Abstract
Purpose
To assess the feasibility of texture analysis for classifying fibrosis stage and necroinflammatory activity grade in patients with chronic hepatitis C on T2-weighted (T2W), T1-weighted (T1W) and Gd-EOB-DTPA-enhanced hepatocyte-phase (EOB-HP) imaging.
Materials and methods
From April 2008 to June 2012, MR images from 123 patients with pathologically proven chronic hepatitis C were retrospectively analyzed. Texture parameters derived from histogram, gradient, run-length matrix, co-occurrence matrix, autoregressive model and wavelet transform methods were estimated with imaging software. Fisher, probability of classification error and average correlation, and mutual information coefficients were used to extract subsets of optimized texture features. Linear discriminant analysis in combination with 1-nearest neighbor classifier (LDA/1-NN) was used for lesion classification. In compliance with the software requirement, classification was performed based on datasets from all patients, the patient group with necroinflammatory activity grade 1, and that with fibrosis stage 4, respectively.
Results
Based on all patient dataset, LDA/1-NN produced misclassification rates of 28.46%, 35.77% and 20.33% for fibrosis staging and 34.15%, 25.20% and 28.46% for necroinflammatory activity grading in T2W, T1W and EOB-HP images. In the patient group with necroinflammatory activity grade 1, LDA/1-NN yielded misclassification rates of 5.00%, 0% and 12.50% for fibrosis staging in T2W, T1W and EOB-HP images respectively. In the patient group with fibrosis stage 4, LDA/1-NN yielded misclassification rates of 5.88%, 12.94% and 11.76% for necroinflammatory activity grading in T2W, T1W and EOB-HP images respectively.
Conclusion
Texture quantitative parameters of MR images facilitate classification of the fibrosis stage as well as necroinflammatory activity grade in chronic hepatitis C, especially after categorizing the input dataset according to the activity or fibrosis degree in order to remove the interference between the fibrosis stage and necroinflammatory activity grade on texture features.
Introduction
Hepatitis C related liver cirrhosis remains an important cause of morbidity and mortality [1], and predisposes to hepatocellular carcinoma [2]. Liver biopsy is routinely used to diagnose and stage the severity of hepatitis in patients with chronic hepatitis C [3], but has the drawbacks of invasiveness, sampling error and inherent risks [4]. A noninvasive accurate method to evaluate liver cirrhosis would reduce the risks and costs for extra examinations and facilitate prompt assessment in clinical practice for hepatitis C related liver cirrhosis.
Texture analysis is a new useful post-processing method for radiological images that provides more than the perceivable texture feature parameters. In addition, all parameters of texture features are expressed as numerical values that quantitatively evaluate images. These texture features include histogram, gradient, gray-level co-occurrence matrix, run-length matrix, autoregressive model and wavelet transform parameters [5]. Histogram, co-occurrence matrix, run-length matrix, and gradient come from statistical approaches; autoregressive model is yielded by model-based methods; wavelet is based on transform method [5]. Simply, these texture parameters reflect the extent of heterogeneity, granularity, randomness and so on. Thus, they represent characteristics that may be associated with the histopathological changes and contribute to the differential diagnosis or development grade assessment. Until now, texture analysis has provided a wide range of applications to detect the progression of disease or evaluate the response to treatment [6–8].
The pathogenesis of liver cirrhosis in chronic hepatitis C is progressive fibrosis, namely fibrous scarring that may lead to architectural distortion [9]. In MR images, cirrhotic liver parenchyma is characterized by a heterogeneous intensity that is determined by the underlying pathological features involving regenerating nodules and fibrotic scars, and these alterations are exacerbated with progression of the cirrhosis. Contrast enhancement may emphasize the heterogeneous liver background to stage the grade of hepatic cirrhosis [10], especially gadolinium ethoxybenzyl diethylenetriaminepentaacetic acid (Gd-EOB-DTPA) because of its liver-specific character and excellent enhancement of liver parenchyma on the hepatobiliary phase of Gd-EOB-DTPA enhanced MR imaging (EOB-HP) [11].
Since texture analysis has hitherto succeeded in separating cirrhotic patients and healthy volunteers [12], the impact of texture features measured on MR images is expected to further define the diagnostic performance of discrimination of various fibrotic stages and activity grades. The aim of this study was to investigate whether the texture features extracted from T1W, T2W and EOB-HP images can differentiate fibrosis stages or necroinflammatory activity grades in patients with chronic hepatitis C.
Materials and Methods
Patients
The approval of this retrospective analysis of the daily clinical data was waived by Kanazawa University Graduate School of Medical Science institutional review board. Written informed consent for the use of daily clinical data, including imaging materials, was obtained from all patients at the time of their admission. Patient records/information was not anonymized and de-identified prior to analysis, because we should check the clinical data of these patients during analysis. We enrolled 158 patients with pathology-proven chronic hepatitis C who underwent Gd-EOB-DTPA-enhanced MR examination from April 2008 to June 2012. Inclusion criteria included the interval between MR examination and biopsy or resection (within 90 days) and the absence of any evident artifacts in the T1W, T2W and EOB-HP images. 35 patients were excluded because the interval between MR examination and biopsy or resection was over 90 days in 28 patients and evident imaging artifacts in T2W images in 7 patients, leaving 123 patients (75 men, mean age 68.4 years, range 52~84 years; 48 women; mean age 70.2 years, range 50~86 years) for analysis. Child-Pugh classification of the patients was as follows: Child-Pugh A (n = 86), Child-Pugh B (n = 33) and Child-Pugh C (n = 4).
The histopathology (liver biopsy, n = 104; resection, n = 19) was interpreted by one of two board-certified pathologists, who were blinded to the imaging results, as part of routine clinical practice. The necroinflammatory activity grades were classified as follows: A1, mild activity; A2, moderate activity; and A3, severe activity. Criteria for the fibrosis stages were as follows: F1, fibrous portal expansion; F2, bridging fibrosis; F3, bridging fibrosis with architectural distortion; and F4, cirrhosis [13].
MRI examinations
T2-weighted images were obtained with frequency-selective fat saturation sequence with the following parameters: TR/TE = 15000/89 ms, flip angle 90°, field of view 40×40 cm, matrix 512×512, slice thickness 4.0 mm. T1-weighted images were obtained with fat-suppressed 3D SPGR T1-weighted sequence with the following parameters: LAVA-XV, GEM, TR/TE = 3.2–4.0/1.6 ms, flip angle 6–15°, field of view 42×42 cm, matrix 192×320 (512 interpolated), slice thickness 4.2 mm. A dose of 0.1 ml/kg of Primovist (0.25 mmol/ml of gadoxetic acid, Bayer Schering Pharma, Berlin) was intravenously injected at a flow rate of 1.0 ml/sec, followed by a 30-ml saline flush. The hepatobiliary phase was obtained 20 min after the injection.
Texture analysis and classification
For each sequence (T1W, T2W and EOB-HP), one slice at the level of the porta hepatis was selected and then loaded in bitmap format in the texture analysis software, MaZda 4.6 (URL: http://www.eletel.p.lodz.pl/programy/mazda/) [14–16]. A circular region of interest (ROI) was placed in the right lobe of the liver parenchyma devoid of large vessels, hepatic lesions, and artifacts by an experienced radiologist with 12 years of clinical experience (Fig. 1). To keep the ROI consistent, each 20-pixel diameter ROI was loaded from a file containing a standard-size ROI box. Altogether 279 descriptors were produced automatically and derived from the gray-level histogram (n = 9, the basic characters reflecting image uniformity) [15], gradient (n = 5, a direction change in grey level intensity representing the image intensity distribution) [15], the run-length matrix (n = 20, calculated from different run angles in four directions including horizontal, vertical, 45°and 135°, and indicating image coarseness) [17–20], the co-occurrence matrix (n = 220, computed from intensities of pairs of pixels and describing the homogeneity) [15,21–22], the autoregressive model (n = 5, the coefficients of neighboring pixels reflecting the coarse-to-fine stratification) [5, 18, 19], and the wavelet transform (n = 20, the spatial frequencies at multiple scales identifying coarseness) [23–25]. The categories of the texture parameters are listed in Table 1 in detail [26].
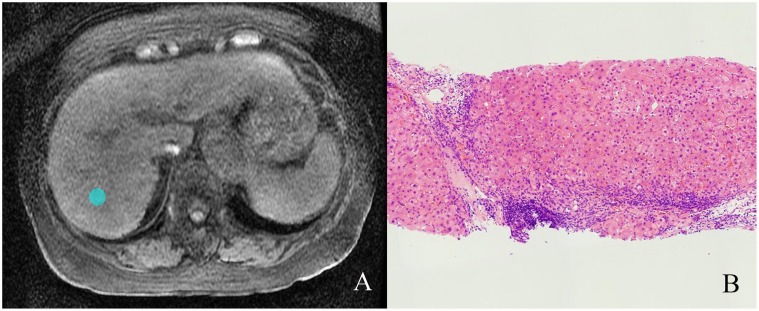
Fig 1. Illustration of region of interest (ROI).
(A) One 20-pixel diameter ROI was placed in the axial T1W images (3.6/1.6; flip angle, 12°) at the level of the porta hepatis in a 76-year-old woman with chronic hepatitis C. (B) Liver biopsy specimen showed stage 4 hepatic fibrosis and grade 2 necroinflammatory activity. (Hematoxylin-eosin stain; original magnification, ×50.)
Table 1. Texture feature list in MaZda software.
| Methods | Texture features |
|---|---|
| Histogram | Mean (histogram’s mean); Variance (histogram’s variance); Skewness (histogram’s skewness); Kurtosis (histogram’s kurtosis); Perc.01% (1% percentile); Perc.10% (10% percentile); Perc.50% (50% percentile); Perc.90% (90% percentile); Perc.99% (99% percentile) |
| Gradient | GrMean (absolute gradient mean); GrVariance (absolute gradient variance); GrSkewness (absolute gradient skewness); GrKurtosis (absolute gradient kurtosis); GrNonZeros (percentage of pixels with nonzero gradient) |
| Run-length matrix | RLNonUni (run length nonuniformity); GLevNonU (grey level nonuniformity); LngREmph (long run emphasis); ShrtREmp (short run emphasis); Fraction (fraction of image in runs) |
| Co-occurrence matrix | AngScMom (angular second moment); Contrast (contrast); Correlat (correlation); SumOfSqs (sum of squares); InvDfMom (inverse difference moment); SumAverg (sum average); SumVarnc (sum variance); SumEntrp (sum entropy); Entropy (entropy); DifVarnc (difference variance); DifEntrp (difference entropy). Features are computed for 5-pixel distance (1, 2, 3, 4, 5) and for 4 various directions (horizontal, 45 degrees, vertical, 135 degrees) |
| Autogressive model | Teta1 (parameter θ1); Teta2 (parameter θ2); Teta3 (parameter θ3); Teta4 (parameter θ4); Sigma (parameter σ) |
| Wavelet transform | WavEn (wavelet energy) feature is computed at 5 scales within four frequency bands: low-pass filtering in both directions (LL) assessed the lowest frequencies, low-pass filtering followed by high-pass filtering (LH) assessed horizontal edges, high-pass filtering followed by low-pass filtering (HL) assessed vertical edges; high-pass filtering in both directions (HH) assessed diagonal details. |
Gray-level intensity normalization was run in the texture analysis application separately for each ROI, with method-limiting image intensities in the range μ-3σ, μ+3σ, where μ is the mean gray-level value and σ the standard deviation, to minimize the influence of contrast and brightness variation, which might otherwise obscure the true image texture [26].
Following the texture parameter calculation, feature reduction was performed. Based on a pooled texture dataset of all patients, the 30 most discriminating texture features were selected automatically from 279 parameters using MaZda program for distinguishing fibrosis stages or necroinflammatory activity grades based on the combination of three methods (Fisher coefficients [Fisher]; classification error probability [POE] combined with average correlation coefficients [ACC]; mutual information coefficients [MI]) as Fisher+POE+ACC+MI separately for T2W, T1W and EOB-HP images [27]. Then the patients were divided into 12 subgroups in terms of both fibrosis stages (F1~4) and necroinflammatory activity grades (A1~3). Since the software requirement is that the sample size for each subgroup must exceed one, but the sample size for F1A2, F1A3, F2A3 and F3A3 subgroup was less than or equal to one, only A1 and F4 groups met this condition. Further texture feature reduction was performed for fibrosis staging of A1 group and necroinflammatory activity grading of F4 group. With the same selection method Fisher+POE+ACC+MI as before, the 30 most discriminating texture features were selected in texture datasets of A1 group and F4 group, separately. A total of 12 groups of 30 parameters were obtained.
B11 application (version 3.4) of MaZda software package was used for texture data analysis and classification. Analyses were performed for a combination of 30 parameters selected automatically with Fisher+POE+ACC+MI methods. Linear discriminant analysis (LDA) was run for these chosen texture feature groups, followed by the 1-nearest neighbor (1-NN) classification used for selecting the most discriminating features (MDF). LDA is a supervised feature transformation method to produce new feature vectors, the so-called MDFs, which are optimized for both maximum between-class scatter and minimum within-class scatter [28]. The classification results of LDA were represented graphically as the relationship between MDFs. The number of MDF axes is less than the output classes by one. When 1-NN classifier is used, these subsets are optimal in the sense that they provide minimal classification error [26]. The classification procedures were run by B11 automatically.
Statistical analysis
For different fibrosis stages or necroinflammatory activity grades, patient age, body mass index (BMI), hepatitis C virus (HCV) RNA titer, alanine transaminase (ALT), aspartate aminotransferase (AST), glucose, total cholesterol, triglyceride, high density lipoprotein (HDL) and low density lipoprotein (LDL) were compared by one-way analysis of variance (ANOVA) followed by Tukey post hoc test; patient sex ratio and HCV serogroup ratio were compared by the Chi-square test or Fisher exact test, with Bonferroni correction.
Results
The stages of fibrosis and grades of necroinflammatory activity observed among the 123 patients on histopathologic analysis are summarized in Table 2. Patient demographic variables according to fibrosis stage and necroinflammatory activity grade are summarized in Table 3. No significant differences were found in age, BMI, HCV RNA titer, ALT, AST, glucose, total cholesterol, triglyceride, HDL, LDL, HCV serogroup ratio or sex ratio among them.
Table 2. Distribution of Various Stages of Fibrosis and Grades of Necroinflammatory Activity.
| Necroinflammatory activity grade | Total | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Fibrosis stage | 1 | 3 | 1 | 0 | 4 |
| 2 | 10 | 3 | 0 | 13 | |
| 3 | 12 | 8 | 1 | 21 | |
| 4 | 15 | 68 | 2 | 85 | |
| Total | 40 | 80 | 3 | 123 | |
Data indicate number of patients.
Table 3. Demographic variables classified by fibrosis stage and necroinflammatory activity grade.
| Parameter | F1 | F2 | F3 | F4 | A1 | A2 | A3 |
|---|---|---|---|---|---|---|---|
| Age (years) | 67.8±9.2 | 67.9±5.0 | 72.5±7.5 | 69.2±8.1 | 68.8±7.5 | 70.0±8.0 | 68.7±9.0 |
| Sex distribution | |||||||
| No. of men | 2 | 9 | 15 | 49 | 24 | 50 | 1 |
| No. of women | 2 | 4 | 6 | 36 | 16 | 30 | 2 |
| BMI (kg/m2) | 22.0±1.9 | 22.7±2.8 | 24.0±2.5 | 23.0±3.1 | 23.5±3.7 | 23.0±2.6 | 22.5±1.9 |
| HCV Serogroup* | (1) | (3) | (4) | (17) | (13) | (12) | (0) |
| No. of type 1 | 2 | 8 | 14 | 59 | 22 | 58 | 3 |
| No. of type 2 | 1 | 2 | 3 | 9 | 5 | 10 | 0 |
| HCV RNA titer (KIU/mL)* | 1473.6±1706.3 (1) | 884.5±807.0 (7) | 1302.5±941.9 (6) | 1279.7±1844.0 (23) | 1731.5±2349.7(12) | 1044.9±1151.9 (25) | 884.9±168.9 (0) |
| ALT (U/L) | 52.0±30.1 | 28.6±11.9 | 53.0±33.1 | 63.9±39.1 | 42.9±31.5 | 65.1±38.2 | 68.7±33.4 |
| AST (U/L) | 42.7±10.8 | 32.3±11.1 | 54.6±33.1 | 65.9±29.2 | 45.9±26.6 | 66.2±30.0 | 69.3±12.7 |
| Glucose (mg/dL) | 95.8±11.3 | 116.5±32.7 | 120.3±22.0 | 128.7±50.0 | 124.4±41.0 | 124.7±46.7 | 138.3±13.6 |
| Total cholesterol (mg/dL) | 160.8±27.3 | 182.5±36.8 | 161.2±37.0 | 154.7±112.7 | 164.1±38.4 | 157.9±115.7 | 119.3±16.7 |
| Triglyceride (mg/ dL) | 96.0±49.1 | 102.7±33.5 | 95.8±37.2 | 93.1±47.5 | 92.7±38.2 | 95.3±47.8 | 103.0±19.3 |
| HDL (mg/dL) * | 46.8±16.2 | 56.3±16.5 | 43.1±12.1 (1) | 42.7±13.6 (2) | 49.0±14.3 (1) | 42.8±13.5 (2) | 26.0±8.7 |
| LDL (mg/dL) * | 94.8±19.6 | 105.7±34.2 | 97.9±30.5 (1) | 94.2±114.7 (2) | 96.1±30.4 (1) | 97.0x118.2 (2) | 72.7±9.3 |
Data are the means± standard deviation. BMI body mass index (the individual’s body weight [in kilograms] divided by the square of his or her height [in meters]).
*Numbers in parentheses are the number of patients with missing data.
Tissue classification by texture analysis
30 texture features were selected with Fisher +POE+ACC+ MI methods in MaZda from 279 original parameters calculated in all patients for classifying the fibrosis stage or necroinflammatory activity grade in T2W, T1W and EOB-HP images, respectively. The misclassification rates, calculated across all images, were 28.46% for fibrosis staging and 34.15% for necroinflammatory activity grading using LDA in combination with 1-NN classification (LDA/1-NN) in T2W imaging depending on the method of feature selection (Fisher+POE+ACC+MI), 35.77% for fibrosis staging, 25.20% for necroinflammatory activity grading in T1W imaging, 20.33% for fibrosis staging and 28.46% for necroinflammatory activity grading in EOB-HP imaging, respectively. The clusters of samples were presented in two- or three-dimensional space of selected features, demonstrating the graphical relationship between the MDFs. The number of MDF axes is less than the number of the output classes by one, and so the sample distributions are visualized in three-dimensional space for classification of the fibrosis stages (F1~4) and in two-dimensional space for classification of necroinflammatory grades (A1~3) (Figs. 2 and 3).
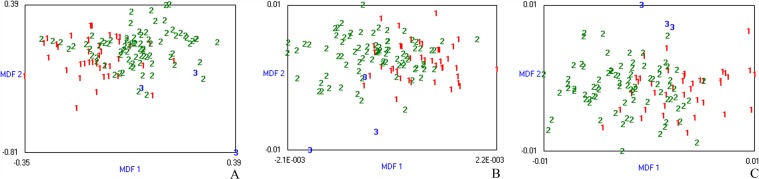
Fig 2. Discrimination of fibrosis stages based on all patient dataset.
Misclassification rates were 28.46%, 35.77% and 20.33% in T2W (A), T1W (B) and EOB-HP (C) images, respectively. The three-dimensional distribution of data vectors is based on the top three of the 30 texture features that were extracted using Fisher coefficients + classification error probability combined with average correlation coefficients + mutual information coefficients (Fisher+POE+ACC+MI) methods, followed by linear discrimination analysis /1-nearest neighbor (LDA/1-NN) classification: fibrosis stage 1 (1), stage 2 (2), stage 3 (3), and stage 4 (4). Most discriminating factor1 (MDF1), MDF 2 and MDF 3 are the most discriminating feature axes used in LDA to represent the classification graphically.
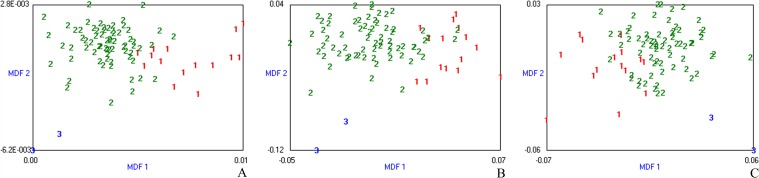
Fig 3. Discrimination of necroinflammatory activity grades based on all patient dataset.
Misclassification rates were 34.15%, 25.20% and 28.46% in T2W (A), T1W (B) and EOB-HP (C) images, respectively. The three-dimensional distribution of data vectors is based on the top three of the 30 texture features that were extracted using Fisher+POE+ACC+MI method, following by LDA/1-NN classification: necroinflammatory activity grade 1 (1), grade 2 (2) and grade 3 (3). MDF1 and MDF 2 are the most discriminating features axes used in LDA to represent the classification graphically.
To avoid interference between fibrosis stage and necroinflammatory activity grade on texture features, the classification in the subgroups of necroinflammatory activity grades or fibrosis stages was further investigated. Only A1 group and F4 group met the condition that the sample size for each subgroup must exceed one. Further classification of A1 group for fibrosis staging and F4 group for necroinflammatory activity grading was run. 30 texture features were extracted using the same Fisher+POE+ACC+MI methods as before in MaZda in T2W, T1W and EOB-HP images, respectively. LDA/1-NN resulted in misclassification rates of 5.00% for fibrosis staging of A1 group and 5.88% for necroinflammatory activity grading of F4 group in T2W images. LDA/1-NN resulted in misclassification rates of 0% for fibrosis staging of A1 group and 12.94% for necroinflammatory activity grading of F4 group in T1W images. LDA/1-NN resulted in misclassification rates of 12.50% for fibrosis staging of A1 group and 11.76% for necroinflammatory activity grading of F4 group in EOB-HP images (Table 4). The separation of clusters reflects better discrimination in A1 or F4 groups for fibrosis staging or necroinflammatory activity grading than that in the all patient dataset (Figs. 4 and 5).
Table 4. Results of texture-based classification of liver fibrosis stages or necroinflammatory activity grades of chronic liver hepatitis C calculated for T2W, T1W and EOB-HP imaging sequences, according to linear discrimination analysis /1-nearest neighbor (LDA/1-NN) classification method.
| T2W (mis %) | T1W (mis %) | EOB-HP (mis %) | |
|---|---|---|---|
| Classification of fibrosis staging in all patients | 28.46 | 35.77 | 20.33 |
| Classification of necroinflammatory activity grading in all patients | 34.15 | 25.20 | 28.46 |
| Classification of fibrosis staging in A1 patient group | 5.00 | 0 | 12.50 |
| Classification of necroinflammatory activity grading in F4 patient group | 5.88 | 12.94 | 11.76 |
Misclassification percentage (mis %) was given in columns.
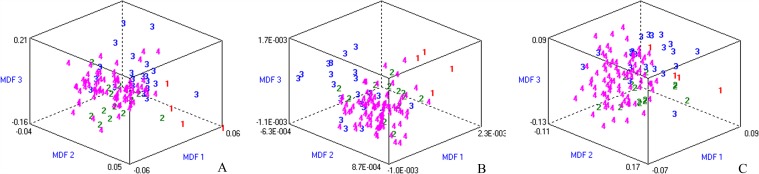
Fig 4. Discrimination of fibrosis stages based on patient group with necroinflammatory activity grade 1.
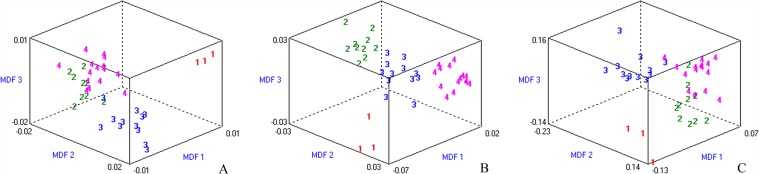
Misclassification rates were 5.00%, 0% and 12.50% in T2W (A), T1W (B) and EOB-HP (C) images, respectively. The three-dimensional distribution of data vectors is based on the top three of the 30 texture features that were extracted using Fisher+POE+ACC+MI method, followed by LDA/1-NN classification: fibrosis stage 1 (1), stage 2 (2), stage 3 (3), and stage 4 (4). MDF1, MDF 2 and MDF 3 are the most discriminating features axes used in LDA to represent the classification graphically.
Fig 5. Discrimination of necroinflammatory activity grades based on patient group with fibrosis stage 4.
Misclassification rates were 5.88%, 12.94% and 11.76% in T2W (A), T1W (B) and EOB-HP (C) images, respectively. The three-dimensional distribution of data vectors is based on the top three of the 30 texture features that were extracted using Fisher +POE+ACC +MI method, following by LDA/1-NN classification: necroinflammatory activity grade 1 (1), grade 2 (2) and grade 3 (3). MDF1 and MDF 2 are the most discriminating features axes used in LDA to represent the classification graphically.
Discussion
Our results demonstrated that texture features can not only differentiate normal liver from cirrhotic liver, but also have the ability to stratify the cirrhosis severity [13]. Furthermore, texture features showed the ability to evaluate the necroinflammatory activity grade as well as fibrosis stage, in contrast to many other methods that are more useful in discriminating the fibrosis stage rather than necroinflammatory activity grade. In the previous literature, the contrast enhancement index on EOB-HP showed a significant correlation with fibrosis stage but no correlation with necroinflammatory activity grades [11], and liver stiffness measurement using transient elastography could accurately reflect the amount of fibrosis but not determine the grade of necroinflammatory activity [29]. The discriminating ability of texture analysis revealed in this study is speculated to relate to the histopathological characteristics. The necroinflammatory process in chronic viral hepatitis, characterized by lymphocyte infiltration and hepatocyte damage [30, 31], is surmised to destroy the homogeneity of the liver parenchyma, which could be reflected by various texture features. The influence of fibrosis over texture could be deduced from the formation of fibrous septa and variously sized nodules [30, 31].
Until now, no widely accepted criterion for inclusion of texture features in evaluating MR images has been available. Past studies on texture analysis may have focused on all or some of the above parameters. In the study of Kato et al [32], the degree of fibrosis was accurately correlated with seven texture features. Given the diversity of derivation approaches of texture features, we expanded the data pool that included 279 texture parameters in this study, and 30 texture parameters were selected as independent variables, being relatively more numerous than those in the previous studies [29, 32–34]. In our opinion, a big data pool may be of benefit to flexible selection and application of texture parameters. Even though texture analysis was based on the same data pool, mostly different texture parameters were selected for different purposes, and these selected texture features could be used to discriminate either the fibrosis stage or necroinflammatory activity grade.
In this study, the misclassification rates for fibrosis staging in A1 group were much less than those for fibrosis staging in all patients in T2W, T1W or EOB-HP images. This suggests that the discriminating ability for classification of fibrosis stages can be improved when the influence of necroinflammatory activity grades is eliminated. Because progression of the fibrosis stage and necroinflammatory activity grade are not necessarily parallel in chronic hepatitis C, patients with the same fibrosis stage may have different necroinflammatory activity grades, and so the dataset containing different necroinflammatory activity grades will decrease the accuracy of classification of fibrosis stages using texture analysis method. Likewise, the accuracy of activity grade classification was also improved after the dataset was stratified in terms of the fibrosis stage. So we hypothesize that the texture features are related to both the fibrosis stage and necroinflammatory activity grade in chronic hepatitis C, and that furthermore, the influences on texture parameters from fibrosis stage and necroinflammatory activity grade may overlap to some extent. The subgroups divided by fibrosis or necroinflammatory activity degrees can be more accurately classified for activity grading or fibrosis staging based on the texture analysis method.
We also note that the differentiation ability for fibrosis staging is as good as that of necroinflammatory activity grading, which extends the application of texture analysis in the evaluation of chronic hepatitis C. This finding is a good proof for the powerful effect of texture analysis, which can be attributed to the enormous dataset calculated by many different methods including histogram, gradient, run-length matrix, co-occurrence matrix, autoregressive model and wavelet transformation methods. In this way the useful texture parameters selected differ according to the classification purpose and produce the optimum composition leading to the best results in either fibrosis or necroinflammatory activity classification.
There are several limitations in this study. First, we did not divide the patient population into a training and a test dataset, because of the small size of some subsets. We designed this study as a preliminary study that focused on the general feasibility of texture-based classification of fibrosis or necroinflammatory activity degrees, rather than constructing a working classification model at present. In addition, the 1-NN classifier using leave-one-out technique implemented in the MaZda program does not require a separate training dataset [35]. Second, the small sample size of patients with F1A2, F1A3, F2A3 and F3A3 was another drawback of this study, and so we only classified the A1 group for fibrosis staging, and F4 group for necroinflammatory activity grading, which was also limited by its retrospective nature.
In conclusion, based on the texture parameters calculated in T2W, T1W and EOB-HP images from patients with chronic hepatitis C, we conclude that texture analysis with the input dataset categorized according to activity or fibrosis degree shows promise as a useful quantitative tool for comprehensively evaluating the progress of fibrosis and necroinflammatory activity.
Supporting Information
(XLS)
(ZIP)
(ZIP)
Data Availability
All relevant data are within the paper and its Supporting Information files. Additional data are available upon request to the corresponding author.
Funding Statement
This study was supported by Japan China Sasakawa Medical Fellowship. The fellowship fund sponsored 1 year of research for Zhuo Wu in Japan, and had the role in study design, data collection and analysis.
References
- 1. Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 2006;45:529–538. [DOI] [PubMed] [Google Scholar]
- 2. Ghany MG, Strader DB, Thomas DL, Seeff LB; American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 2009;49:1335–1374. 10.1002/hep.22759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manning DS, Afdhal NH. Diagnosis and quantitation of fibrosis. Gastroenterology 2008;134:1670–1681. 10.1053/j.gastro.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 4. West J, Card TR. Reduced mortality rates following elective percutaneous liver biopsies. Gastroenterology 2010;139:1230–1237. 10.1053/j.gastro.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 5. Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59:1061–1069. [DOI] [PubMed] [Google Scholar]
- 6. Mahmoud-Ghoneim D, Cherel Y, Lemaire L, de Certaines JD, Maniere A. Texture analysis of magnetic resonance images of rat muscles during atrophy and regeneration. Magn Reson Imaging 2006;24:167–171. [DOI] [PubMed] [Google Scholar]
- 7. Harrison LC, Luukkaala T, Pertovaara H, Saarinen TO, Heinonen TT, Järvenpää R, et al. Non-Hodgkin lymphoma response evaluation with MRI texture classification. J Exp Clin Cancer Res 2009;28:87 10.1186/1756-9966-28-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amin A, Mahmoud-Ghoneim D. Zizyphus spina-christi protects against carbon tetrachloride-induced liver fibrosis in rats. Food Chem Toxicol 2009;47:2111–2119. 10.1016/j.fct.2009.05.038 [DOI] [PubMed] [Google Scholar]
- 9. Wong VS, Wight DG, Palmer CR, Alexander GJ. Fibrosis and other histological features in chronic hepatitis C virus infection: a statistical model. J Clin Pathol 1996;49:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Semelka RC, Chung JJ, Hussain SM, Marcos HB, Woosley JT. Chronic hepatitis: correlation of early patchy and late linear enhancement patterns on gadolinium-enhanced MR images with histopathology initial experience. J Magn Reson Imaging 2001;13:385–391. [DOI] [PubMed] [Google Scholar]
- 11. Watanabe H, Kanematsu M, Goshima S, Kondo H, Onozuka M, Moriyama N, et al. Staging hepatic fibrosis: comparison of gadoxetate disodium-enhanced and diffusion-weighted MR imaging—preliminary observations. Radiology 2011;259:142–150. 10.1148/radiol.10100621 [DOI] [PubMed] [Google Scholar]
- 12. Jirák D, Dezortová M, Taimr P, Hájek M. Texture analysis of human liver. J Magn Reson Imaging 2002;15:68–74. [DOI] [PubMed] [Google Scholar]
- 13. Ichida F, Tsuji T, Omata M, Ichida T, Inoue K, Kamimura T, et al. New Inuyama classification new criteria for histological assessment of chronic hepatitis. Int Hepatol. Commun 1996;6:112–119. 10.1016/S0928-4346(96)00325-8 [DOI] [Google Scholar]
- 14. Szczypiński PM, Strzelecki M, Materka A, Klepaczko A. MaZda—a software package for image texture analysis. Comput Methods Programs Biomed 2009; 94:66–76. 10.1016/j.cmpb.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 15.Szczypiński PM, Strzelecki M, Materka A. MaZda—a Software for Texture Analysis, Proc. of International Symposium on Information Technology Convergence (ISITC), 2007 November 23–24, Republic of Korea: 245–249. Available: http://ieeexplore.ieee.org/xpl/ Accessed 26 June 2014.
- 16. Strzelecki M, Szczypiński PM, Materka A, Klepaczko A. A software tool for automatic classification and segmentation of 2D/3D medical images, Nucl Instrum Methods Phys Res A. 2013;702:137–140. 10.1016/j.nima.2012.09.006 [DOI] [Google Scholar]
- 17. Hu Y, Dennis T. Textured image segmentation by context enhanced clustering. IEE Proc Vis Image Signal Process 1994;141:413–421. [Google Scholar]
- 18. Kashyap R, Chellappa R. Estimation and choice of neighbors in spatial interaction models of images. IEEE Trans Inf Theory. 1983;29:60–72. [Google Scholar]
- 19. Mayerhoefer ME, Schima W, Trattnig S, Pinker K, Berger-Kulemann V, Ba-Ssalamah A. Texture-based classification of focal liver lesions on MRI at 3.0 Tesla: a feasibility study in cysts and hemangiomas. J Magn Reson Imaging 2010;32:352–359. 10.1002/jmri.22268 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y. MRI texture analysis in multiple sclerosis. Int J Biomed Imaging. 2012:762804 10.1155/2012/762804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen P, Pavlidis T. Segmentation by texture using a co-occurrence matrix and a split-and-merge algorithm. Comput Graph Image Proc 1979;10:172–182. [Google Scholar]
- 22. Haralick R, Shanmugam K, Dinstein I. Texture features for image classification. IEEE Trans. Systems Man Cybernetics 1973; SMC-3:610–621. 10.1109/TSMCB.2010.2060325 [DOI] [PubMed] [Google Scholar]
- 23. Mallat S. Multifrequency channel decompositions of images and wavelet models. IEEE Trans Acoust 1989;37:2091–2110. [Google Scholar]
- 24. Laine A, Fan J. Texture classification by wavelet packet signatures. IEEE Trans Pattern Anal Mach Intell 1993;15:1186–1191. [Google Scholar]
- 25. Kassner A, Thornhill RE. Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol 2010;31:809–816. 10.3174/ajnr.A2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Materka A, Strzelecki M, Szczypinski PM. MaZda manual. 2006. Available: http://www.eletel.p.lodz.pl/programy/mazda/download/mazda_manual.pdf Accessed 12 April 2012.
- 27. Harrison LC, Raunio M, Holli KK, Luukkaala T, Savio S, Elovaara I, et al. MRI texture analysis in multiple sclerosis: toward a clinical analysis protocol. Acad Radiol 2010;17:696–707. 10.1016/j.acra.2010.01.005 [DOI] [PubMed] [Google Scholar]
- 28. Fukunaga K. Introduction to statistical pattern recognition. 2nd ed New York: Academic Press;1991. [Google Scholar]
- 29. Marcellin P, Ziol M, Bedossa P, Douvin C, Poupon R, de Lédinghen V, et al. Non-invasive assessment of liver fibrosis by stiffness measurement in patients with chronic hepatitis B. Liver Int 2009;29:242–247. 10.1111/j.1478-3231.2008.01802.x [DOI] [PubMed] [Google Scholar]
- 30. Guido M, Mangia A, Faa G, Gruppo Italiano Patologi Apparato Digerente (GIPAD), Società Italiana di Anatomia Patologica e Citopatologia Diagnostica/International Academy of Pathology, Italian division (SIAPEC/IAP). Chronic viral hepatitis: the histology report. Dig Liver Dis 2011;43 Suppl 4:S331–343. 10.1016/S1590-8658(11)60589-6 [DOI] [PubMed] [Google Scholar]
- 31. Rahn SB. Liver biopsy interpretation in chronic hepatitis. J Insur Med 2001;33:110–113. [PubMed] [Google Scholar]
- 32. Kato H, Kanematsu M, Zhang X, Saio M, Kondo H, Goshima S, et al. Computer-aided diagnosis of hepatic fibrosis: preliminary evaluation of MRI texture analysis using the finite difference method and an artificial neural network. AJR Am J Roentgenol 2007;189:117–122. [DOI] [PubMed] [Google Scholar]
- 33. Duan J, Hu C, Luo S, Zhao X, Wang T. Microcomputed tomography with diffraction-enhanced imaging for morphologic characterization and quantitative evaluation of microvessel of hepatic fibrosis in rats. PLoS One 2013;21;8:e78176 10.1371/journal.pone.0078176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herlidou S, Grebe R, Grados F, Leuyer N, Fardellone P, Meyer ME.Influence of age and osteoporosis on calcaneus trabecular bone structure: a preliminary in vivo MRI study by quantitative texture analysis. Magn Reson Imaging 2004;22:237–243. [DOI] [PubMed] [Google Scholar]
- 35. Hajek M, Dezortova M, Materka A, Lerski R. Texture analysis for magnetic resonance imaging Pragure: Med4Publishing; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(ZIP)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Additional data are available upon request to the corresponding author.