Abstract
Vaccination with live attenuated rubella virus induces a strong immune response in most individuals. However, small numbers of subjects never reach or maintain protective antibody levels, and there is a high degree of variability in immune response. We have previously described genetic polymorphisms in HLA and other candidate genes that are associated with interindividual differences in humoral immunity to rubella virus. To expand our previous work, we performed a genome-wide association study (GWAS) to discover single-nucleotide polymorphisms (SNPs) associated with rubella virus–specific neutralizing antibodies. We identified rs2064479 in the HLA-DPB1 genetic region as being significantly associated with humoral immune response variations after rubella vaccination (P = 8.62 × 10−8). All other significant SNPs in this GWAS were located near the HLA-DPB1 gene (P ≤ 1 × 10−7). These findings demonstrate that polymorphisms in HLA-DPB1 are strongly associated with interindividual differences in neutralizing antibody levels to rubella vaccination and represent a validation of our previous HLA work.
Keywords: genome-wide association study, polymorphism, genetic, humoral, neutralizing antibody, immunity, measles-mumps-rubella vaccine
Rubella is usually mild, unless it occurs during pregnancy and infection spreads to the fetus. During the first trimester, up to 90% of cases of rubella virus infection can lead to fetal defects, including stillbirth [1]. Newborn infants diagnosed with congenital rubella syndrome can present with multiple ophthalmic, auditory, cardiac, and craniofacial defects [2]. On average, there are 100 000 worldwide cases of congenital rubella syndrome reported annually [3]. Humans are the only known host for rubella virus, making the disease a logical target for global eradication. However, incomplete vaccination strategies have led to recent outbreaks in Poland, Romania, and Japan [4–6]. These outbreaks are concerning because of the potential risk of subsequent exposure to mother and fetus.
The rubella virus vaccine strain currently licensed in the United States is RA 27/3. It is administered in a 2-dose series as a component of the measles-mumps-rubella (MMR II) vaccine. Seroprotective levels are as high as 98% after the second dose [7, 8]. Protective levels of humoral immunity are observed 20 years after vaccination [9]. Although vaccination may lead to lifetime protection, there is evidence of waning immunity, and we have previously reported a broad spectrum of interindividual differences in humoral responses to rubella vaccination, including subjects with antibody responses below the protective threshold [10–13].
Our laboratory has focused on explaining the genetic contributions to variations in rubella vaccine–induced immunity [14]. We have demonstrated that HLA genes play an important role in immune responses to rubella vaccine, accounting for up to 20% of the overall genetic variation observed in rubella virus–specific antibody levels [15]. In regard to humoral immunity, we have identified associations between HLA class I and II alleles, as well as polymorphisms in TNFA/TNFRSF1B, IL2B, RARB, DDX58, TRIM5, TRIM22, IRF9, and OAS2 [15–18], with interindividual differences in response to rubella vaccination. The biological relevance of the HLA-DPB1 locus for immune response to rubella vaccination is not well understood. We have reported several HLA allelic (DPB1*0401) and haplotypic (DRPB1*04-DQB1*03-DPB1*03 and DRB1*15/16-DQB1*06-DPB1*03) associations with rubella vaccine–induced antibodies that were verified in separate study cohorts [15]. We also demonstrated that HLA-DPB1 (*0401) homozygosity was significantly associated with rubella virus antibody levels [19]. Here, we extend our previous work and report the first genome-wide association study (GWAS) in children and younger adults who received live rubella virus vaccine. We identified a significant association between rs2064479 in the HLA-DPB1 gene and the levels of neutralizing antibody response. This work validates the growing database that demonstrates differences in responses to vaccination and viral infection associated with genetic polymorphisms in this HLA class II locus.
METHODS
Study Participants
The study cohort was a large population-based sample of 1174 healthy children and younger adults (age, 11–22 years) from all socioeconomic strata in Rochester, Minnesota. The total cohort consists of 3 separate recruitment efforts, and detailed descriptions of these cohorts have been published elsewhere [18, 20–24]. For 1101 children, a parent agreed to let their child join the current rubella vaccine study, and from these children we obtained a blood sample. All 1101 participants had written records of having received 2 doses of MMR II vaccine (Merck). The Institutional Review Board of the Mayo Clinic approved the study. Written informed consent was obtained from each adult subject and from the parents of all children who participated in the study.
Rubella Virus-Specific Neutralizing Antibodies
The description for assaying the levels of neutralizing antibodies against live rubella virus is nearly identical to that in our previous report [11]. Briefly, serial dilutions of subjects' sera were incubated with the rubella vaccine virus strain HPV77. After incubation, the virus/sera mixture was added to Vero cells cultured in a flat-bottomed 96-well plate and incubated for 72 hours at 37°C in 5% CO2. Cells were then fixed in cold methanol for 10 minutes, blocked with phosphate-buffered saline (PBS) supplemented with 5% skim milk (Difco; BD, New Jersey) and 0.1% Tween 20 for 30 minutes, and washed 3 times with PBS supplemented with 0.05% Tween 20 (PBS-T). Fixed cells were incubated with anti-E1 glycoprotein (Centers for Disease Control and Prevention, Atlanta, Georgia) for 30 minutes and washed 3 times with PBS-T. The secondary goat anti-mouse horseradish peroxidase–conjugated antibody (Invitrogen; Carlsbad, California) was added for 30 minutes. Plates were washed again, and antibody conjugate was visualized by adding NeA-Blue TMB substrate (Clinical Science Products; Mansfield, Massachusetts) for 10 minutes. The ODs were read at 450 nm/630 nm on an Eon microplate spectrophotometer (BioTek; Winooski, Vermont). The Loess method of statistical interpolation was used to estimate the median infectious dose from observed values [25]. The intraclass correlation coefficient for rubella virus–specific neutralizing antibody (NT50) measurements was 0.89.
GWAS
The genome-wide SNP typing method used for this study is essentially identical to that used in our previously published reports [26–28]. Briefly, DNA was extracted from each subject's blood specimen, using the Gentra Puregene Blood kit (Gentra Systems; Minneapolis, Minnesota) and quantified by Picogreen (Molecular Probes; Carlsbad, California). The genome-wide SNP typing was performed using the Infinium Omni 1 M-Quad SNP array (Illumina; San Diego, California). DNA samples underwent amplification, fragmentation, and hybridization onto each BeadChip, which were imaged on an Illumina BeadArray reader. Genotype calls based on clustering of the raw intensity data were made using BeadStudio 2 software. The resulting genotype data on SNPs were exported into SAS for analysis. Quality-control checks included genotyping reproducibility, sex checks, cryptic relatedness to identify similar/identical subjects, removal of SNPs when typing failed in samples for >1% of subjects, removal of subjects when typing failed for >1% of SNPs, elimination of monomorphic SNPs, removal of duplicate samples, and a Hardy–Weinberg Equilibrium check (SNPs with P < 1 × 10−7 were flagged as having poor genotyping quality). We assessed population substructure by means of the principal components approach implemented in EIGENSTRAT, using SNPs spanning the genome that were in low linkage disequilibrium (LD; defined as an r2 value of < 0.1) and had HWE P values of >1 × 10−3 [29]. We removed all subjects whose genetic background was farther than 15% of the way between the predominant White cluster and the other genetic clusters identified by the first two principal axes of genetic admixture.
Statistical Analysis
Demographic and vaccination history data were summarized for all study participants, using counts and percentages for categorical features and medians and interquartile ranges for quantitative features, including neutralizing antibody levels. Linear regression analyses were performed for each SNP to assess its association with neutralizing antibody levels. In these regression analyses, the primary test of significance evaluated an ordinal association between the genotypes of each SNP and log-transformed neutralizing antibody levels, with adjustment for sex, ages at enrollment and at vaccination, time between immunization and blood sample collection, assay batch, and population stratification eigenvectors. We assessed the degree to which the significance levels were inflated due to unmeasured confounding and adjusted our level of significance according to the estimated inflation factor [30]. For SNPs most strongly associated with neutralizing antibody levels, we summarized the genotype counts and the per-genotype medians and interquartile ranges of the interpolated NT50 values and derived a LocusZoom of the P values in the genomic region harboring the primary genetic signal [31].
RESULTS
The cohort consisted of 1052 subjects with appropriate data. As the participants were predominantly White (85.3%), all analyses focused on the 897 subjects who were members of this racial group. From this group, we obtained neutralizing antibody and genotype information on a sample size of 897. The sex distribution was 490 males (54.6%) and 407 females (45.4%). The median age at enrollment was 15.0 years (interquartile range [IQR], 13.0–17.0 years), the median age at first vaccination was 15.0 months (IQR, 15.0–16.0 months), the median age at second vaccination was 10.0 years (IQR, 5.0–12.0 years), and median time since the last vaccination was 6.4 years (IQR, 4.6–8.5 years). The median interpolated NT50 was 55.4 (IQR, 34.4–91.3).
Genotyping assays performed using the Omni 1 M were attempted for 1063 potential participants. Only 10 (0.94%) of these attempted samples had SNP call rates of <99%. After removing these subjects and other subjects with inconsistent data on sex (3) or other characteristics (8), a total of 1052 potential subjects were available for analysis. Of the 934 149 candidate SNPs with call rates of >99%, 793 644 had minor allele frequencies of >1% and were included in the primary analysis. A total of 5656 of these SNPs were flagged as having HWE P values of <1 × 10−7. Of these SNPs, there were 70 742 with HWE P values of > 1 × 10−3 and with pairwise r2 values of < 0.1 that were used in the assessment of population stratification. After removing subjects with >15% genetic admixture beyond the primary White genetic cluster, 897 subjects remained for analysis. Quality control measures were repeated on this subset, leaving 752 869 SNPs with minor allele frequencies of >1% for analysis. Three race-specific eigenvectors were obtained to control for residual genetic differences within this White subset.
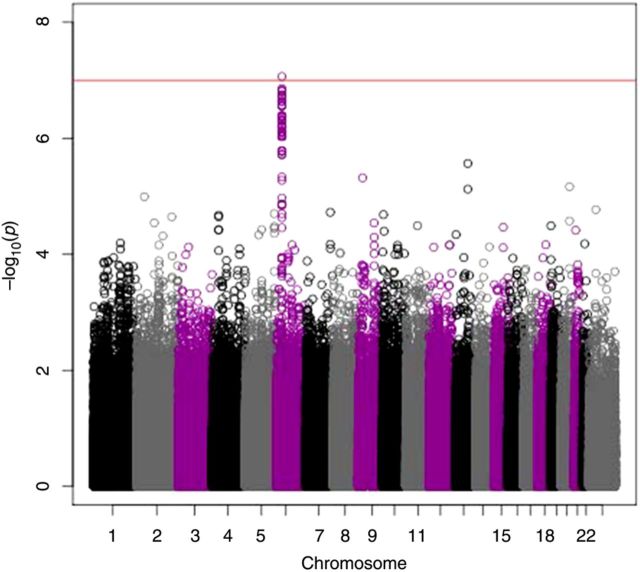
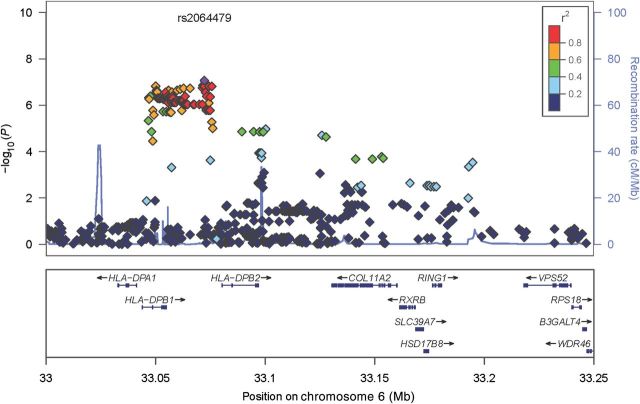
For this study, we set the threshold of significance at P ≤ 1 × 10−7 [32]. Our GWAS revealed that the most significant SNP (rs2064479) associated with variations in rubella virus–specific neutralizing antibody response was located in chromosome 6 (Figure 1), within a cluster of neighboring SNPs that approached significance. Table 1 lists the characteristics of all significant SNPs with P values of ≤ 2.82 × 10−7. There were 118 SNPs within the HLA-DPB1 genetic region with P values of ≤ 1 × 10−5. To obtain a graphical illustration of all SNPs in LD with rs2064479, we generated a LocusZoom plot representing the genetic region on 6p21.3, showing LD structure, SNP-associated P values, and the recombination rate (Figure 2). There is an obvious clustering of associated SNPs around the HLA-DPB1 gene. There were 81 SNPs in high LD (r2 ≤ 0.8) with rs2064479. Of these, 28 were located in the 3’ untranslated region, downstream (hereafter, the “3’ UTR/downstream region”), of the HLA-DPB1 gene (data not shown).
Figure 1.
Genome-wide Manhattan plot of significant single-nucleotide polymorphisms (SNPs) associated with a neutralizing antibody response to rubella virus identified a majority of hits in chromosome 6, including rs2064479 (P = 8.62 × 10−8). The horizontal line near the top of the plot marks P values of <1 × 107. The functional location of this SNP maps to the HLA-DPB1 gene.
Table 1.
Single-Nucleotide Polymorphisms (SNPs) Associated With Variations in Neutralizing Antibody Response to Rubella Virus
| SNP ID | Chr | Genea | Locationb | Genotype | No.c | Interpolated NT50, Median (IQR) | P Valued |
|---|---|---|---|---|---|---|---|
| rs2064479 | 6 | HLA-DPB1 | Intergenic | GG | 485 | 61.3 (37.7–100.7) | 8.62 × 10−8 |
| GA | 336 | 51.5 (31.15–79.6) | |||||
| AA | 66 | 43.3 (32.6–77.5) | |||||
| rs3128925 | 6 | HLA-DPB1 | Intergenic | CC | 519 | 60.4 (37.7–99.1) | 1.39 × 10−7 |
| CA | 317 | 51.0 (31.2–80.3) | |||||
| AA | 51 | 40.1 (32.6–75.1) | |||||
| rs9277359 | 6 | HLA-DPB1 | Intronic | CC | 444 | 60.8 (39.5–100.6) | 1.48 × 0−7 |
| CA | 364 | 51.8 (31.4–82.8) | |||||
| AA | 80 | 43.3 (31.6–79.6) | |||||
| rs2064478 | 6 | HLA-DPB1 | Intergenic | GG | 519 | 60.4 (37.7–99.1) | 1.52 × 10−7 |
| GA | 318 | 51.0 (31.2–80.4) | |||||
| AA | 51 | 40.1 (32.6–75.1) | |||||
| rs3117230 | 6 | HLA-DPB1 | Intergenic | AA | 519 | 60.4 (37.7–99.1) | 1.52 × 10−7 |
| AG | 318 | 51.0 (31.2–80.4) | |||||
| GG | 51 | 40.1 (32.6–75.1) | |||||
| rs3117233 | 6 | HLA-DPB1 | Intergenic | GG | 519 | 60.4 (37.7–99.1) | 1.52 × 10−7 |
| GA | 318 | 51.0 (31.2–80.4) | |||||
| AA | 51 | 40.1 (32.6–75.1) | |||||
| rs3117239 | 6 | HLA-DPB1 | Intergenic | GG | 519 | 60.4 (37.7–99.1) | 1.52 × 10−7 |
| GA | 318 | 51.0 (31.2–80.4) | |||||
| AA | 51 | 40.1 (32.6–75.1) | |||||
| rs9277357 | 6 | HLA-DPB1 | Intronic | AA | 443 | 60.8 (39.5–100.6) | 1.56 × 10−7 |
| AG | 365 | 51.5 (31.5–83.8) | |||||
| GG | 80 | 40.9 (30.15–75.3) | |||||
| rs3117240 | 6 | HLA-DPB1 | Intergenic | AA | 519 | 60.4 (37.7–99.1) | 1.77 × 10−7 |
| AG | 317 | 51.0 (31.2–80.4) | |||||
| GG | 51 | 40.1 (32.6–75.1) | |||||
| rs2064473 | 6 | HLA-DPB1 | Intergenic | GG | 602 | 59.5 (36.7–98.0) | 1.86 × 10−7 |
| GA | 251 | 50.8 (30.9–75.3) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs3117211 | 6 | HLA-DPB1 | Intergenic | CC | 602 | 59.5 (36.7–98.0) | 1.86 × 10−7 |
| CA | 251 | 50.8 (30.9–75.3) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs3117218 | 6 | HLA-DPB1 | Intergenic | GG | 602 | 59.5 (36.7–98.0) | 1.86 × 10−7 |
| GA | 251 | 50.8 (30.9–75.3) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs3130189 | 6 | HLA-DPB1 | Intergenic | AA | 602 | 59.5 (36.7–98.0) | 2.06 × 10−7 |
| AG | 250 | 50.7 (30.9–75.5) | |||||
| GG | 36 | 39.4 (31.5–69.0) | |||||
| rs2281389 | 6 | HLA-DPB1 | Intergenic | AA | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| AG | 250 | 50.8 (31.1–75.5) | |||||
| GG | 35 | 39.4 (31.0–71.0) | |||||
| rs3128965 | 6 | HLA-DPB1 | Intergenic | GG | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| GA | 250 | 50.8 (31.1–75.5) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs3128966 | 6 | HLA-DPB1 | Intergenic | GG | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| GA | 250 | 50.8 (31.1–75.5) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs9277379 | 6 | HLA-DPB1 | Intronic | AA | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| AC | 250 | 50.8 (31.1–75.5) | |||||
| CC | 35 | 39.4 (31.0–71.0) | |||||
| rs9277381 | 6 | HLA-DPB1 | Intronic | GG | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| GA | 250 | 50.8 (31.1–75.5) | |||||
| AA | 35 | 39.4 (31.0–71.0) | |||||
| rs9277384 | 6 | HLA-DPB1 | Intronic | CC | 603 | 59.5 (36.6–98.0) | 2.29 × 10−7 |
| CG | 250 | 50.8 (31.1–75.5) | |||||
| GG | 35 | 39.4 (31.0–71.0) | |||||
| rs3097650 | 6 | HLA-DPB1 | Intergenic | GG | 444 | 60.7 (38.9–100.6) | 2.69 × 10−7 |
| GA | 364 | 52.3 (31.5–82.8) | |||||
| AA | 80 | 43.3 (31.6–79.6) | |||||
| rs3128961 | 6 | HLA-DPB1 | Intronic | GG | 444 | 60.7 (38.9–100.6) | 2.69 × 10−7 |
| GA | 364 | 52.3 (31.5–82.8) | |||||
| AA | 80 | 43.3 (31.6–79.6) | |||||
| rs9277386 | 6 | HLA-DPB1 | Intronic | AA | 444 | 60.7 (38.9–100.6) | 2.69 × 10−7 |
| AG | 364 | 52.3 (31.5–82.8) | |||||
| GG | 80 | 43.3 (31.6–79.6) | |||||
| rs9277378 | 6 | HLA-DPB1 | Intronic | AA | 444 | 60.7 (38.9–100.6) | 2.72 × 10−7 |
| AG | 363 | 52.1 (31.5–82.4) | |||||
| GG | 80 | 43.3 (31.6–79.6) | |||||
| rs1042544 | 6 | HLA-DPB1 | UTR | AA | 432 | 61.1 (38.4–101.1) | 2.76 × 10−7 |
| AG | 374 | 52.6 (31.6–83.8) | |||||
| GG | 82 | 43.3 (31.0–77.5) | |||||
| rs3130210 | 6 | HLA-DPB1 | Intergenic | CC | 519 | 60.4 (38.4–101.1) | 2.82 × 10−7 |
| CA | 318 | 51.0 (31.2–80.4) | |||||
| AA | 50 | 42.7 (33.4–75.2) |
Abbreviations: Chr, chromosome; ID, identification number; IQR, interquartile range; NT50, neutralizing titer; UTR, untranslated region.
a Gene or genetic region.
b Predicted function.
c Number of subjects for each genotype.
d Adjusted for demographic and clinical variables, as well as for inflation of significance, as described in “Methods” section. All identified SNPs with P ≤ 2.64 × 10−6 were associated with the HLA-DPB1 gene. The top 25 SNPs are listed above (P ≤ 2.82 × 10−7).
Figure 2.
LocusZoom plot of single-nucleotide polymorphisms (SNPs) in high linkage disequilibrium (LD) with rs2064479. The LocusZoom plot allows for a visual depiction of the high-level LD of multiple SNPs with index SNP rs2064479 and the clustering of these SNPs near HLA-DPB1. A total of 82 SNPs were within an r2 value of ≥0.8. Of those SNPs in high LD, 28 had a predicted function in the 3’ untranslated region, downstream, of HLA-DPB1.
To determine whether SNPs located in the 3’ UTR/downstream region of the HLA-DPB1 gene that are in high LD with rs2064479 may be located in microRNA (miRNA) binding sites, we queried the PolymiRTS (Polymorphism in microRNAs and their TargetSites) database [33]. Ten SNPs were identified as potentially influencing miRNA binding within the 3’ UTR/downstream region of the HLA-DPB1 gene (Table 2).
Table 2.
High–Linkage Disequilibrium (LD) Single-Nucleotide Polymorphisms (SNPs) in Potential microRNA (miRNA) Binding Sites of HLA-DPB1
| SNP ID | r2a | miRNA ID | Context + Score Changeb |
|---|---|---|---|
| rs3117228 | 0.822797 | hsa-miR-16-2-3p | 0.22 |
| hsa-miR-195-3p | 0.21 | ||
| rs9277533 | 0.822797 | hsa-miR-3607-5p | −0.077 |
| hsa-miR-568 | −0.004 | ||
| hsa-miR-6516-3p | −0.118 | ||
| rs9277538 | 0.822797 | hsa-miR-432-3p | −0.087 |
| rs9277539 | 0.926541 | hsa-miR-3165 | −0.109 |
| hsa-miR-8071 | −0.157 | ||
| rs9277542 | 0.822797 | hsa-miR-6730-5p | 0.024 |
| rs9277547 | 0.822797 | hsa-miR-4493 | −0.035 |
| hsa-miR-4499 | 0.006 | ||
| hsa-miR-605-3p | −0.121 | ||
| hsa-miR-6126 | −0.094 | ||
| hsa-miR-873-3p | −0.037 | ||
| hsa-miR-124-3p | −0.005 | ||
| hsa-miR-3714 | 0.009 | ||
| hsa-miR-3910 | −0.055 | ||
| hsa-miR-506-3p | −0.012 | ||
| rs9277549 | 0.822797 | hsa-miR-552-3p | 0.067 |
| hsa-miR-764 | 0.023 | ||
| hsa-miR-6789-3p | −0.183 | ||
| rs9277550 | 0.822797 | hsa-miR-6077 | 0.032 |
| hsa-miR-7-5p | 0.005 | ||
| hsa-miR-1185-5p | −0.066 | ||
| hsa-miR-3679-5p | −0.066 | ||
| hsa-miR-4534 | 0.046 | ||
| hsa-miR-4802-5p | −0.028 | ||
| hsa-miR-6894-5p | −0.111 | ||
| hsa-miR-7154-3p | −0.043 | ||
| hsa-miR-765 | −0.026 | ||
| hsa-miR-766-5p | −0.032 | ||
| hsa-miR-8082 | 0.028 | ||
| rs9277554 | 0.822797 | hsa-miR-1271-3p | No change |
| hsa-miR-4297 | 0.03 | ||
| hsa-miR-550a-3-5p | 0.003 | ||
| hsa-miR-550a-5p | 0.003 | ||
| hsa-miR-550b-2-5p | −0.02 | ||
| hsa-miR-5581-5p | 0.027 | ||
| hsa-miR-4724-3p | 0.032 | ||
| rs9277555 | 0.964201 | hsa-miR-345-5p | −0.033 |
The PolymiRTS (Polymorphism in microRNAs and their TargetSites) database allows a query of SNPs in potential miRNA seed and binding sites.
Abbreviation: ID, identification number.
a Correlation value between 2 loci.
b More-negative values indicate that a given SNP has an increased likelihood of disrupting miRNA binding. Ten SNPs in high LD with rs2064479 (r2 > 0.8) may influence miRNA binding sites in the 3’ untranslated region of HLA-DPB1.
We have previously characterized genome-wide transcription patterns between individuals with low and high antibody responses to rubella vaccine [34]. In the current study, we did not find any associations between differential gene expression and GWAS findings. This may be due to the difference in the antibody measure used to characterize these cohorts. Haralambieva et al used a rubella virus–specific total immunoglobulin G assay, while we used the levels of neutralizing antibodies for this association study. Also, the genome-wide transcriptional pattern work was also performed in a rather small cohort (n = 30). This may not have been a large enough cohort to detect the HLA associations.
DISCUSSION
To our knowledge, this is the first GWAS to investigate genetic influences on interindividual variations in rubella virus–specific neutralizing antibody response after vaccination. In a cohort of vaccinated individuals, we identified rs2064479, an intergenic SNP in the HLA-DPB1 genetic region, as having genome-wide significance in association with antibody levels. Interestingly, all SNPs that were significant (P ≤ 1 × 10−7) were located near the HLA-DPB1 gene. Further analyses revealed 82 SNPs in high LD (r2 ≥ 0.8) with rs2064479. Twenty-eight of these SNPs were located in the 3’ UTR/downstream region of the HLA-DPB1 gene, and 10 of those may be involved in miRNA binding sites.
The highly polymorphic nature of the HLA genes and their essential role in host response and antigen-specific adaptive immunity help to explain the large amount of data linking specific HLA variants to differences in susceptibility to infection and vaccine response. Specific variants in the HLA-DPB1 gene have been associated with clinical outcomes in hepatitis B virus (HBV) and hepatitis C virus infections and in vaccine responses against hepatitis B virus, rubella virus, measles virus, influenza virus, and malarial parasites [21, 35–41].
Although our most significant HLA-DPB1 finding (rs2064479) has yet to be identified as a causal variant with functional consequences in other genetic studies, it is in high LD (r2 = 0.82) with rs9277534, which has a predicted function in the 3’ UTR of HLA-DPB1. The 496 GG genotype of the latter SNP is associated with susceptibility to HBV persistence and an increase in HLA-DP protein and gene expression [36]. The authors hypothesized in that study that susceptibility to HBV persistence is due to high levels of HLA-DPB1 expression and not to differences in peptide repertoire. Our data revealed that the GG genotype of rs9277534 (minor allele) is associated with an allele dose-related decrease in rubella virus neutralizing antibodies, compared with the AA genotype (AA = 60.83, and GG = 43.35; P = 5.01 × 10−7). The mechanism behind the significant differences in antibody levels observed in association with rs2064479 remains unclear. The next step is to perform additional fine-mapping panels to identify the true causal variant. SNPs in the 3’ UTR/downstream region of the HLA C gene may interfere with the ability of regulatory miRNAs to bind and are associated with control of human immunodeficiency virus [29]. The multiple SNPs we identified in predicted miRNA binding targets may influence rubella vaccine–induced humoral immunity through a similar mechanism.
The strength of our study is the use of a well-characterized viral vaccine. Rubella virus contains a stable genome with very similar immune responses across strains [42, 43]. We also chose this well-defined study cohort because of the written documentation of having received 2 doses of MMR II vaccine and a lack of circulating virus in Rochester. These data allow us to assume that the measured differences in antibody response against rubella virus are from vaccination and not from infection.
We acknowledge that the cohort might be considered small for a GWAS. However, recruitment and vaccine administration in a larger cohort is not economically or logistically feasible. The overwhelming number of SNPs that displayed a trend toward significance associated with regions near HLA-DPB1, and the potential of certain SNPs to influence miRNA binding sites, increases our confidence that this is a valid finding. If fine-mapping analyses and a proposed validation GWAS identify the causal variant as associated with HLA-DPB1, then functional studies will be designed to measure differences in HLA-DP protein and gene expression in variants. These data will contribute immensely to studies aimed at elucidating genetic variants associated with differences in immunity to live viral vaccines.
Notes
Acknowledgments. We thank the study subjects and their families, for their willingness to participate in our study; the Mayo Clinic nurses and study coordinators, for their efforts in subject recruitment; Dr Julie Cunningham and the Mayo Medical Genome Facility Genotyping Core, for their assistance with genotyping efforts; Beth Larrabee and Nathaniel Warner, for their contributions to the statistical analyses; and Caroline Vitse, for her contribution to the editing and layout of the manuscript.
This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (award AI033144) and the National Institute of Allergy And Infectious Diseases of the National Institutes of Health (award AI048793).
Potential conflicts of interest. G. A. P. is the chair of a safety evaluation committee for novel investigational vaccine trials being conducted by Merck Research Laboratories and offers consultative advice on vaccine development to Merck, CSL Biotherapies, Avianax, Sanofi Pasteur, Dynavax, Novartis Vaccines and Therapeutics, PAXVAX, Emergent Biosolutions, Adjuvance, and Vaxness. G. A. P. and I. G. O. hold patents related to vaccinia and measles peptide research. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Miller E, Cradock-Watson JE, Pollock TM. Consequences of confirmed maternal rubella at successive stages of pregnancy. Lancet. 1982;2:781–4. doi: 10.1016/s0140-6736(82)92677-0. [DOI] [PubMed] [Google Scholar]
- 2.Plotkin SA, Reef SE, Cooper LZ, Alford CA., Jr . Rubella. In: Remington JS, Klein JO, Wilson CB, Nizet V, Maldonado Y, editors. Infectious diseases of the fetus and newborn infant. 7th ed. Philadelphia: Elsevier; 2011. pp. 861–98. [Google Scholar]
- 3.Robertson SE, Featherstone DA, Gacic-Dobo M, Hersh BS. Rubella and congenital rubella syndrome: global update. Rev Panam Salud Publica. 2003;14:306–15. doi: 10.1590/s1020-49892003001000005. [DOI] [PubMed] [Google Scholar]
- 4.Paradowska-Stankiewicz I, Czarkowski MP, Derrough T, Stefanoff P. Ongoing outbreak of rubella among young male adults in Poland: increased risk of congenital rubella infections. Euro Surveill. 2013;18:pii:20485. [PubMed] [Google Scholar]
- 5.Janta D, Stanescu A, Lupulescu E, Molnar G, Pistol A. Ongoing rubella outbreak among adolescents in Salaj, Romania, September 2011-January 2012. Euro Surveill. 2012;17:pii:20089. [PubMed] [Google Scholar]
- 6.Minakami H, Kubo T, Unno N. Causes of a nationwide rubella outbreak in Japan, 2012–2013. J Infect. 2014;68:99–101. doi: 10.1016/j.jinf.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Freestone DS, Reynolds GM, McKinnon JA, Prydie J. Vaccination of schoolgirls against rubella.Assessment of serological status and a comparative trial of Wistar RA 27/3 and Cendehill strain live attenuated rubella vaccines in 13-year-old schoolgirls in Dudley. Brit J Prev Soc Med. 1975;29:258–61. doi: 10.1136/jech.29.4.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottiger M, Heller L. Experiences from vaccination and revaccination of teenage girls with three different rubella vaccines. J Biol Stand. 1976;4:107–14. doi: 10.1016/0092-1157(76)90020-2. [DOI] [PubMed] [Google Scholar]
- 9.Davidkin I, Jokinen S, Broman M, Leinikki P, Peltola H. Persistence of measles, mumps, and rubella antibodies in an MMR-vaccinated cohort: a 20-year follow-up. J Infect Dis. 2008;197:950–6. doi: 10.1086/528993. [DOI] [PubMed] [Google Scholar]
- 10.LeBaron CW, Forghani B, Matter L, et al. Persistence of rubella antibodies after 2 doses of measles-mumps-rubella vaccine. J Infect Dis. 2009;200:888–99. doi: 10.1086/605410. [DOI] [PubMed] [Google Scholar]
- 11.Lambert ND, Pankratz VS, Larrabee BR, et al. High-throughput assay optimization and statistical interpolation of rubella-specific neutralizing antibody titers. Clin Vaccine Immunol. 2014;21:340–6. doi: 10.1128/CVI.00681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwood NP, Ovsyannikova IG, Vierkant RA, O'Byrne MM, Poland GA. A qualitative and quantitative comparison of two rubella virus-specific IgG antibody immunoassays. Viral Immunol. 2010;23:353–7. doi: 10.1089/vim.2010.0026. [DOI] [PubMed] [Google Scholar]
- 13.Lambert ND, Haralambieva IH, Ovsyannikova IG, Larrabee BR, Pankratz VS, Poland GA. Characterization of humoral and cellular immunity to rubella vaccine in four distinct cohorts. Immunol Res. 2013;58:1–8. doi: 10.1007/s12026-013-8475-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson RM, Ovsyannikova IG, Poland GA. Genetic basis for variation of vaccine response: our studies with rubella vaccine. Paediatr Child Health. 2009;(supp 2):S156–9. doi: 10.1016/j.paed.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ovsyannikova IG, Jacobson RM, Vierkant RA, O'Byrne MM, Poland GA. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine. 2009;27:6926–31. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhiman N, Haralambieva IH, Kennedy RB, et al. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics. 2010;62:197–210. doi: 10.1007/s00251-010-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ovsyannikova IG, Haralambieva IH, Dhiman N, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis. 2010;201:207–13. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haralambieva IH, Dhiman N, Ovsyannikova IG, et al. 2′-5′-Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Hum Immunol. 2010;71:383–91. doi: 10.1016/j.humimm.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy RB, Ovsyannikova IG, Vierkant RA, Jacobson RM, Poland GA. Effect of human leukocyte antigen homozygosity on rubella vaccine-induced humoral and cell-mediated immune responses. Hum Immunol. 2010;71:128–35. doi: 10.1016/j.humimm.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Hum Immunol. 2004;65:1506–15. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. Human leukocyte antigen class II alleles and rubella-specific humoral and cell-mediated immunity following measles-mumps-rubella-II vaccination. J Infect Dis. 2005;191:515–9. doi: 10.1086/427558. [DOI] [PubMed] [Google Scholar]
- 22.Ovsyannikova IG, Dhiman N, Haralambieva IH, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010;127:207–21. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovsyannikova IG, Haralambieva IH, Vierkant RA, O'Byrne MM, Jacobson RM, Poland GA. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--a replication study and examination of novel polymorphisms. Hum Hered. 2011;72:206–23. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30:2146–52. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleveland WS, Gross E, Shyu WM. Local regression models. In: Chambers JM, Hastie TJ, editors. Statistical models. Wadsworth & Brooks/Cole; 1992. [Google Scholar]
- 26.Kennedy RB, Ovsyannikova IG, Pankratz VS, et al. Genome-wide genetic associations with IFNgamma response to smallpox vaccine. Hum Genet. 2012;131:1433–51. doi: 10.1007/s00439-012-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy RB, Ovsyannikova IG, Shane PV, Haralambieva IH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum Genet. 2012;131:1403–21. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovsyannikova IG, Kennedy RB, O'Byrne M, Jacobson RM, Pankratz VS, Poland GA. Genome-wide association study of antibody response to smallpox vaccine. Vaccine. 2012;30:4182–9. doi: 10.1016/j.vaccine.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 31.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panagiotou OA, Ioannidis JP. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41:273–86. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haralambieva IH, Oberg AL, Ovsyannikova IG, et al. Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination. PLoS One. 2013;8:e62149. doi: 10.1371/journal.pone.0062149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–5. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 36.Thomas R, Thio CL, Apps R, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–85. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mbarek H, Ochi H, Urabe Y, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Gen. 2011;20:3884–92. doi: 10.1093/hmg/ddr301. [DOI] [PubMed] [Google Scholar]
- 38.Urabe Y, Ochi H, Kato N, et al. A genome-wide association study of HCV-induced liver cirrhosis in the Japanese population identifies novel susceptibility loci at the MHC region. J Hepatol. 2013;58:875–82. doi: 10.1016/j.jhep.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 39.Png E, Thalamuthu A, Ong RT, Snippe H, Boland GJ, Seielstad M. A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum Mol Gen. 2011;20:3893–8. doi: 10.1093/hmg/ddr302. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson RM, Ovsyannikova IG, Vierkant RA, Shane PV, Poland GA. Human leukocyte antigen associations with humoral and cellular immunity following a second dose of measles-containing vaccine: Persistence, dampening, and extinction of associations found after a first dose. Vaccine. 2011;29:7982–91. doi: 10.1016/j.vaccine.2011.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens HA, Brown AE, Chandanayingyong D, et al. The presence of the HLA class II allele DPB1*0501 in ethnic Thais correlates with an enhanced vaccine-induced antibody response to a malaria sporozoite antigen. Eur J Immunol. 1995;25:3142–7. doi: 10.1002/eji.1830251123. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Ushijima H, Frey TK. Genomic analysis of diverse rubella virus genotypes. J Gen Virol. 2007;88:932–41. doi: 10.1099/vir.0.82495-0. [DOI] [PubMed] [Google Scholar]
- 43.Best JM, Thomson A, Nores JR, O'Shea S, Banatvala JE. Rubella virus strains show no major antigenic differences. Intervirology. 1992;34:164–8. doi: 10.1159/000150277. [DOI] [PubMed] [Google Scholar]