Abstract
AIM: To investigate GATA5, SFRP2, and ITGA4 methylation in plasma DNA as noninvasive biomarkers for colorectal cancer (CRC) or adenomas.
METHODS: There were 57 CRC patients, 30 adenomas patients, and 47 control patients enrolled in this study. Methylation-specific polymerase chain reaction was used to determine the promoter methylation status of GATA5, SFRP2, and ITGA4 genes in plasma DNA, and their association with clinical outcome in CRC. The predictive ability of GATA5, SFRP2, and ITGA4 methylation, individually or in combination, to detect CRC or adenomas was further analyzed.
RESULTS: Hypermethylated GATA5 was detected in plasma in 61.4% (35/57) of CRC cases, 43.33% (13/30) of adenoma cases, and 21.28% (10/47) of control cases. The hypermethylation of SFRP2 was detected in 54.39% (31/57), 40.00% (12/30), and 27.66% (13/47) in plasma samples from CRC, adenomas, and controls, respectively. ITGA4 methylation was detected in 36.84% (21/57) of plasma samples of CRC patients and in 30.00% (9/30) of plasma samples from patients with colorectal adenomas, and the specificity of this individual biomarker was 80.85% (9/47). Moreover, GATA5 methylation in the plasma was significantly correlated with larger tumor size (P = 0.019), differentiation status (P = 0.038), TNM stage (P = 0.008), and lymph node metastasis (P = 0.008). SFRP2 and ITGA4 methylation in plasma significantly correlated with differentiation status (SFRP2, P = 0.012; ITGA4, P = 0.007), TNM stage (SFRP2, P = 0.034; ITGA4, P = 0.021), and lymph node metastasis (SFRP2, P = 0.034; ITGA4, P = 0.021). From the perspective of predictive power and cost-performance, using GATA5 and SFRP2 together as methylation markers seemed the most favorable predictor for CRC (OR = 8.06; 95%CI: 2.54-25.5; P < 0.01) and adenomas (OR = 3.35; 95%CI: 1.29-8.71; P = 0.012).
CONCLUSION: A combination of GATA5 and SFRP2 methylation could be promising as a marker for the detection and diagnosis of CRC and adenomas.
Keywords: Colorectal cancer; GATA binding protein 5; Secreted frizzled-related protein 2; Integrin, alpha 4; Hypermethylation; Methylation-specific PCR
Core tip: Hypermethylated GATA5 was identified as a novel plasma gene in colorectal cancer (CRC) and adenomas, and it showed high potential as a biomarker in plasma-based DNA testing. Furthermore, this study suggests that a combination of GATA5 and SFRP2 methylation could be used as a promising marker for the detection, diagnosis, and prognosis of CRC and adenomas.
INTRODUCTION
Colorectal cancer (CRC) is the third most prevalent cancer and the fourth leading cause of cancer-related mortality worldwide. Over 1 million new cases are diagnosed annually worldwide, and approximately 50% of these patients will die of this disease[1]. The mean 5-year survival rate for CRC is estimated to be less than 10% if the cancer is detected at stage IV, but can be as high as 90% for stage I cases[2,3]. Therefore, identifying and treating CRC in its early stage or with pre-malignant lesions is of great importance in reducing disease-specific mortality. Currently, colonoscopy is the gold standard for CRC diagnosis[4,5]. However, this procedure is invasive and uncomfortable to many patients, who are therefore reluctant to undergo colonoscopy. A significant advance in screening could be realized by using blood-based indicators, which could be sensitive and specific for identifying patients at risk of CRC development. These patients may benefit from early and/or more frequent surveillance for CRC[6,7].
Recently, epigenetic mutations of specific genes as marker candidates for the early detection of cancer have received considerable attention[8,9]. Aberrant methylation of CpG islands in the promoter regions of genes are commonly associated with transcriptional silencing of tumor suppressor genes and have been found to be crucial in the early phases of CRC carcinogenesis[10,11]. It is widely accepted that CRC develops following progressive accumulation of genetic and epigenetic alterations during the transformation of normal mucosa to a precursor adenoma and ultimately to carcinoma. Since it takes 7-10 years for an adenoma to progress to a carcinoma, there is a window of opportunity for detecting and resecting advanced adenomas or early-stage CRC; mass screening aids in this detection[12,13]. Studies have demonstrated that there are higher levels of freely circulating methylated DNA in the peripheral blood of CRC patients than in healthy control patients, and DNA methylation often occurs very early during CRC carcinogenesis[14,15]. Several genes such as DAPK1, SEPT9, RUNX3, or vimentin have been reported to be methylated in the serum/plasma of CRC patients and can potentially be used as epigenetic biomarkers in a noninvasive manner for early detection of CRC[16-19]. The detection of circulating methylated DNA in the serum or plasma represents one of the most promising methods for the early detection and diagnosis of CRC and adenomas.
For the present study, we propose a panel of genes that have been reported to be frequently methylated in CRC and adenoma tissues. However, few studies have investigated the methylation of these genes in DNA from plasma samples of CRC patients, in parallel with samples from healthy individuals and from patients with adenomas. The genes evaluated in this study were GATA-binding protein 5 (GATA5), secreted frizzled-related protein gene 2 (SFRP2), and integrin, alpha 4 (ITGA4). The methylation-specific polymerase chain reaction (MSP) technique was used to analyze the specificity and sensitivity of this method for detecting CRC and adenomas and to evaluate the clinical diagnostic significance of these DNA methylation-based plasma markers.
MATERIALS AND METHODS
Patients and plasma samples
Fifty seven CRC patients, 30 patients with adenomas, and 47 control patients with endoscopically normal colons were enrolled in this study at Li Huili Hospital (Ningbo, China) between April 2012 and April 2013. The mean age of the patients in the CRC, adenoma, and control groups was 56.64 ± 8.27, 57.00 ± 11.27, and 61.40 ± 12.41 years, respectively. The ratio of male to female patients was 34:23 in the CRC group, 19:11 in the adenoma group, and 27:20 in the control group. There were no significant differences in age and gender between the CRC group, adenoma group, and control group (data not shown). None of the enrolled patients had previously received preoperative chemotherapy or radiation therapy. This study was approved by the ethics committee of Li Huili Hospital (Ningbo, China), and informed consent was obtained from all participants. All the patients were diagnosed with CRC based on pathological and/or cytological evidence. Tumor stage was determined according to the tumor node metastasis (TNM) criteria of the Union for International Cancer Control/American Joint Committee on Cancer, 2010[20]. The plasma samples were collected prior to treatment in the patient and control groups. The plasma samples were immediately isolated by centrifugation at 1000 × g for 10 min and stored at -80 °C until use for DNA extraction.
DNA isolation
DNA was isolated from each plasma sample (200 μL) using the QIAamp DNA Blood mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Plasma DNA was dissolved in a total volume of 80 μL of elution buffer (EB) and stored at -20 °C until use in the experiments.
Sodium bisulfite conversion
Sodium bisulfite conversion and DNA recovery were performed using the Qiagen Epitect Plus DNA bisulfite kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA was then resuspended in 30 μL of EB and stored at -20 °C.
MSP
The methylation of GATA5, SFRP2, and ITGA4 promoters in the bisulfite-modified DNA was increased using MSP, and primer pairs were designed to discriminate between methylated and unmethylated alleles. The primer sequences used are shown in Table 1.
Table 1.
Summary of primer sequences and annealing temperatures used for methylation-specific polymerase assays
| Gene | Primer | Sequence (5'-3') | Annealing temperature (°C) | Ref. |
| GATA5 | MF | TTAGAAATCGAGGAAATCGC | 54 | |
| MR | GTAAACCCCCTCGTTACGTA | |||
| UF | TGTTTAGAAATTGAGGAAATTGT | 48 | ||
| UR | CCCATAAACCCCCTCATTACATA | |||
| SFRP2 | MF | TTTTTGTAGGGGCGTTTTTATAAC | 58 | [21] |
| MR | TATCGATATACTCCCCAATACCG | |||
| UF | AGATTTTTGTAGGGGTGTTTTTATAAT | 54 | ||
| UR | ACCTATCAATATACTCCCCAATACCA | |||
| ITGA4 | MF | TAGAGTTATTTCGCGTTTTGCG | 56 | [22] |
| MR | CTTCGAATACTCGCGCTACTT | |||
| UF | GTTTAGAGTTATTTTGTGTTTTGTG | 50 | ||
| UR | AAAACTTCAAATACTCACACTACT |
M: Methylated; U: Unmethylated; F: Forward; R: Reverse.
Each 50 μL reaction mixture consisted of 2 μL of bisulfite-modified DNA template, 10 μL of 1 × KAPA2G buffer (Kapa Biosystems, Woburn, MA, United States), 1 μL of 10 mmol/L dNTP mix (Kapa Biosystems), 1 μL of each primer (50 mmol/L), and 0.5 units of KAPA2GTM Robust Hotstart DNA polymerase (Kapa Biosystems). The thermocycler conditions included a single cycle at 95 °C for 5 min; 10 cycles of 95 °C for 30 s, Tm + 8 °C (decreasing by 0.8 °C for each cycle) for 60 s, and 72 °C for 30 s; 38 cycles of 95 °C for 30 s, Tm for 60 s, and 72 °C for 30 s; and a final extension step for 10 min at 72 °C. The PCR products were then electrophoresed on a 2.5% agarose gel and visualized under ultraviolet illumination (ChemiDoc XRS; Bio-Rad, Hercules, CA, United States). Each experiment was performed in triplicate to validate the results. The researchers who performed all the assays were blinded to all the clinical information.
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, United States) was used for all the statistical analyses. The sensitivity and specificity with 95%CIs of plasma DNA assays were calculated. To compare the characteristics of different groups of patients, the χ2 test or Fisher’s exact test were used. Odds ratios (ORs) with the corresponding 95%CIs were used to assess the association between these methylation genes. P < 0.05 was considered statistically significant.
RESULTS
Frequencies of GATA5, SFRP2, ITGA4 promoter methylation in the plasma of individuals with CRC or adenomas
The MSP assay was used to detect the methylation of DNA extracted from peripheral blood plasma. Methylation of GATA5, SFRP2, and ITGA4 was observed in 61.4% (35/57), 54.39% (31/57), and 36.84% (21/57), respectively, of CRC patients (Figure 1). The presence of GATA5, SFRP2, and ITGA4 methylation was detected in 43.33% (13/30), 40.00% (12/30), and 30.00% (9/30), respectively, of patients with adenomas. GATA5, SFRP2, and ITGA4 methylation were detected in 21.28% (10/47), 27.66% (13/47), and 19.15% (9/47), respectively, of the healthy controls (Figure 1). The methylation frequency of all three genes was significantly higher in CRC plasma samples than in normal plasma samples (GATA5, P < 0.01; SFRP2, P < 0.01; ITGA4, P = 0.048) (Figure 1). The difference in the levels of GATA5 methylation in plasma samples from patients with adenomas compared with that for normal controls was statistically significant, while no statistically significant difference was identified between SFRP2 or ITGA4 methylation levels in the plasma DNA of patients with adenoma and of normal controls (GATA5, P = 0.039; SFRP2, P = 0.259; ITGA4, P = 0.273). Additionally, there was no significant difference in the levels of the three genes in the plasma of CRC and adenoma patients (GATA5, P = 0.107; SFRP2, P = 0.202; ITGA4, P = 0.426). Representative agarose gel electrophoresis results of the MSP for the three genes are shown in Figure 2.
Figure 1.
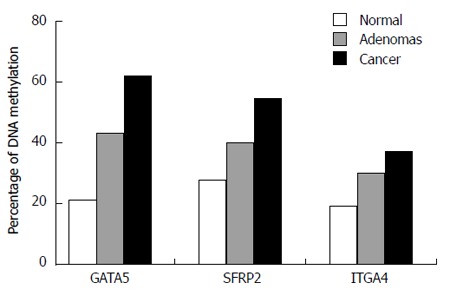
Frequency of detecting methylated DNA in the plasma of colorectal cancer, adenomas, and control samples.
Figure 2.
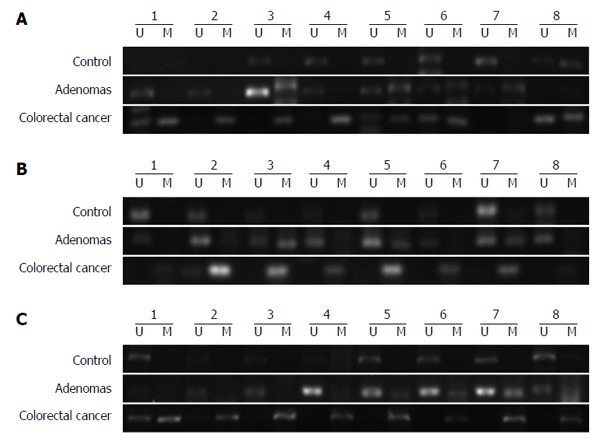
Representative methylation-specific polymerase results of GATA5 (A), SFRP2 (B) and ITGA4 (C) aberrant methylation in colorectal cancer, adenomas, and control patients. U: Results obtained using unmethylated primers; M: Results obtained using methylated primers.
Correlation between DNA methylation and clinicopathological features in plasma
GATA5 methylation in the plasma correlated significantly with larger tumor size (P = 0.019), differentiation status (P = 0.038), TNM stage (P = 0.008), and lymph node metastasis (P = 0.008). SFRP2 and ITGA4 methylation in plasma correlated significantly with differentiation status (SFRP2, P = 0.012; ITGA4, P = 0.007), TNM stage (SFRP2, P = 0.034; ITGA4, P = 0.021), as well as lymph node metastasis (SFRP2, P = 0.034; ITGA4, P = 0.021). There was no significant trend in the spread to distant metastasis for all three genes (GATA5, P = 0.151; SFRP2, P = 0.168; ITGA4, P = 0.620), probably owing to the small number of CRC patients with distant metastasis. In adenomas, the methylation status of the three analyzed genes was independent of adenoma size (GATA5, P = 0.431; SFRP2, P = 0.201; ITGA4, P = 1.000), number of adenomas (GATA5, P = 0.113; SFRP2, P = 0.130; ITGA4, P = 1.000), and intraepithelial neoplasia (GATA5, P = 0.643; SFRP2, P = 0.184; ITGA4, P = 0.329). Complete information regarding the distribution of markers and the clinicopathologic characteristics of the CRC and adenoma samples is shown in Table 2.
Table 2.
Clinicopathological features and DNA hypermethylation in plasma samples of 57 colorectal cancer patients and 30 patients with adenomas
| Parameters | n |
GATA5 |
SFRP2 |
ITGA4 |
||||||
| M | U | P value | M | U | P value | M | U | P value | ||
| Colorectal cancer | ||||||||||
| Gender | ||||||||||
| Male | 34 | 20 | 14 | 0.627 | 21 | 13 | 0.174 | 10 | 24 | 0.157 |
| Female | 23 | 15 | 8 | 10 | 13 | 11 | 12 | |||
| Age, yr | ||||||||||
| ≤ 60 | 28 | 16 | 12 | 0.516 | 16 | 12 | 0.681 | 8 | 20 | 0.203 |
| > 60 | 29 | 19 | 10 | 15 | 14 | 13 | 16 | |||
| Tumor size, cm | ||||||||||
| < 5 | 42 | 22 | 20 | 0.019 | 21 | 21 | 0.266 | 13 | 29 | 0.123 |
| ≥ 5 | 15 | 13 | 2 | 10 | 5 | 8 | 7 | |||
| Differentiation | ||||||||||
| Well | 6 | 2 | 4 | 0.038 | 2 | 4 | 0.012 | 0 | 6 | 0.007 |
| Moderately | 36 | 20 | 16 | 16 | 20 | 11 | 25 | |||
| Poorly | 15 | 13 | 2 | 13 | 2 | 10 | 5 | |||
| TNM stage | ||||||||||
| I-II | 33 | 15 | 18 | 0.008 | 14 | 19 | 0.034 | 8 | 25 | 0.021 |
| III-IV | 24 | 20 | 4 | 17 | 7 | 13 | 11 | |||
| Lymph node metastasis | ||||||||||
| N0 | 33 | 15 | 18 | 0.008 | 14 | 19 | 0.034 | 8 | 25 | 0.021 |
| N1–3 | 24 | 20 | 4 | 17 | 7 | 13 | 11 | |||
| Distant metastasis | ||||||||||
| M0 | 53 | 31 | 22 | 0.151 | 27 | 26 | 0.168 | 19 | 34 | 0.620 |
| M1 | 4 | 4 | 0 | 4 | 0 | 2 | 2 | |||
| Location | ||||||||||
| Colon | 19 | 10 | 9 | 0.336 | 12 | 7 | 0.347 | 6 | 13 | 0.560 |
| Rectum | 38 | 25 | 13 | 19 | 19 | 15 | 23 | |||
| Adenomas | ||||||||||
| Tumor size, cm | ||||||||||
| ≤ 1 | 14 | 5 | 9 | 0.431 | 5 | 9 | 0.201 | 4 | 10 | 1.000 |
| > 1 | 16 | 8 | 8 | 7 | 9 | 5 | 11 | |||
| Tumor number | ||||||||||
| 1 | 26 | 13 | 13 | 0.113 | 12 | 14 | 0.13 | 8 | 18 | 1.000 |
| ≥ 2 | 4 | 0 | 4 | 0 | 4 | 1 | 3 | |||
| Intraepithelial neoplasia | ||||||||||
| Low | 24 | 11 | 13 | 0.673 | 8 | 16 | 0.184 | 6 | 18 | 0.329 |
| High | 6 | 2 | 4 | 4 | 2 | 3 | 3 | |||
Effect of combining measurement of plasma GATA5, SFRP2, and ITGA4 methylation for CRC and adenomas determination
ORs were determined to estimate the predictive index of methylation status of the three genes in the plasma for detecting CRC and adenomas. The ORs for GATA5, SFRP2 and ITGA4 methylation and the combined test results are shown in Table 3. The OR of GATA5 methylation was the most optimal of the three genes for predicting the presence of CRC (OR = 5.89; 95%CI: 2.44-14.18; P < 0.01) and adenomas (OR = 2.83; 95%CI: 1.04-7.73; P = 0.039). The OR for predicting CRC (OR = 8.06; 95%CI: 2.54-25.5; P < 0.01) was higher for combined detection of GATA5 and SERP2 methylation than for GATA5 methylation alone (OR = 5.89; 95%CI: 2.44-14.18; P < 0.01). Although the sensitivity decreased relative to that for single gene detection, the specificity for CRC increased when the two genes were used together. If both GATA5 and SFRP2 methylation markers were detected, the OR of the adenomas (OR = 3.35; 95%CI: 1.29-8.71; P = 0.012) was higher than that obtained using GATA5 (OR = 2.83; 95%CI: 1.04-7.73; P = 0.039) or SFRP2 (OR = 1.74; 95%CI: 0.66-4.60; P = 0.259) methylations individually. Even though the specificity (65.96%) for predicting adenomas was lower than when using GATA5 methylation alone (78.72%), the sensitivity was markedly increased by 20%. Overall, the combined detection of GATA5 and SFRP2 methylation appeared to be the most effective predictor of CRC and adenomas in terms of analytic validity, clinical validity, and clinical utility.
Table 3.
Comparison of predictive ability of GATA5, SFRP2, ITGA4 methylation, when used alone or combined, in colorectal cancer and adenomas
| Sensitivity (95%CI) | Specificity (95%CI) | Odds ratio (95%CI) | P value | |
| Adenomas | ||||
| GATA5 | 43.33% (25.46%-62.57%) | 78.72% (64.34%-89.30%) | 2.83 (1.04-7.73) | 0.039 |
| SFRP2 | 40.00% (22.66%-59.40%) | 72.34% (57.36%-84.38%) | 1.74 (0.66-4.60) | 0.259 |
| ITGA4 | 30.00% (14.37%-49.40%) | 80.85% (66.74%-90.85%) | 1.81 (0.62-5.26) | 0.273 |
| GATA5 or SFRP2 | 63.33% (43.86%-80.07%) | 65.96% (50.69%-79.14%) | 3.35 (1.29-8.71) | 0.012 |
| GATA5 or ITGA4 | 60.00% (40.60%-77.34%) | 65.96% (50.69%-79.14%) | 2.91 (1.13-7.50) | 0.025 |
| SFRP2 or ITGA4 | 55.67% (37.43%-74.53%) | 61.70% (46.38%-75.49%) | 1.48 (0.92-2.39) | 0.114 |
| GATA5 or SFRP2 or ITGA4 | 70.00% (50.60%-85.27%) | 55.32% (40.12%-69.83%) | 1.57 (1.06-2.33) | 0.030 |
| GATA5 and SFRP2 | 26.77% (12.28%-45.89%) | 91.49% (79.62%-90.00%) | 3.13 (1.03-9.51) | 0.032 |
| GATA5 and ITGA4 | 10.00% (2.11%-26.53%) | 82.46% (70.09%-91.25%) | 0.41 (0.10-1.64) | 0.231 |
| SFRP2 and ITGA4 | 13.33% (3.76%-30.72%) | 85.11% (71.69%-93.80%) | 0.88 (0.23-3.31) | 1.000 |
| GATA5 and SFRP2 and ITGA4 | 6.67% (0.82%-22.07%) | 93.62% (82.46%-98.66%) | 1.04 (0.19-5.89) | 1.000 |
| Colorectal cancer | ||||
| GATA5 | 61.40% (47.57%-74.00%) | 78.72% (64.34%-89.30%) | 5.89 (2.44-14.18) | < 0.01 |
| SFRP2 | 54.39% (40.66%-67.64%) | 72.34% (57.36%-84.38%) | 3.12 (1.37-7.12) | < 0.01 |
| ITGA4 | 36.84% (24.45%-50.66%) | 80.85% (66.74%-90.85%) | 2.46 (1.00-6.09) | 0.048 |
| GATA5 or SFRP2 | 73.68% (60.34%-84.46%) | 65.96% (50.69%-79.14%) | 5.43 (2.33-12.61) | < 0.01 |
| GATA5 or ITGA4 | 73.68% (60.34%-84.46%) | 65.96% (50.69%-79.14%) | 5.43 (2.33-12.61) | < 0.01 |
| SFRP2 or ITGA4 | 70.18% (56.60%-81.57%) | 61.70% (46.38%-75.49%) | 3.79 (1.67-8.59) | < 0.01 |
| GATA5 or SFRP2 or ITGA4 | 80.70% (68.09%-89.95%) | 55.32% (40.12%-69.83%) | 5.18 (2.16-12.41) | < 0.01 |
| GATA5 and SFRP2 | 42.86% (29.71%-50.00%) | 91.49% (79.62%-90.00%) | 8.06 (2.54-25.5) | < 0.01 |
| GATA5 and ITGA4 | 6.52% (1.37%-17.90%) | 82.46% (70.09%-91.25%) | 0.33 (0.08-1.27) | 0.094 |
| SFRP2 and ITGA4 | 21.05% (11.38%-33.89%) | 85.11% (71.69%-93.80%) | 1.52 (0.55-4.25) | 0.419 |
| GATA5 and SFRP2 and ITGA4 | 15.79% (7.48%-27.87%) | 93.62% (82.46%-98.66%) | 2.75 (0.7-10.82) | 0.217 |
DISCUSSION
The notion of using molecular tests to detect genetic and epigenetic abnormalities in blood DNA have been regarded as simple and noninvasive methods for CRC and adenomas screening in a large-scale population. Here, we show that the combined detection of GATA5 and SFRP2 methylation in plasma maybe an effective method for screening CRC and pre-cancerous adenomas.
Epigenetic alterations, leading to genetic silencing, often occur early in the progression of CRC, even in precancerous lesions. This suggests that detection of DNA methylation in plasma is helpful in the early diagnosis of tumors, evaluation of tumor metastasis and prognosis, and guiding clinical treatment[23]. Among a wide range of commonly methylated genes in CRC, only a few have undergone clinical trials for detection of CRC and are commercially available, such as SEPT9 (ColoVantage®) and vimentin (ColoSureTM)[17,24]. ColoVantage® test is a blood-based SEPT9 methylated DNA assay with an overall sensitivity of 90% (45/50) for methylated SEPT9 in the plasma and specificity of 88% (11/94) in the case of CRC, but this test only detected 12% (12/104) of adenomas[17]. ColoSureTM test is a fecal-based vimentin methylation assay, and a meta-analysis demonstrated its sensitivity ranging from 38%-88% and specificity ranging from 73%-100% for CRC in the feces, but vimentin methylation tests in blood are not commercially available yet[24]. However, our preliminary study revealed that the methylation of SEPT9 and vimentin in the plasma was extremely low in CRC patients using the MSP method (data not shown).
The majority of promising blood-based DNA methylation biomarkers are not commercially available, but are currently in research, development, or in clinical trials. A panel of biomarkers is also currently being investigated, since combinations of robust biomarkers achieve greater sensitivities than individual markers, and thus have great potential in diagnosis. In the current study, we evaluated three methylation markers, GATA5, SFRP2, and ITGA4 and found that 80.70% (46/57) of patients with CRC, 70.00% (21/30) of patients with adenomas, and 44.68% (21/47) of normal controls exhibited at least one methylated gene in their plasma samples.
GATA5, a zinc-finger transcription regulatory factor and a member of the GATA family of proteins (GATA1 to GATA6), is known to be functionally involved in cell lineage specification and cell differentiation during embryonic development of the heart, lung, urogenital tract, and gut epithelium[25,26]. GATA5 is thought to be a potential tumor suppressor in gastrointestinal tissues[27], a guiding factor in intestinal epithelial cell differentiation[28], and a mediator of carcinogenesis in CRC. Hellebrekers et al[27] showed GATA5 methylation in 79% (61/77) of CRC tissues and in 13% (13/100) of normal colon tissue samples from controls who did not have cancer. GATA5 methylation was also independent of clinicopathologic features. Currently, no report has illustrated a link between GATA5 methylation in plasma and CRC or adenoma. In our study, GATA5 methylation in plasma displayed the highest sensitivity among three genes for noninvasive detection of CRC and intestinal adenoma. The results showed that methylation of GATA5 was detected in 61.4% (35/57) of CRC patients, 43.33% (13/30) of patients with adenoma, and in 21.28% (10/47) of the control patients in plasma samples. Although the sensitivity and specificity were lower than those in previous studies involving CRC tissues, these results still have a high clinical value in the diagnosis of CRC and adenomas in plasma. Unlike previous reports, our results revealed that GATA5 methylation in the plasma was significantly correlated with larger tumor size (P = 0.019), differentiation status (P = 0.038), TNM stage (P = 0.008), and lymph node metastasis (P = 0.008), suggesting that GATA5 methylation may also be useful in determining the prognosis of CRC. However, there was no significant difference in the levels of the GATA5 gene in the plasma of CRC and adenoma patients. GATA5 methylation may be involved in the very early development of CRC, even in precursor adenomas. Furthermore, the MSP method is a type of qualitative method, rather than quantitative method (such as Q-MSP), and could not provide exact values, only the number with methylation or not, leading to statistical deviation. Thus, we need to expand our sample size or use the Q-MSP combined pyrosequencing method to validate the results in future.
Another interesting gene SFRP2, which is activated via the Wnt/β-catenin signaling pathway, was also investigated as a novel early detection marker in the plasma. Tang et al[29] investigated SFRP2 methylation in DNA from feces and serum of patients with CRC, adenoma, or controls. Sensitivity for methylated SFRP2 in fecal DNA was higher (46% in patients with adenomas; 84% in patients with CRC) than that for serum DNA (6% in patients with adenomas; and 67% in patients with CRC). However, serum SFRP2 methylation levels showed markedly higher specificity in CRCs (94%) than SFRP2 methylation levels in fecal DNA (54%). Moreover, serum SFRP2 methylation was significantly associated with poor differentiation grade (P = 0.019), serosal/subserosal invasion (P < 0.001), lymph node metastasis status (P < 0.001), and TNM stage (P < 0.001) of CRC. In the current study, SFRP2 methylation was detected in plasma samples of 54.39% of CRC patients and 40.00% of colorectal adenoma patients, and the specificity of this single biomarker was 72.34%. Similarly, SFRP2 methylation in plasma were significantly correlated with the differentiation status (P = 0.012), TNM stage (P = 0.034), and lymph node metastasis (P = 0.034).
Integrins are a superfamily of transmembrane glycoproteins that are involved in cell proliferation, differentiation, adhesion, and migration[30]. The altered expression of ITGA4 has shown a correlation with transformation or metastasis in several human cancers[31,32]. ITGA4 has been identified as a novel gene, which is methylated frequently in CRC. Methylated ITGA4 of tissue is present in 75% of colon adenomas (n = 27), 92% of colon adenocarcinomas (n = 69), and 6% of colon mucosa (n = 32). Methylated ITGA4 in the fecal sample was found in 69% (9/13) of patients with colon adenomas and in 21% (6/28) of patients with no polyps[33], but ITGA4 methylation changes in blood have not been previously investigated. In our study, ITGA4 methylation was detected in 36.84% (21/57) of CRC, 30% (9/30) of adenomas, and 19.15% (7/47) of controls. ITGA4 alone could not be considered a unique marker for cancer detection, because its sensitivity and specificity are relatively low.
Methylated GATA5, SFRP2, and ITGA4 can be used together as diagnostic markers for CRC and adenomas and for screening individuals at risk. We used the MSP assay to evaluate the reliability, sensitivity, and specificity of GATA5, SFRP2 and ITGA4 methylation to detect CRC and adenomas. On the basis of the MSP assay, we found that the three genes (GATA5, SFRP2, and ITGA4) exhibited high methylation levels in CRC and adenoma plasma samples. When analysis of GATA5 and SFRP2 genes was used together for an assay involving peripheral blood samples, the respective sensitivities and specificities were 42.86% and 91.49% for CRC detection (simultaneous detection of two genes, OR = 8.06; P < 0.01), and 63.33% and 65.96% for adenoma detection (detection of one of the two genes, OR = 3.35; P = 0.012. The combined detection of methylated GATA5 and SFRP2 in plasma will be tested in subsequent studies to evaluate their clinical performance.
To this end, further studies with larger amounts of plasma from CRC and adenoma patients will be needed to validate GATA5, SFRP2, and ITGA4 as biomarkers for population-based screening of CRC and pre-neoplastic disease. GATA5 methylation in plasma is the most frequently detected among the three genes across all CRC stages and precancerous lesions. Its combination with SFRP2 may also be useful for monitoring patients with CRC and adenomas. Since the sensitivity of the MSP detection method is low, the pyrosequencing technique will need to be used to verify whether GATA5 and SFRP2 provide a feasible and reliable noninvasive screening tool for CRC and adenomas with the use of blood samples. In the future, the 5-year survival rate of these patients will be tracked, and blood from these patients will be periodically collected to explore the clinical value and significance of GATA5 and SFRP2 in CRC prognosis.
COMMENTS
Background
Gastric cancer is one of the most frequently diagnosed malignancies in China. The search for better non-invasive biomarkers for colorectal cancer (CRC) remains ongoing. Free circulating, tumor-derived methylated DNA is a good target as a plasma marker for early detection. GATA5, SFRP2 and ITGA4 are potential tumor suppressor genes, and loss of GATA5, SFRP2 or ITGA4 expression are considered a critical step in the genesis and development of CRC.
Research frontiers
Aberrant methylation of CpG islands at the promoter regions of genes are commonly associated with transcriptional silencing of tumor suppressor genes and have been found to be better non-invasive biomarkers for the detection of CRC. Studies on the correlation of DNA methylation and clinicopathological characteristics could help clinicians to optimize selection of suitable candidates for CRC screening.
Innovations and breakthroughs
To date, no report has illustrated a link between GATA5, ITGA4 methylation in plasma and CRC or adenoma, also no report has analyzed the relationship between the levels of GATA5 and ITGA4 methylation in plasma and their clinicopathological characteristics. The current study showed that GATA5 methylation in plasma displayed the highest sensitivity among three genes for noninvasive detection of CRC and intestinal adenoma. GATA5 methylation in the plasma significantly correlated with larger tumor size differentiation status, TNM stage, and lymph node metastasis. Furthermore, using GATA5 and SFRP2 in plasma together as methylation markers seemed the most favorable predictor for CRC and adenomas.
Applications
Hypermethylated GATA5 was identified as a novel plasma gene in CRC and adenomas, and it showed high potential as a biomarker in plasma-based DNA testing. Furthermore, assessment of SFRP2 genes will be useful to raise the sensitivity or specificity and could be used as a promising marker for the detection and diagnosis of CRC and adenomas.
Terminology
GATA5, a zinc-finger transcription regulatory factor, is known to be functionally involved in cell lineage specification and cell differentiation during embryonic development of the heart, lung, urogenital tract, and gut epithelium. SFRP2, activated via the Wnt/β-catenin signaling pathway, was also investigated as a novel early detection marker in plasma. ITGA4, has shown correlation with transformation or metastasis in several human cancers, including in CRC.
Peer-review
In this study, the authors investigated the feasibility of detecting aberrant methylation of GATA5, SFRP2, and ITGA4 promoters in plasma DNA as noninvasive biomarkers for CRC or adenomas, and to evaluate the clinical utility of these markers.
Footnotes
Supported by Social Development Foundation of Ningbo, No. 2011C50022; Natural Science Foundation of Ningbo, No. 2012A610212; and the Scientific Innovation Team Project of Ningbo, No. 2013B82010.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 11, 2014
First decision: October 14, 2014
Article in press: December 1, 2014
P- Reviewer: Jiang WJ, Wang YD, Zhang GJ S- Editor: Qi Y L- Editor: Cant MR E- Editor: Wang CH
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, Dash C, Giardiello FM, Glick S, Levin TR, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–160. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 5.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected] Am J Gastroenterol. 2009;104:739–750. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 6.Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, Kim TS, Kim NK, Chung HC, An S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Cassinotti E, Melson J, Liggett T, Melnikov A, Yi Q, Replogle C, Mobarhan S, Boni L, Segato S, Levenson V. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int J Cancer. 2012;131:1153–1157. doi: 10.1002/ijc.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Y, Zhao H, Li H, Li X, Yang S. DNA methylation as an early diagnostic marker of cancer (Review) Biomed Rep. 2014;2:326–330. doi: 10.3892/br.2014.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 11.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140–148. doi: 10.1136/gut.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philipp AB, Nagel D, Stieber P, Lamerz R, Thalhammer I, Herbst A, Kolligs FT. Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer. BMC Cancer. 2014;14:245. doi: 10.1186/1471-2407-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H. High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett. 2003;194:99–105. doi: 10.1016/s0304-3835(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 17.Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, Fang JC, Samowitz WS, Heichman KA. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18:1225–1230. [PubMed] [Google Scholar]
- 19.Shirahata A, Hibi K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Res. 2014;34:4121–4125. [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Kato K, Iida S, Uetake H, Takagi Y, Yamashita T, Inokuchi M, Yamada H, Kojima K, Sugihara K. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer. 2008;122:603–608. doi: 10.1002/ijc.23143. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Jung EJ, Lee HS, Kim MA, Kim WH. Comparative analysis of DNA methylation between primary and metastatic gastric carcinoma. Oncol Rep. 2009;21:1251–1259. doi: 10.3892/or_00000348. [DOI] [PubMed] [Google Scholar]
- 23.Zitt M, Zitt M, Müller HM. DNA methylation in colorectal cancer--impact on screening and therapy monitoring modalities? Dis Markers. 2007;23:51–71. doi: 10.1155/2007/891967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ned RM, Melillo S, Marrone M. Fecal DNA testing for Colorectal Cancer Screening: the ColoSure™ test. PLoS Curr. 2011;3:RRN1220. doi: 10.1371/currents.RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 26.Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 27.Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med. 2011;34:E88–E95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 30.Hughes AL. Evolution of the integrin alpha and beta protein families. J Mol Evol. 2001;52:63–72. doi: 10.1007/s002390010134. [DOI] [PubMed] [Google Scholar]
- 31.Do SI, Ko E, Kang SY, Lee JE, Nam SJ, Cho EY, Kim DH. Aberrant DNA methylation of integrin α4 in human breast cancer. Tumour Biol. 2014;35:7079–7084. doi: 10.1007/s13277-014-1952-7. [DOI] [PubMed] [Google Scholar]
- 32.Uhm KO, Lee JO, Lee YM, Lee ES, Kim HS, Park SH. Aberrant DNA methylation of integrin alpha4: a potential novel role for metastasis of cholangiocarcinoma. J Cancer Res Clin Oncol. 2010;136:187–194. doi: 10.1007/s00432-009-0646-9. [DOI] [PubMed] [Google Scholar]
- 33.Ausch C, Kim YH, Tsuchiya KD, Dzieciatkowski S, Washington MK, Paraskeva C, Radich J, Grady WM. Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clin Chem. 2009;55:1559–1563. doi: 10.1373/clinchem.2008.122937. [DOI] [PubMed] [Google Scholar]


