schematic of pssDNA substrates with microhomology outlined (left). Non-denaturing gel showing MMEJ reactions with the indicated pssDNA (middle). Model of MMEJ (right). * = 32P, * = MMEJ products. c, MMEJ of pssDNA modeled after telomeres. (Top) pssDNA with telomere repeats underlined and microhomology outlined. (Bottom) Non-denaturing gel showing a time course of MMEJ. d, Schematic of solid-phase MMEJ assay (left). Denaturing gel of DNA purified from supernatant and pellet fractions following MMEJ reactions in the presence (lane 2 and 4) and absence (lanes 1 and 3) of Polθ. 10% of supernatant was analyzed (right). * = 32P, * = MMEJ products.
Since MMEJ facilitates telomere fusions, we examined whether Polθ joins substrates modeled after telomeres which also contain internal microhomology (Fig. 4c)4,8. Again, we observed high molecular weight products indicative of MMEJ (Fig. 4c). The distribution of high and low molecular weight products was nearly identical to that generated from pssDNA-2 in Figure 1c, demonstrating low efficiency of MMEJ due to limited microhomology (2 bp). As a result of inefficient MMEJ, the major lower molecular weight byproduct is due to ‘snap-back’ replication (Supplementary Fig. 2d). We confirmed that the high molecular weight products are due to MMEJ by performing the solid-phase assay illustrated in Figure 1e with the telomere substrate (Fig. 4d). The slightly longer than expected MMEJ products (i.e. >74 bp) are likely due to the ability of Polθ to promote nucleotide insertions along repetitive sequences due to primer slippage as shown in previous studies and in Figure 6d20. Together, the data presented in Figure 4 demonstrate the ability of Polθ to perform MMEJ when microhomology is positioned internally.
Figure 6. Insertion loop 2 promotes microhomology-mediated end-joining, DNA binding and polymerase complexes.
a, Schematic of Polθ with polymerase domains and insertion loops indicated. b, Superposition of Bacillus Pol I structure (blue; PDB code 4DQQ)31 in complex with primer-template (orange) and Polθ model (grey; residues 1944–2590) assembled by Swiss Model server30 using Bacillus Pol I:primer-template structure (PDB code 4DQQ)31 as a template. c, Non-denaturing gel showing MMEJ reactions with Polθ WT (lane 2) and Polθ L2 (lane 3). d, Denaturing gel showing primer-template extension with Polθ WT (lane 2), Polθ L2 (lane 3), and Polδ (lane 4). (e,f) EMSA with Polθ WT (left) and Polθ L2 (right) on pssDNA-4 (e) and primer-template (f). g, Plot of % DNA bound calculated from EMSA in panels e and f. % bound = intensity of upper band/sum of the intensities of upper and lower bands.
Polθ promotes DNA synapse formation and strand annealing
We next tested whether Polθ promotes DNA synapse formation separately from its replication activity. Here, fluorescence resonance energy transfer (FRET) was used to probe Polθ dependent formation of DNA synapses in the absence of dNTPs (Fig. 5a). We found that fluorescence intensity increased as a function of Polθ concentration, whereas no increase was observed when the donor (Cy3) substrate was omitted (Fig. 5a, left panel). Hence, these data demonstrate that Polθ promotes DNA synapse formation separately from its replication activity. Next, the assay was repeated using pssDNA substrates with and without microhomology to determine whether microhomology promotes synapse formation. Remarkably, Polθ promoted DNA synapses in the absence of microhomology, yet the presence of microhomology increased the extent of synapses as indicated by higher fluorescence (Fig. 5a, right panel). These data indicate that the dissociation rate of DNA synapses or distance between substrates is decreased by overhang base-pairing.
Figure 5. Polθ promotes DNA synapse formation and strand annealing separately from its replication function.
a, Schematic of DNA synapse assay (left). (Left panel) Plot of relative fluorescence intensity following Polθ synapse formation in the presence of Cy5 pssDNA with (grey) and without (black) Cy3 pssDNA. (Right panel) Plot of relative fluorescence intensity following Polθ synapse formation in the presence of Cy3 and Cy5 pssDNA with (grey) and without (black) 4 bp of microhomology. RU = relative units. Error bars, s.d. (n = 3 independent experiments). (b,c) Schematic of annealing assay (left). (Middle) Non-denaturing gel showing ssDNA (b) and pssDNA (c) annealing in the presence (lane 3) and absence (lane 2) of Polθ. ssDNA (b) and pssDNA (c) marker (lane 1). (Right) Plot of % annealing. % annealing = (intensity of upper band)/(sum of the intensities of upper and lower bands). Error bars, s.d.. (n = 3 independent experiments).
The ability of Polθ to promote DNA synapses in the absence of microhomology suggests that MMEJ includes two initial steps: DNA synapse formation, which brings the overhangs into close proximity regardless of microhomology, and; Microhomology annealing, which promotes base-pairing between substrates that is necessary for overhang extension. We considered whether Polθ contributes to microhomology annealing or whether this process occurs spontaneously once the overhangs are in close proximity. For the former case, the polymerase would have to exhibit strand annealing activity. Indeed, Polθ facilitated annealing of ssDNA (Fig. 5b) and pssDNA (Fig. 5c). Together, these data demonstrate that Polθ promotes DNA synapse formation and strand annealing which likely contribute to MMEJ.
Insertion loop 2 is essential for MMEJ and Polθ multimers
Polθ includes three insertion loops that are not present in other A-family Pols, but are highly conserved in vertebrate Polθ (Fig. 6a; Supplemental Note)17,18. Interestingly, loop 2 is necessary for Polθ ssDNA annealing and extension suggesting it may contribute to MMEJ (Supplementary Fig. 5b,c)19. Structural modeling using homologous Bacillus Pol I in complex with DNA as a template predicts that loop 2 is positioned between the palm and thumb domains and lies close to the 3’ terminus of the primer, suggesting it may bridge opposing overhangs (Fig. 6b; Supplementary Fig. 6)30,31. Indeed, a loop 2 deletion mutant (Polθ L2) failed to perform MMEJ (Fig. 6c), but is active on a primer-template (Fig. 6d). Hence, loop 2 is essential for MMEJ which suggests it may enable polymerase binding to pssDNA in a particular conformation. Remarkably, Polθ L2 failed to bind pssDNA (Fig. 6e,g), but bound a primer-template, albeit with lower affinity than Polθ WT (Fig. 6f,g). This explains why Polθ L2 is inactive in MMEJ and less active than Polθ WT on a primer-template (Fig. 6d)18. Notably, Polθ WT shifted DNA to the well, whereas Polθ L2 did not (Fig. 6f), suggesting that loop 2 promotes polymerase complexes. Indeed, Polθ WT formed dimers and to a lesser extent multimers, whereas Polθ L2 behaved solely as a monomer (Supplementary Fig. 5d). Hence, loop 2 promotes Polθ complexes that are active and exhibit a high affinity for pssDNA which is essential for MMEJ. These data demonstrate the importance of loop 2 in conferring structural and functional characteristics necessary for Polθ MMEJ.
DISCUSSION
Although some of the genetic requirements for MMEJ have been determined, the central mechanism of this pathway has remained undefined, especially in mammalian cells4. Recent genetic studies in mice, however, have confirmed a role for Polq in mammalian MMEJ, demonstrating that the polymerase promotes class-switch recombination and confers resistance to DSB inducing agents16. Considering that a consistent role for Polθ in MMEJ has been shown in mice, flies and worms, the polymerase appears to have evolved to perform an important alternative end-joining function in higher eukaryotes13–16.
Here, we demonstrate that purified human Polθ is highly efficient in MMEJ of DNA containing 3’ overhangs with microhomology, and show that MMEJ is dependent on Polθ expression in human cells. Our data indicate that MMEJ activity is specific to Polθ since multiple other Pols from the X, Y, A and B families fail to perform this function in vitro. This suggests that Polθ possesses a unique structural configuration that has been selected to perform MMEJ. Indeed, Polθ includes three insertion loop domains that are highly conserved among vertebrate Polθ, but are not present in other A-family Pols (Supplemental Note).
We found that loop 2 is essential for MMEJ in vitro which suggests that this domain may interact with and potentially coordinate the positioning of 3’ overhangs during DNA synapse formation and microhomology annealing (Fig. 7). Consistent with this, structural modeling predicts that loop 2 lies in close proximity to the 3’ terminus of the primer (Supplementary Fig. 6). Furthermore, this domain includes multiple positively charged residues, such as lysine and arginine, suggesting that it interacts with DNA (Supplemental Note). Indeed, we found that loop 2 promotes DNA binding, especially in the case of pssDNA which demonstrates that it confers a structural configuration that favors pssDNA binding.
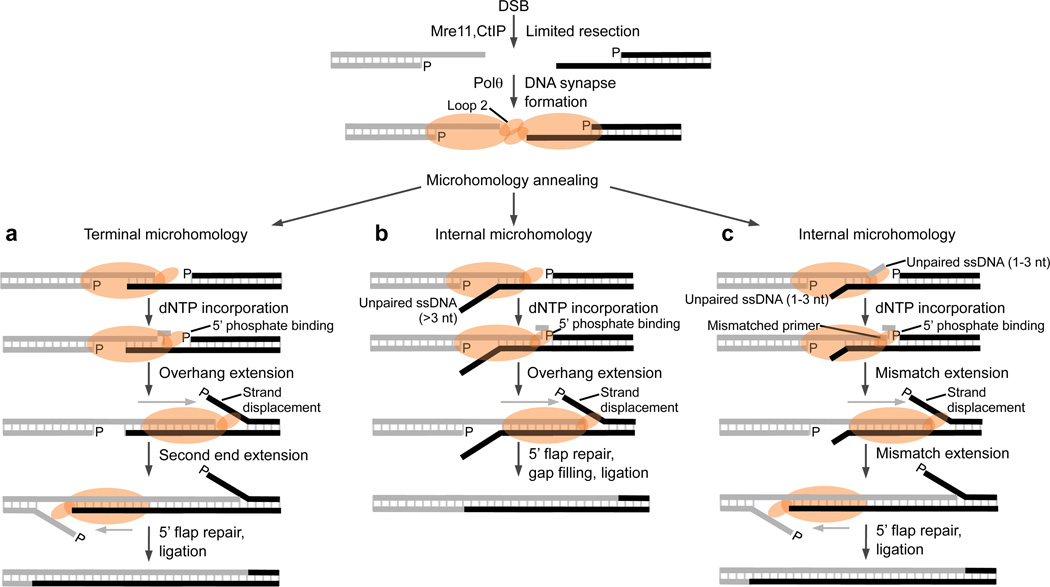
Figure 7. Models of Polθ MMEJ.
Following limited resection of a DSB by Mre11 and CtIP, Polθ dimers promoted by loop 2 facilitate DNA synapse formation (top). Polθ promotes annealing of terminal (a) or internal (b,c) microhomology following DNA synapse formation. Polθ extends the annealed overhang by using the opposing overhang as a template in trans which stabilizes the DNA synapse. Overhang extension is facilitated by Polθ binding to the 5’ terminal phosphate on the opposing DNA and results in strand displacement. a, Polθ extends both overhangs in the case of terminal microhomology. b, Polθ only extends the terminally paired overhang when internal microhomology is located relatively far from the 3’ terminus on the opposing strand. c, Polθ performs mismatch extension of overhangs that contain internal microhomology relatively close to their 3’ terminus. Last, 5’ flap repair is presumably required prior to ligation in each case.
We found that loop 2 also promotes high-order structures of the polymerase. For example, wild-type Polθ formed dimers, and to a lesser extent multimers, as indicated by gel filtration (Supplementary Fig. 5d). In contrast, Polθ lacking loop 2 acted solely as a monomer (Supplementary Fig. 5d), and as a potential result of this failed to perform MMEJ (Fig. 6c). We therefore propose a model whereby Polθ dimers facilitate DNA synapse formation (Fig. 7, top). Consistent with this model, previous structural studies demonstrate dimerization of an end-joining polymerase as a mechanism of DNA synapse formation32. Considering that Polθ facilitated annealing of complementary overhangs separately from its replication activity, we propose that the polymerase promotes microhomology annealing after bringing the overhangs into close proximity (Fig. 7). Since loop 2 was necessary for strand annealing (Supplementary Fig. 5c), it is likely involved in forming minimally paired overhangs. This function would explain why loop 2 is required for Polθ ssDNA extension since such activity requires a minimally paired primer (Supplementary Fig. 2)16.
Although it is unclear how Mre11 and CtIP perform the limited resection step required for MMEJ, microhomology exposed by this activity may be positioned internally or at the 3’ terminus of overhangs (Fig. 7). Remarkably, we found that Polθ promotes MMEJ of pssDNA containing terminal or internal microhomology. In the case of terminal microhomology, our data support a mechanism whereby the polymerase extends each overhang by using the opposing overhang as a template in trans which stabilizes the DNA synapse (Fig. 7a). Since the rate of overhang extension was increased in the presence of a 5’-terminal phosphate, Polθ probably possesses a 5’-terminal phosphate interacting motif like X-family Pols involved in NHEJ and base excision repair (Fig. 7)27. In the case of internal microhomology, we present two models based on our findings. In the first example, where one end contains internal microhomology relatively far (>3 nt) from the 3’ terminus, Polθ is limited to extending the paired end containing terminal microhomology (Fig. 7b). In the second example, in which both ends contain internal microhomology relatively close (1–3 nt) to the 3’ termini, the polymerase performs mismatch extension of each end, potentially generating mutations in addition to a DNA deletion (Fig. 7c). Lastly, since Polθ promotes a certain degree of strand displacement synthesis, 5’ flap repair is likely required prior to ligation.
In summary, our data reveal a central mechanism of MMEJ promoted by Polθ. Considering that POLQ also encodes for a N-terminal helicase domain and a large central domain, it will be interesting to determine whether these components contribute to the mechanism or regulation of MMEJ17. Finally, since upregulation of Polθ corresponds to a poor clinical outcome for breast cancer patients33, it will be important to determine whether MMEJ contributes to cancer progression and chemotherapy resistance in these tumors.
ONLINE METHODS
MMEJ
100 nM 5’-32P radio-labeled pssDNA or ssDNA pre-incubated at 37° C in buffer A (25 mM Tris-HCl pH 8.8, 10% glycerol, 1 mM DTT, 0.01% NP-40, 10 mM MgCl, 0.1 mg/ml BSA) was mixed with 100 nM Polθ for 15 min followed by the addition of 500 µM dNTPs for another 30 min at 37° C in a total volume of 20 µl. For analysis in non-denaturing gels, reactions were terminated by the addition of 4 µl of non-denaturing stop buffer (0.5 M Tris-HCl pH 7.5, 10 mg/ml proteinase K, 80 mM EDTA, 1.5% SDS) followed by a further 15–30 min incubation at 37° C. For analysis in denaturing gels, reactions were terminated by the addition of 20 µl of 2X denaturing stop buffer (90% formamide, 50 mM EDTA). Radio-labeled DNA was resolved in non-denaturing or denaturing (urea) polyacrylamide gels and visualized by autoradiography. Concentrations are listed as final. Figure 2c was performed as above, however, pssDNA-4 was conjugated with Cy3 and BHQ, and strand displacement was determined by Cy3 fluorescence intensity using a Clariostar (BMG Labtech) plate reader.
MMEJ solid-phase assay
MMEJ was performed with and without Polθ as described in methods summary, however, 4 reaction volumes were pooled, equimolar concentrations (50 nM) of radio-labeled and biotinylated pssDNA substrates were used, and the pooled reactions (80 µl) were incubated with 160 µl of magnetic streptavidin beads (Thermo Scientific Pierce) for 30 min rather than the addition of stop buffer. The supernatant containing soluble DNA was removed using a magnetic separation rack (NEB), then the beads were washed three times with 200 µl of 200 mM NaCl. Last, the pellet fraction containing biotinylated DNA was removed from the beads by the addition of 30 µl of 1X denaturing stop buffer (45 mM formamide, 25 mM EDTA) followed by boiling and removal of supernatant using a magnetic separation rack. DNA from the supernatant fraction was ethanol precipitated then resuspended in 30 µl TE (10 mM TrisHCl pH 8.0, 1 mM EDTA). Pellet and 50% of supernatant fractions were analyzed in a denaturing urea polyacrylamide gel. For Figure 4d, 10 reactions were pooled, the pellet was washed with 50 mM NaCl, and 10% of the supernatant was analyzed.
I-Sce-I induced MMEJ
U2OS-EJ2-GFP cells with MMEJ reporter24 (1 × 105) were plated on 6 well plate and transfected 24 hr later with 2.5 µg pCMV-3xNLS-I-SceI or 0.5 µg control vector pCMV-3xNLS using Lipofectamine 2000 (Invitrogen). GFP+ frequencies were measured 3 days post transfection by FACS using GUAVA flow cytometer (Millipore) in triplicates and corrected for transfection efficiency and background events. Transfection efficiency was measured simultaneously by parallel transfection with 0.05 µg wt GFP expression vector (pCMV-3xNLS-GFP). For siRNA experiments, cells were transfected with 100 ρmol siRNA (Polθ SMARTpool (Dharmacon) or scrambled control) + 0.3 µg of pCMV-3xNLS-I-SceI (or control vector, GFP expression vector) per well.
Western Blotting
A portion of U2OS-Alt-EJ cells24 from the I-Sce-I induced MMEJ reporter assays performed with siRNA was used for Western blotting analysis. Whole-cell lysates were prepared by lysing the cells in RIPA buffer (25 mM Tris–HCl, 125 mM NaCl, 1% Nonidet-P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS) and a Protease Inhibitor Cocktail (Pierce)). Equal amounts (20 µg) of cell lysates were separated by SDS–polyacrylamide gel electrophoresis then transferred to membrane and immunoblotted with antibodies against the polymerase domain of Polθ (Sigma, Cat no. AV49203, Experimental dilution 1:1000. Antibody was validated in Supplemental Data Set 1 and in previous studies21) and β-Actin (Abcam, Clone no. mAbcam 8226, Experimental dilution 1:5000. Antibody validation is available on Abcam website). Blots were stained with an enhanced chemi luminescence detection kit (ECL-Plus, Amersham Biosciences).
DNA synapse formation
Equimolar concentrations (50 nM) of Cy3 and Cy5 labeled pssDNA substrates with or without 4 bp of microhomology were mixed with the indicated amounts of Polθ for 30 min in 40 µl of buffer A at room temp. FRET (540 nm, excitation; 675 nm, emission) was then measured using a Clariostar (BMG Labtech) plate reader. Experiments were performed in triplicate and data was normalized and plotted with standard deviation.
Strand annealing
5’-32P radio-labeled ssDNA (RP40, 20 nM) or pssDNA (RP40/RP364, 1 nM) substrates were incubated with or without 100 nM Polθ WT or Polθ L2 in 20 µl of buffer C (50 mM Tris-HCl pH 7.5, 1 mM DTT, 0.1 mg/mL BSA, 10% glycerol, 0.5 mM MgAc) at 37° C for 5 min. Complementary ssDNA (RP40C, 20 nM) or pssDNA (RP365/RP364, 1 nM) was then added for 30 s. Annealing was terminated by the addition of excess cold ssDNA (RP40, 200 nM) or pssDNA (RP40/RP364, 30 nM) then 4µl of non-denaturing stop buffer (0.5 M Tris-HCl pH 7.5, 10 mg/ml proteinase K, 80 mM EDTA, 1.5% SDS) followed by a further 15–30 min incubation at 37° C. DNA was resolved in non-denaturing 12% polyacrylamide gels and analyzed by phosphorimager (Fujifilm FLA 7000).
Structural modeling
Modeling was performed using Swiss Model server (http://swissmodel.expasy.org/)2 using Polθ residues 1890–2590. Bacillus Pol I in complex with primer-template in closed conformation (PDB code: 4DQQ) was used as a template for modeling3. SWISS-MODEL was used in first approach mode using default parameters. Structures were visualized using Swiss-PDBViewer4. Images were generated using PyMOL software5. Superposition of Polθ model and Bacillus Pol I:DNA structure: The Ca2+ bound form of Pol I was used as a template onto which residues 1944–2590 of the Polθ model was superimposed. Using least-squares fitting option in SPDBV2, all carbon atoms superimpose with an r.m.s. 0.22Å.
Primer extension
The indicated Pol (100 nM) was incubated with 5’-32P radio-labeled primer-template (RP25/RP266, 100 nM) along with 0.5 mM dNTPs in 10 µl of buffer A for 30 min at 37° C. Reactions were terminated by the addition of 10 µl of denaturing stop buffer and DNA was resolved in denaturing (urea) polyacrylamide gels and analyzed by autoradiography.
EMSA
12 nM Cy3 conjugated primer-template (RP25Cy3/RP266), pssDNA (RP348Cy3/RP343P), or ssDNA (RP348Cy3) was mixed with the indicated amounts of Polθ WT or Polθ L2 in 20 µl of buffer (25 mM Tris-HCl pH 7.5, 30 mM NaCl, 0.01% NP-40, 1 mM DTT, 0.1 mg/ml BSA, 0.5 mM MgCl and 10% glycerol) on ice (primer-template) or at room temp (pssDNA) for 60 min. Reactions were resolved in non-denaturing 5% polyacrylamide gels containing 2.5% glycerol and DNA was visualized using a MultiImage III fluorescence imager (Alpha Innotech) and FluorChem Q software by monitoring Cy3 fluorescence.
Gel filtration
Polθ in protein storage buffer (50 mM HEPES pH 8.8, 300 mM NaCl, 1 mM DTT, 0.02% NP-40, 10% (v/v) glycerol) was injected into a HiLoad 16/600 Superdex 200 pg column (GE Health Sciences) equilibrated with protein storage buffer using AKTA L1 Pure (GE Health Sciences) with automated fraction collector and a multi-channel peristaltic pump. Protein peaks were plotted versus elution volume using a UV monitor detecting at 280 nm.
Native gel analysis
1 µM Polθ WT or Polθ L2 was incubated in 25 mM Tris-HCl pH 8.8, 0.01% NP-40, 1 mM DTT, 10% glycerol, 676 mM NaCl, 0.2% Tween-40 at room temp for 1 hr. Protein solutions were then resolved in a 4–15% Mini-PROTEAN TGX native gel (Bio-Rad) in Tris-Glycine Buffer, pH 8.9. Protein was then visualized by silver staining.
Sequence alignment
The indicated amino-acid sequences of the polymerase domain of Polθ from the indicated vertebrates and other indicated A-family Pols were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/; European Bioinformatics Institute) default settings.
Cell lines and cell culture
U2OS cell line (EJ2-GFP)1 was a kind gift from J. Stark. U2OS cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% Fetal bovine serum (FBS).
Proteins
Polθ WT and Polθ L2 were purified as described6. Klenow fragment was purchased from New England Biolabs. Polδ was purified as described34. Polκ was provided by A.K. Aggarwal. Polµ was purchased from Enzymax. Polη was purified as described35.
DNA
Templates. pssDNA-6 (RP344/RP343), pssDNA-4 (RP348/RP343), pssDNA-2 (RP340/RP343), pssDNA-0 (RP347/RP343), ssDNA-6 (RP344), ssDNA-4 (RP348), ssDNA-2 (RP340), ssDNA-0 (RP347). Fig. 1e: RP348/RP343, RP348B/RP343. Fig. 2a,b: pssDNA-4 (RP348/RP343). Fig. 2c: RP348Cy3/RP343P-BkFQ. Fig. 3a: RP366/RP343, RP348/RP343, RP362/RP343, RP363/RP343. Fig. 3c: RP340/RP343, RP357/RP343. Fig. 4a: pssDNA-A (RP399/RP343); pssDNA-B (RP348/RP343). Fig. 4b: pssDNA-C (RP396/RP343); pssDNA-D (RP397/RP343). Fig. 4c: RP331/RP332, RP331B/RP332. Fig. 5a: RP362Cy3/RP343P, RP363/RP343Cy5. Fig. 5b: RP40, RP40C. Fig. 5c: RP365/RP364, RP40/RP364. Fig. 6: Radio-labeled and Cy3 conjugated pssDNA-4 (RP348/RP343) and primer-template (RP25/RP266). Supplementary Fig. 1: a, pssDNA-4 (RP348/RP343), pssDNA-2 (RP340/RP343); b, RP370/RP343; c, pssDNA-4A (RP348/RP343), pssDNA-4B (RP356/RP353). Supplementary Fig. 2: b, RP360, RP316; c, RP348; d, RP331,RP332. Supplementary Fig. 3: primer-template (RP25/RP266). Supplementary Fig. 4: pssDNA-E, RP396/RP343; pssDNA-F, RP402/RP343.
Supplementary Fig. 5: a, RP316Cy3; b, ssDNA-4 (RP348); c, RP40, RP40C.
All pssDNA templates were 5’-phosphorylated on the shorter strand using T4 polynucleotide kinase (NEB) and ATP or purchased with a 5’-phosphate, except for RP331/RP332 and where indicated in the text and figures. pssDNA substrates were annealed by mixing a ratio of 1:1.5 of long to short strands then boiling and allowing to slowly cool to room temp. DNA was 32P-5’ radio-labeled using T4 polynucleotide kinase (NEB) and 32P-γ-ATP (Perkin Elmer).
DNA oligonucleotides (Integrated DNA Technologies)(5’-3’): RP348, CACTGTGAGCTTAGGGTTAGAGCCGG; RP348B, Biotin-CACTGTGAGCTTAGGGTTAGAGCCGG; RP343P, P-CTAAGCTCACAGTG; RP344, CACTGTGAGCTTAGGGTTAGCCCGGG; RP343, CACTGTGAGCTTAGGGTTAGCCCGGG; RP370, CACTGTGAGCTTAGGGTTAGGAATTC; RP368, CACTGTGAGCTTAGAGAAAA; RP343Cy5, Cy5-CTAAGCTCACAGTG; RP348Cy3, Cy3-CACTGTGAGCTTAGGGTTAGAGCCGG; RP362Cy3, Cy3-CACTGTGAGCTTAGAGCCGG; RP366, CACTGTGAGCTTAGATTGGTTAGAGCCGG; RP365, TTATCATCGATATTAATACGACTCAAATATTCTCATCCCTCCCTCCTCCCTATAG; RP40, TTATCATCGATATTAATACGACTCACTATAGGGAGGAGGGAGGGATGAGAATATT; RP40C, AATATTCTCATCCCTCCCTCCTCCCTATAGTGAGTCGTATTAATATCGATGATAA; RP364, TGAGTCGTATTAATATCGATGATAA; RP362, CACTGTGAGCTTAGAGCCGG; RP363, CACTGTGAGCTTAGATTCTAGGTTAGAGCCGG; RP343P-BkFQ, P-CTAAGCTCACAGTG-BkFQ; RP357, CACTGTGAGCTTAGGGTTAGAGAGAT; RP356, TGTGTCAGAGGTTTTCACCGTCATCA; RP353, TGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGG; RP340, CACTGTGAGCTTAGGGTTAGAGATCG; RP347, CACTGTGAGCTTAGGGTTAGAGATAC; RP331, ACTGTGAGCTTAGGGTTAGGGTTAGGGTTAGGGTTAG; RP331B, Biotin-ACTGTGAGCTTAGGGTTAGGGTTAGGGTTAGGGTTAG; RP332, CTAACCCTAACCCTAAGCTCACAGT; RP396, CACTGTGAGCTTAGTTAGGGAGGGGT; RP397, CACTGTGAGCTTAGTTTACCCTCCTA; RP316Cy3, Cy3-TTTTTTTTTTTTTTTTTTTTTTTTTTTTT; RP402, CACTGTGAGCTTAGTTTACCCTCCTAT
Supplementary Material
ACKNOWLEDGEMENTS
Research was funded by National Institutes of Health grant (4R00CA160648-03) awarded to R.T.P and Temple University School of Medicine start-up funds to R.T.P. We thank S. Wallace (University of Vermont) for Polθ WT and Polθ L2 expression vectors, J. stark (Beckman Research Institute City of Hope) for U2OS cell line EJ2-GFP24 and A.K. Aggarwal (Mt. Sinai Hospital) for Pol κ.
Footnotes
AUTHOR CONTRIBUTIONS R.T.P., T.K., G.C., S.M.M. and A.Y.O. performed the experiments. R.T.P. designed the experiments and wrote the paper.
References
- 1.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truong LN, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 5.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 6.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 7.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Molecular cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones RE, et al. Escape from Telomere-Driven Crisis Is DNA Ligase III Dependent. Cell Rep. 2014;8:1063–1076. doi: 10.1016/j.celrep.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nature structural & molecular biology. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 12.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nature structural & molecular biology. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koole W et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 14.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yousefzadeh MJ, et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic acids research. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic acids research. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic acids research. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. A Role for DNA Polymerase theta in Promoting Replication through Oxidative DNA Lesion, Thymine Glycol, in Human Cells. The Journal of biological chemistry. 2014;289:13177–13185. doi: 10.1074/jbc.M114.556977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad R, et al. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic acids research. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. The Journal of biological chemistry. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 25.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA repair. 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014 doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Diaz M, et al. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Molecular cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 28.Moon AF, et al. The X family portrait: structural insights into biological functions of X family polymerases. DNA repair. 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brissett NC, et al. Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase. Cell Rep. 2013;5:1108–1120. doi: 10.1016/j.celrep.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic acids research. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Wu EY, Hellinga HW, Beese LS. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. The Journal of biological chemistry. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brissett NC, et al. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 33.Lemee F, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fazlieva R, et al. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37:2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acharya N, et al. Complex formation of yeast Rev1 with DNA polymerase eta. Mol Cell Bio. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 34.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 35.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic acids research. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Wu EY, Hellinga HW, Beese LS. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. The Journal of biological chemistry. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 39.Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.