Abstract
We have shown that Borrelia burgdorferi gene product BB0323 is essential for cell fission and pathogen persistence in vivo. Here we describe characterization of a conserved hypothetical protein annotated as BB0238, which specifically interacts with the N-terminal region of BB0323. We show that BB0238 is a subsurface protein, and similar to BB0323, exists in the periplasm and as a membrane-bound protein. Deletion of bb0238 in infectious B. burgdorferi did not affect microbial growth in vitro or survival in ticks, but the mutant was unable to persist in mice or transmit from ticks—defects that are restored on genetic complementation. Remarkably, BB0238 and BB0323 contribute to mutual posttranslational stability, because deletion of one causes dramatic reduction in the protein level of the other partner. Interference with the function of BB0238 or BB0323 and their interaction may provide novel strategies to combat B. burgdorferi infection.
Keywords: Borrelia burgdorferi, BB0238, BB0323, protein–protein interaction, posttranslational stability, pathogen persistence
Lyme borreliosis is a prevalent vector-borne disease [1–3] caused by Borrelia burgdorferi sensu lato [4, 5], which persists in a complex life cycle involving Ixodes scapularis ticks and vertebrate hosts [6–8]. The microbe is transmitted to the mammalian dermis via tick bite and disseminates to a variety of internal organs, causing an array of clinical complications that range from arthritis and carditis to neurological disorders [9, 10]. Antibiotic treatment usually cures the infection, although some patients display chronic disease, which may be unrelated to persistent infection [11]. A vaccine to prevent Lyme disease in humans is unavailable [12].
Spirochetes are unique among bacteria; for example, in addition to a peptidoglycan layer, flagella also help determine the cell shape [13]. B. burgdorferi possess periplasmic flagella attached at either end of the cell running between the outer membrane (OM) and protoplasmic cylinder (PC), which is surrounded by an inner membrane and peptidoglycan layer [14]. Given that spirochetes possess periplasmic flagella that actively rotate, how the OM connects to the PC and maintains its stability remains puzzling [15]. In addition, limited information is available about OM biogenesis in B. burgdorferi, especially how this structure is stabilized and tethered to the PC. In most gram-negative bacteria, a family of proteins spanning the periplasmic space, called the Tol-Pal complex, is responsible for OM-PC interaction supporting membrane stability and cell fission [16–19].
Although the B. burgdorferi genome lacks identifiable orthologs of Tol-Pal complex members, earlier studies showed that a membrane protein, BB0323, supports OM stability, cell fission, and infection [20, 21]. Notably, BB0323 undergoes proteolytic processing, initially by a broad-spectrum borrelial protease, BB0104 (BbHtrA) [15, 22–24], and later by another C-terminal protease, CtpA [25], ultimately yielding distinct N- and C-terminal polypeptides [15]. Such multistep posttranslational maturation of BB0323, particularly involving multiple proteases, is intriguing and possibly relevant to the tight quality control of proteins, such as BB0323, that are critical for OM stability, fission, and virulence. Incidentally, our previous findings suggesting that BbHtrA is a BB0323 processing enzyme also indicated the possible existence of additional BB0323-interacting borrelial proteins, including BB0238 [15]. In this report, we characterize BB0323–BB0238 interaction and show that the proteins stabilize each other and support infectivity. A better understanding of the biological significance of the BB0323–BB0238 complex may shed new light on spirochete biology and infection and help in the development of possible therapeutic strategies to combat Lyme disease.
MATERIALS AND METHODS
B. burgdorferi, Mice, and Ticks
B. burgdorferi infectious isolate B31-A3 [26] was grown in Barbour–Stoenner–Kelly-H (BSK-H) medium containing 6% rabbit serum at 34°C. Generation of bb0323 mutants (bb0323−), complemented isolates (bb0323 com), and C-terminal BB0323-truncation mutants (bb0323ΔC and ΔLysM) was described elsewhere [15, 21]. The I. scapularis ticks were reared in the laboratory, as described elsewhere [21]. C3H/HeN mice 4–6 weeks old were purchased from the National Institutes of Health. Animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and Institutional Biosafety Committee of the University of Maryland, College Park.
Coimmunoprecipitation, 2-Dimensional Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis, and Liquid Chromatography–Tandem Mass Spectrometry
About 1010 spirochete cells were lysed with BugBuster Reagent (Novagen) and protease inhibitor (Sigma), incubated initially with BB0323 antiserum for 4 hours and later with Protein G(Sigma-Aldrich) beads overnight at 4°C. The beads were then centrifuged and washed, and bound proteins were eluted in a buffer (7 mol/L urea, 2 mol/L thiourea, 20 mmol/L dithiothreitol, 0.5% immobilized pH gradient buffer [GE Healthcare] containing 0.5% Triton X-100), resolved using 2-dimensional electrophoresis, and stained with SYPRO Ruby (Invitrogen). Samples for liquid chromatography–tandem mass spectrometry analysis were prepared by tryptic in-gel digestion of excised protein bands, as described elsewhere [27].
Generation of Recombinant Proteins and Antiserum
The bb0238 gene was cloned into pGEX-6P-1 (Amersham Pharmacia Biotech) by using specific primers shown in Supplementary Table 1, and the recombinant protein without the N-terminal leader sequence was produced in Escherichia coli. Expression and purification of the with glutathione S-transferase (GST)–fused proteins, with or without the tag, were performed as described elsewhere [15]. To generate polyclonal antiserum, the proteins (without the GST tag) produced in E. coli were emulsified in complete Freund adjuvant and injected into groups of 5 mice (10 µg/animal). The animals were boosted twice at 3-week intervals with the same dose of antigen in incomplete Freund adjuvant, and the serum samples were collected 2 weeks after the second boost. Assessment of the titer and specificity of the antiserum were performed as described elsewhere [15].
Western Blotting and Far-Western Blotting
These assays were performed using standard procedures, as detailed elsewhere [15, 28]. Antibody incubations were performed at room temperature for 1 hour using the following dilutions: anti-BB0238 (1:3000), anti-BB0323 N-terminal (1:3000), anti-BB0238 C-terminal (1:10 000), anti-FlaB (1:2000), anti-OspA (1:500), and anti-GST (1:5000) antibodies and horseradish peroxidase–conjugated secondary antibodies (1:5000–10 000). For far-Western analyses, proteins (0.5 µg per lane) were fractionated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and blocked in Tris-buffered saline pH 7.5 with 0.05% Tween 20 (TBST) and 2% skim milk at room temperature for 1 hour. Membranes were incubated overnight at 4°C with GST-fused proteins (2 µg/mL) in blocking buffer and washed with TBST, and bound proteins were identified by immunoblotting using anti-GST and horseradish peroxidase–conjugated secondary antibodies, as noted above. Protein signals were visualized using chemiluminescence Western Blotting Detection Reagent (GE Healthcare).
Proteinase K Accessibility Assay, Isolation of OM, and Triton X-114 Phase Partitioning Assay
The proteinase K digestion assay was performed as described elsewhere [21, 28]. Briefly, 108 spirochetes were incubated in phosphate-buffered saline (PBS) in the absence or presence of proteinase K (Sigma) at a concentration of 200 µg/mL for 20 minutes, treated with phenylmethylsulfonyl fluoride (Sigma), centrifuged, and resuspended in PBS for subsequent immunoblot analysis using antibodies against BB0238, FlaB, or OspA. The OM vesicles and PC were extracted from spirochetes using sucrose density gradient centrifugation, as detailed elsewhere [27], and subjected to immunoblotting using BB0238 or OspA antibodies. The preparation of periplasmic and spheroplasmic proteins is described elsewhere [15]. Triton X-114 phase partitioning assays were performed as described elsewhere [28]. Briefly, spirochetes were resuspended in 800 µL of PBS (pH 7.4), sonicated, and extracted with 2% (final concentration) Triton X-114. After separation by centrifugation, the aqueous and detergent phases were washed with ice-cold acetone to precipitate the proteins, centrifuged at 13 000g for 15 minutes at 4°C [28], and finally immunoblotted using OspA, BB0323, and BB0238 antibodies.
Yeast 2-Hybrid Assay
A yeast 2-hybrid assay was performed using a commercially available kit (Matchmaker Gold Yeast Two-Hybrid System; Clontech Laboratories). Briefly, open reading frames representing bb0323 and bb0238 were amplified with polymerase chain reaction (PCR) using B. burgdorferi DNA and cloned into bait (pGBKT7) or prey (pGADT7) vectors using the primers indicated in Supplementary Table 1, and protein expression was confirmed using antibodies. Protein–protein interaction in yeasts was evaluated by growth in selective media and an X-α-galactosidase filter assay, as detailed in the manufacturer's protocol.
PCR Analysis
The primers used in PCR and quantitative reverse-transcription PCR (qRT-PCR) are shown in Supplementary Table 1. For qRT-PCR, RNA was isolated using TRIzol reagent (Invitrogen), treated with DNase I (NEB), reverse transcribed to complementary DNA (AffinityScript), and analyzed by qRT-PCR using iQ SYBR Green Supermix (Bio-Rad) as described elsewhere [29]. For assessment of bb0238 expression in hosts, mice (3 animals per group) were infected with spirochetes (105 cells per mouse), and tissues were collected at 2 weeks and pooled by type. To assess transcript levels in infected arthropods, uninfected nymphs that parasitized infected mice, unfed infected nymphs, and fed infected nymphs that engorged on naive mice were collected as detailed elsewhere [29]. For quantitative analysis of gene expression, the target transcripts were normalized to the number of flaB transcripts. For quantitative measurement of B. burgdorferi burden in infected tissues or ticks, flaB transcripts were normalized to mouse or tick β-actin levels.
Generation and Characterization of bb0238-Mutant and Complemented Isolates of B. burgdorferi
BB0238-deficient B. burgdorferi was generated via homologous recombination by replacing the entire bb0238 open reading frame with an antibiotic cassette, flaB promoter-fused kanamycin (flaB-Kan), as detailed elsewhere [21, 28, 29]. A bb0238-deletion clone retaining the same set of plasmids as the wild-type isolate was used for further experiments. Genetic complementation of bb0238 mutant was achieved by reinsertion of a wild-type copy of the bb0238 gene in the B. burgdorferi chromosome, as described elsewhere [21, 28, 29]. Spirochetes were also processed for transmission electron microscopy and growth analysis as described elsewhere [21].
Phenotypic Analysis of Genetically Manipulated B. burgdorferi
Spirochete burdens in mice and ticks were assessed using qRT-PCR analysis of flaB transcripts and normalized against murine or tick β-actin levels, as described elsewhere [21, 29]. Groups of mice (3 animals per group) were injected with the mutant or wild-type spirochetes (105 cells per mouse) through needle inoculation intradermally in the lower right quadrant of the abdomen upstream from the joint. Skin, heart, joint, and bladder samples were isolated 2 weeks after infection. The B. burgdorferi levels in individual mouse tissues were assessed by qRT-PCR analysis, as described above. Portions of ear and heart tissues were also cultured in BSK-H medium and analyzed at weekly intervals for ≥3 weeks using a dark field microscope.
For assessment of infection in ticks, nymphs were infected with wild-type or mutant B. burgdorferi via a microinjection procedure [21, 30]. Infected ticks were either fed on naive mice (5 ticks per mouse) within 24 hours or kept in the incubator for 14 days, and the borrelial burdens in unfed ticks were analyzed with qRT-PCR. To assess spirochete transmission from ticks to mice, infected nymphs were allowed to feed to repletion. The engorged ticks were collected as postfed nymphs and subjected to qRT-PCR analysis to determine borrelial burden. At day 14 after the tick feeding, mice were killed, and various tissues were collected and subjected to qRT-PCR analysis for determination of pathogen levels.
Protein Turnover Assay
Stability of BB0323 was analyzed by a turnover assay, as described for another borrelial protein [31]. Briefly, wild-type and bb0238-mutant cultures were incubated in BSK-H medium containing spectinomycin (100 µg/mL) at 34°C for 24 hours. Spirochetes were removed after 4, 8, and 24 hours and processed for Western blotting using anti-BB0323.
Statistical Analysis
Results are expressed as means and standard errors of the mean. The significance of the difference between the mean values of the groups was evaluated with the 2-tailed Student t test.
RESULTS
BB0238 Specifically Interacts With N-terminal Region of BB0323
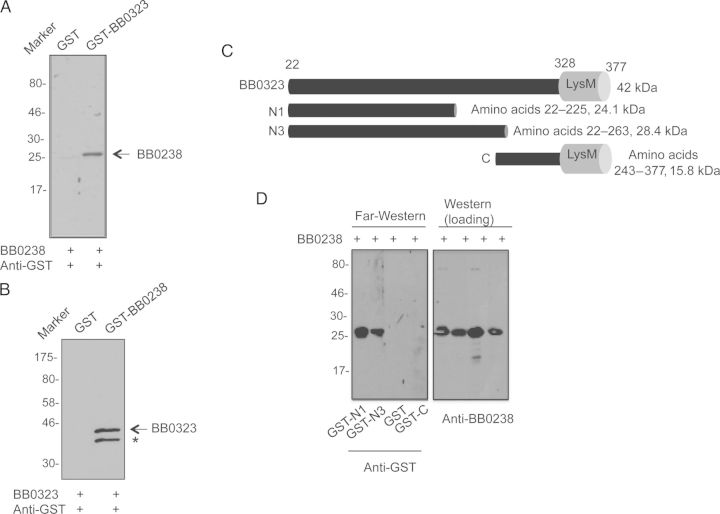
Previously it was demonstrated that a membrane protein, BB0323, supports B. burgdorferi fission and infection, and BB0238 was one of the few candidate proteins identified by BB0323 affinity chromatography [15]. To further assess their interaction, recombinant proteins and murine antibodies were generated and used in far-Western blot assays. BB0238 and BB0323 were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and incubated with GST-BB0323 or GST-BB0238, respectively, or a GST control. Finally, the bound proteins were detected using GST antibodies (Figure 1A and 1B). The result showed that these proteins interact with each other in vitro. In addition, their interaction within a cellular environment was demonstrated using a yeast 2-hybrid assay (Supplementary Figure 1A). As BB0323 is processed into distinct N- and C-terminal polypeptides [15], we next assessed which region binds to BB0238. Subsequent far-Western experiments using truncated BB0323 representing the N- and C-terminal polypeptides (Figure 1C) indicated that BB0238 interacts with the N-terminal BB0323 (Figure 1D).
Figure 1.
A, B, BB0238 interacts with BB0323. Far-Western blot analyses demonstrate specific binding of BB0238 to BB0323 (A) or binding of BB0323 to BB0238 (B). The recombinant proteins (either BB0238 or BB0323) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and incubated with glutathione S-transferase (GST)–fused proteins (either BB0323 or BB0238) or GST (control). Bound proteins were detected by anti-GST antibodies conjugated to horseradish peroxidase. Arrows denote the position of the band specifically detected by the recombinant protein; asterisk, either a nonspecific band or a degraded form of BB0323. Migration of protein standards is shown to the left in kilodaltons. C, Schematic representation of various truncated recombinant BB0323 proteins representing N- and C-terminal polypeptides. Truncated polypeptides were produced with or without GST tags. D, BB0238 binds to the N-terminal BB0323. Left panel denotes far-Western analysis; recombinant BB0238 proteins were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and incubated with GST-fused N1 (GST-N1), GST-fused N3 (GST-N3), GST, or GST-fused C-terminal polypeptide (GST-C). Bound proteins were detected with immunoblotting using anti-GST antibodies. Only N-terminal polypeptides exhibit detectable interaction in the assay. Equal protein loading is indicated by immunoblotting using BB0238 antibody (right panel).
Localization and Expression of BB0238
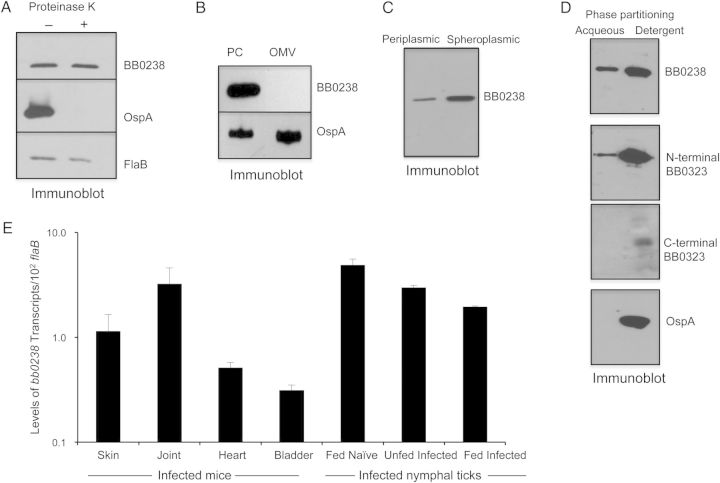
We next assessed subcellular distribution of BB0238. Intact spirochetes were subjected to a controlled proteinase K digestion and then immunoblot analysis using antiserum against BB0238 or known surface (OspA) or subsurface (FlaB) antigens as controls. These results suggested that BB0238 is not surface-exposed (Figure 2A). Further cell fractionation and immunoblot analysis demonstrated that BB0238 is undetectable in the OM (Figure 2B) but detectable in the periplasm (Figure 2C). The Triton X-114 phase partitioning assay, however, demonstrated that BB0238 primarily exists in the detergent phase enriched in membrane-bound proteins (Figure 2D). Of note, proteins that are functionally associated with BB0238, such as BB0323 and BbHtrA, were also detectable in the periplasm [15] and, similar to BB0238, are predominantly present in the detergent-enriched borrelial fractions (Figure 2D).
Figure 2.
BB0238 is a subsurface protein that exists in the periplasm and as a membrane-bound form and is produced throughout the tick-mouse infection cycle of Borrelia burgdorferi. A, BB0238 is not exposed on the spirochete surface. B, burgdorferi cells were incubated with (+) or without (−) proteinase K for the removal of protease-sensitive surface proteins and then processed for immunoblot analysis using either BB0238 or control surface-exposed (OspA) and subsurface (FlaB) protein antibodies. B, BB0238 is undetectable in the isolated outer membrane vesicle fraction. B. burgdorferi lysates were separated into protoplasmic cylinder (PC) and outer membrane vesicle (OMV) fractions by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted with antiserum against BB0238 or outer membrane protein (OspA). C, Detection of BB0238 in the periplasmic and spheroplasmic fractions of B. burgdorferi cells. Periplasmic or spheroplasmic proteins were extracted from cultured spirochetes and immunoblotted using BB0238 antibodies. D, Triton X-114 phase partitioning of B. burgdorferi proteins. Spirochete lysates were subjected to Triton X-114 phase partitioning of aqueous and detergent phases and immunoblotted using antibodies against BB0238, N-terminal BB0323, C-terminal BB0323, or a control (OspA) protein. E, Consistent expression of bb0238 during the enzootic life cycle of the spirochete. Total tissue RNA was isolated from representative stages of murine and tick infection, and bb0238 transcripts were measured using quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and presented as copies of bb0238 transcript per copy of flaB transcript. Error bars show the means and standard errors of the mean from ≥3 independent qRT-PCR analyses from individual ticks.
We next assessed bb0238 expression in vivo; transcripts were measured in various tissues of mice at 2 weeks of infection and in naïve nymphal ticks that acquired spirochetes from the infected mice. To enable examination of gene expression during borrelial persistence after intermolt stages in ticks and subsequent transmission, a fraction of these fed infected nymphal were allowed to molt and analyzed as unfed nymphs or as fed nymphs after engorgement on naive mice, respectively. Assessment of bb0238 transcript levels by qRT-PCR analysis indicated that the gene is constitutively expressed in all mouse tissues as well as in various stages of tick infection (Figure 2E).
BB0238 Facilitates Infection and Transmission
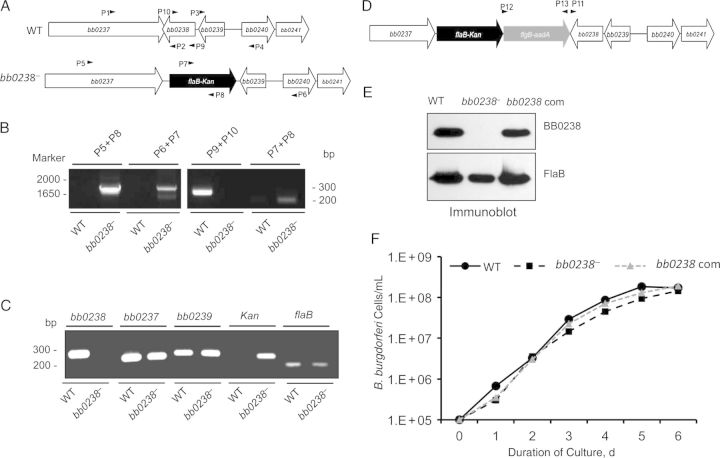
We next assessed the role of BB0238 in the pathogen life cycle by creating bb0238-deletion mutants and complemented isolates from infectious wild-type spirochetes (Figure 3A–E). Genetic manipulation of bb0238 imposed no obvious defects for spirochete survival in culture, as mutants and wild-type spirochetes displayed similar growth patterns (Figure 3F). Transmission electron microscopic analysis suggested no obvious morphological deformities or cell fission defects in BB0238-deficient spirochetes compared with parental isolates (data not shown).
Figure 3.
Construction and analysis of the bb0238 mutant and complemented isolates of Borrelia burgdorferi. A, Schematic drawings of wild-type (WT) and bb0238-mutant (bb0238−) isolates at the bb0238 locus. Genes bb0237–bb0241 (white arrows) and the kanamycin-resistance cassette driven by B. burgdorferi flaB promoter (flaB-kan) (black arrow) are indicated. Primers P1–P4 (arrowheads) were used to amplify 5′ and 3′ arms for homologous recombination, and regions flanking the bb0238 locus were ligated on either side of the flaB-kan cassette to obtain the mutagenic construct, as detailed in the text. B, Integration of the mutagenic construct, flaB-kan, in the intended genomic locus. Primers 5–10 (arrowheads) were employed for polymerase chain reaction (PCR) analysis, using DNA isolated from WT or mutant B. burgdorferi and subjected to agarose gel electrophoresis. The combination of primers used for PCR is indicated at the top, and migration of the DNA ladder is shown on the left. C, Reverse-transcription PCR assessment of bb0238 ablation and the polar effects of mutagenesis. Total RNA was isolated from WT or bb0238 mutant (bb0238−), converted to complementary DNA, and used to amplify regions within bb0238, flaB, the kanamycin-resistance cassette, and genes surrounding the bb0238 locus (bb0237 and bb0239); the products were visualized on a gel. D, Schematic illustration of the bb0238-complemented construct for chromosomal integration of the bb0238 in the original gene locus of the spirochete chromosome. Construct was used for integration into B. burgdorferi genomic DNA via homologous recombination. E, Production of BB0238 by the complemented B. burgdorferi. Spirochete lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted with anti-BB0238 (top) or anti-FlaB (bottom) antibodies. F, Growth curves for WT and genetically manipulated spirochetes. B. burgdorferi were diluted to a density of 105 (or 1.E+05) cells/mL and grown at 34°C in BSK-H medium. Samples were counted under a dark field microscope every 24 hours using a Petroff-Hausser cell counter. Differences between the numbers of WT and bb0238− or bb0238-complemented strains were not significant at all times (P > .05).
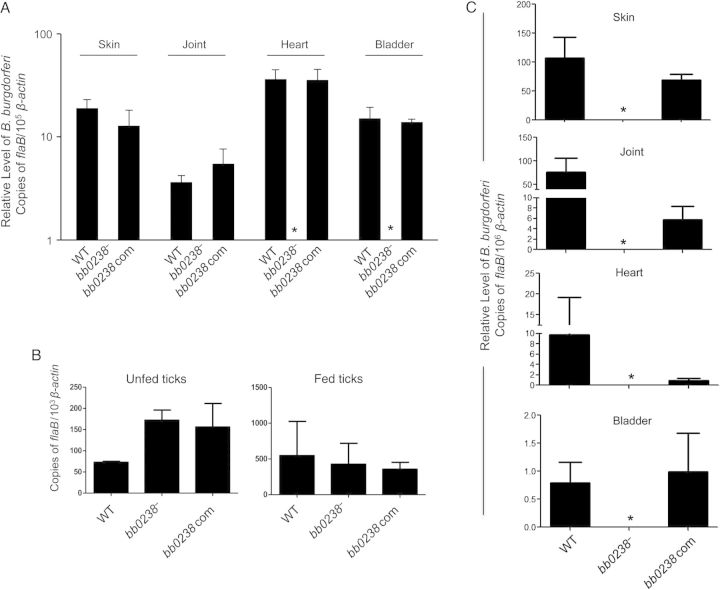
We next examined the role of BB0238 in B. burgdorferi infection of mice and ticks. The qRT-PCR analysis of pathogen levels in the murine hosts 2 weeks after needle inoculation (105 spirochetes per mouse, 3 animals per group) indicated that bb0238 mutants are undetectable, although both bb0238-complemented isolates and wild-type spirochetes persisted in all tested tissues (Figure 4A). The culture analyses further showed that the bb0238 mutants remained undetectable in all of the ear and heart samples collected from 6 mice. In contrast, most of the wild-type (4 of 6 mice) or bb0238-complemented isolate (2 of 3 mice) cultures remained positive. To further explore the role of bb0238 in spirochete persistence within the vector and transmission to naive hosts, we generated infected nymphs via microinjection. After infection, one group of nymphs remained as unfed ticks for 2 weeks, while another group was allowed to parasitize naive mice. Both unfed and fed ticks as well as mice at 2 weeks after feeding were assessed for B. burgdorferi levels by qRT-PCR analysis. Whereas all 3 isolates were able to survive in the unfed and fed repleted ticks (Figure 4B), only wild-type and bb0238-complemented isolates were detectable in all tested murine tissues (Figure 4C). These experiments strongly suggest that although BB0238 function is not necessary for spirochete persistence in ticks, it is essential for B. burgdorferi survival in the murine host and their transmission from infected ticks to naive hosts.
Figure 4.
Deletion of bb0238 attenuates spirochete ability to infect mice and transmit from ticks to mice. A, bb0238 mutants failed to persist in mice. C3H/HeN mice were inoculated intradermally with equal numbers of wild-type (WT), bb0238 mutant (bb0238−), or bb0238-complemented (bb0238 com) Borrelia burgdorferi (105 spirochetes per mouse, 3 animals per group). Infection was assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis of pathogen burden in skin, joint, heart, and bladder samples obtained 2 weeks after infection. Asterisks indicate undetectable levels of bb0238 mutants. B, B. burgdorferi burdens in artificially infected ticks. Ixodes scapularis nymphs were microinjected with equal numbers of WT, bb0238 mutant, or bb0238-complemented B. burgdorferi and then placed on mice (5 ticks per mouse). B. burgdorferi levels in unfed and fed repleted ticks were analyzed with qRT-PCR, measuring the number of copies of B. burgdorferi flaB transcripts and normalizing against tick β-actin transcripts. C, bb0238 is required for spirochete transmission from ticks to mice. Ticks were microinjected with equal numbers WT, bb0238 mutant, or bb0238-complemented B. burgdorferi and allowed to engorge on mice (3 animals per group). Spirochete burdens in different tissues were analyzed with qRT-PCR after 2 weeks of infection by measuring copies of flaB and were normalized against mouse β-actin levels. Error bars represent means and standard errors of the mean for relative levels of pathogen in tissues from 3 independent animal infection experiments.
BB0238 and BB0323 Stabilizes Each Other
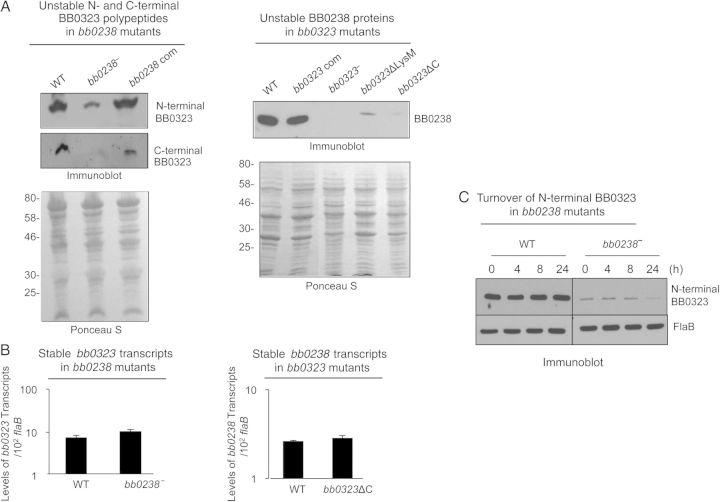
To further assess how deficiency of either of the interacting partners, BB0238 or BB0323, confers loss of infectivity, we examined the level of both proteins in the mutants. We performed immunoblot analysis to examine levels of BB0323 N- and C-terminal polypeptides in bb0238 mutants. Conversely, we also examined levels of BB0238 in bb0323 mutants. The results indicated that BB0323 polypeptides or BB0238 proteins are detectable, but their levels are dramatically reduced in bb0238 or bb0323 mutants, respectively (Figure 5A); although Ponceau S–labeled total protein profiles appeared to be similar in mutants and parental isolates (Figure 5A). Strikingly, bb0238 or bb0323 transcript levels were not altered in the corresponding mutants compared with wild-type spirochetes (Figure 5B), suggesting that the reduction of protein levels is a posttranslational event.
Figure 5.
BB0238 and BB0323 contribute to mutual posttranslational stability. A, Deletion of bb0238 or bb0323 in spirochetes reduces protein levels of the respective partner, as revealed by Western blot analysis. Comparable amounts of lysates from wild-type (WT), bb0238 mutants (bb0238−), bb0238-complemented isolates (bb0238 com), bb0323 mutants (bb0323−), BB0323 C-terminal–deletion mutants (bb0323ΔC), and BB0323 LysM domain–deletion mutants (bb0323ΔLysM) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with specific antibodies against BB0238 or N- or C-terminal polypeptides of BB0323. Protein loading is indicated by staining with Ponceau S (bottom panels). Migration of protein standards is shown to the left in kilodaltons (left panel). B, Targeted loss of bb0238 or bb0323 in Borrelia burgdorferi did not alter transcript levels of the respective partner. The level of bb0323 transcripts in WT and bb0238 mutants (top panel) or bb0238 transcripts in WT and bb323 mutants (bb0323ΔC) (bottom panel) was measured with quantitative reverse-transcription polymerase chain reaction. C, Posttranslational stability of BB0323 in bb0238 mutant. Protein synthesis was inhibited by the addition of 100 µg/mL spectinomycin to growing culture of spirochetes, and levels of N-terminal BB0323 were determined with immunoblot analyses. At indicated time points, spirochetes were collected, and lysates were separated by SDS-PAGE and immunoblotted using anti-N-terminal BB0323 and anti-FlaB (loading control).
To further assess protein turnover, we treated bb0238 mutant cells with an antibiotic that interrupts bacterial protein synthesis, spectinomycin, and the stability of BB0323 was monitored by Western blot analysis. Results show that while the levels of BB0323 in wild-type cells were unaffected 24 hours after antibiotic treatment, their levels in bb0238 mutants reflected a dramatic decrease by 24 hours, suggesting that BB0323 is unstable in bb0238-mutant cells. In contrast, as in wild-type cells, the level of FlaB in bb0238 mutants remained unaltered for 24 hours (Figure 5C).
Finally, in an independent coimmunoprecipitation study using BB0323 antibodies, a 27-kDa protein, which was identified by an liquid chromatography–mass spectrometric approach as BB0238, could readily be immunoprecipitated from wild-type B. burgdorferi (Supplementary Figure 1B); however, the protein is absent in similar coimmunoprecipitation efforts using bb0323 mutants lacking C-terminal polypeptides (bb0323ΔC, Supplementary Figure 2) and producing insignificant levels of BB0238 (Figure 5A). Taken together, these results suggest that BB0238 and BB0323 in B. burgdorferi are essential for each other's optimal stability.
DISCUSSION
Although extensive research over the past few decades has enriched our understanding of the biology of Lyme disease pathogens [32], many questions remain, such as how the OM connects to the PC with the presence of rotating periplasmic flagellar bundles. Although B. burgdorferi lacks orthologs of Tol-Pal proteins [18, 33], which connect the OM to the cell body in most gram-negative bacteria, targeted deletion of one of the borrelial genes, bb0323, resulted in a phenotype that is strikingly similar to the effects of removing certain Tol-Pal proteins [33, 34]. However, unlike the Tol-Pal proteins, BB0323 is processed into distinct N- and C-terminal polypeptides that support host infectivity in a manner that appears to be independent from an indispensable role in the fission process, as spirochetes carrying a C-terminal-truncated version of BB0323 display normal OM and fission yet remain noninfectious in mice [15]. Here we report identification of a novel periplasmic BB0323-interacting protein, BB0238, that is redundant for OM integrity and cell fission yet indispensable for host infectivity. Interaction of BB0238 and BB0323 seems critical for maintenance of posttranslational stability of both proteins. BB0238 might interact with additional proteins besides BB0323, which remains a subject of future investigation.
The existence of BB0238 and BB0323 in the same subcellular compartment of B. burgdorferi, such as in the periplasm, and as membrane-bound forms, further supports their functional association, as indicated by several independent in vitro and in vivo assays. However, we do not know whether formation of such a complex is necessary for host infectivity or whether instability in the absence of either partner (in deletion mutants) is due to a lack of direct interaction. Although bacteria may use small-protein modifiers, such as Pup proteins [35], or classic chaperones to control protein stability [36], limited information is available on possible mechanisms of maintaining posttranslational stability of borrelial proteins. In many gram-negative pathogens, type III–secreted proteins, including translocators, often require interacting proteins, such as specific chaperones, for proper storage and efficient secretion [37–39]. Nevertheless, to the best of our knowledge, BB0323 and BB0238 are a unique example of interacting proteins in which posttranslational stability of both proteins is mutually dependent. Findings have suggested that protein–protein interactions influence protein stability, especially of folded forms [40]. Therefore, it is tempting to speculate that folded BB0323-BB0238 proteins interact in the periplasm or another subcellular compartment pertinent to their stability and function, supporting spirochete infection.
BB0238 is a 27-kDa conserved borrelial protein that harbors an N-terminal tetratricopeptide repeat (TPR) domain, which is known to mediate protein–protein interactions in a variety of biological systems [41]. Not only does BB0238 directly interact with BB0323, but both proteins also follow a similar pattern of expression in murine hosts and ticks. Notably, although BB0238 supports stability of BB0323, it is not absolutely necessary for the existence of the latter protein. In the absence of BB0238, spirochetes can still process BB0323 and maintain a low level of N- and C-terminal polypeptides. Remarkably, in bb0238 mutants, small amounts of N-terminal polypeptide are sufficient to support cell fission, but a low level of C-terminal polypeptide, similar to their absence in bb0323 C-terminal-truncation mutants (bb0323ΔC), was unable to sustain infectivity. This further strengthens our speculation that BB0323 is involved in multiple functions [15], where low levels of N-terminal polypeptide can support normal cell fission but an absence (or a decreased level) of C-terminal polypeptide renders spirochetes noninfectious in the murine host.
Why BB0238 plays a nonessential role in the BB0323-mediated cell fission process yet is required for spirochete infectivity remains enigmatic. Similarly, although BB0323 and BB0238 are required for membrane stability, it is also puzzling why their expression levels, which fall below that of FlaB, differ dramatically during various stages of growth and infectivity. We speculate that their stoichiometry or that of other functionally associated molecules, such as BbHtrA, which mediates processing of BB0323 [15], helps maintain a critically low level of BB0323 or processed polypeptides sufficient for fission, whereas a higher level is required for host infectivity. Nevertheless, our study underscores that inhibition of BB0238 and BB0323 interaction or their function may provide novel strategies to interfere with B. burgdorferi infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Faith Kung for her assistance with this study. We also thank Philip Stewart and Patricia Rosa for the generous gift of a C-terminal BB0323 antibody.
Financial support. This work was supported by funding from the National Institute of Allergy and Infectious Diseases (award R01AI080615 to U. P).
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Piesman J, Eisen L. Prevention of tick-borne diseases. Annu Rev Entomol. 2008;53:323–43. doi: 10.1146/annurev.ento.53.103106.093429. [DOI] [PubMed] [Google Scholar]
- 2.Piesman J, Gern L. Lyme borreliosis in Europe and North America. Parasitology. 2004;129(suppl)):S191–220. doi: 10.1017/s0031182003004694. [DOI] [PubMed] [Google Scholar]
- 3.Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour AG, Hayes SF. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–9. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JF. Epizootiology of Borrelia in Ixodes tick vectors and reservoir hosts. Rev Infect Dis. 1989;11(suppl 6):S1451–9. doi: 10.1093/clinids/11.supplement_6.s1451. [DOI] [PubMed] [Google Scholar]
- 7.de Silva AM, Tyson KR, Pal U. Molecular characterization of the tick-Borrelia interface. Front Biosci. 2009;14:3051–63. doi: 10.2741/3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barthold SW, Diego C, Philipp MT. Animal models of borreliosis. In: Samuels DS, Radolf JD, editors. Borrelia: molecular biology, host interaction and pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. pp. 353–405. [Google Scholar]
- 10.Norris SJ, Coburn J, Leong JM, Hu LT, Höök M. Pathobiology of Lyme disease Borrelia. In: Samuels DS, Radolf JD, editors. Borrelia: molecular biology, host interaction and pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. pp. 299–331. [Google Scholar]
- 11.Radolf J. Posttreatment chronic Lyme disease—what it is not. J Infect Dis. 2005;192:948–9. doi: 10.1086/432736. [DOI] [PubMed] [Google Scholar]
- 12.Marconi RT, Earnhart CG. Lyme disease vaccines. In: Samuels DS, Radolf JD, editors. Borrelia: molecular biology, host interaction and pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. pp. 467–86. [Google Scholar]
- 13.Motaleb MA, Corum L, Bono JL, et al. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A. 2000;97:10899–904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charon NW, Cockburn A, Li C, et al. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol. 2012;66:349–70. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kariu T, Yang X, Marks CB, Zhang X, Pal U. Proteolysis of BB0323 results in two polypeptides that impact physiologic and infectious phenotypes in Borrelia burgdorferi. Mol Microbiol. 2013;88:510–22. doi: 10.1111/mmi.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–8. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol. 2002;184:754–9. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–25. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–7. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- 20.Stewart PE, Hoff J, Fischer E, Krum JG, Rosa PA. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl Environ Microbiol. 2004;70:5973–9. doi: 10.1128/AEM.70.10.5973-5979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Yang X, Kumar M, Pal U. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis. 2009;200:1318–30. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman JL, Crowley JT, Toledo AM, Benach JL. The HtrA protease of Borrelia burgdorferi degrades outer membrane protein BmpD and chemotaxis phosphatase CheX. Mol Microbiol. 2013;88:619–33. doi: 10.1111/mmi.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gherardini FC. Borrelia burgdorferi HtrA may promote dissemination and irritation. Mol Microbiol. 2013;90:209–13. doi: 10.1111/mmi.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell TM, Johnson BJ. Lyme disease spirochaetes possess an aggrecan-binding protease with aggrecanase activity. Mol Microbiol. 2013;90:228–40. doi: 10.1111/mmi.12276. [DOI] [PubMed] [Google Scholar]
- 25.Ostberg Y, Carroll JA, Pinne M, Krum JG, Rosa P, Bergstrom S. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J Bacteriol. 2004;186:2074–84. doi: 10.1128/JB.186.7.2074-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias AF, Stewart PE, Grimm D, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–50. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Promnares K, Qin J, et al. Characterization of multiprotein complexes of the Borrelia burgdorferi outer membrane vesicles. J Proteome Res. 2011;10:4556–66. doi: 10.1021/pr200395b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Lenhart TR, Kariu T, Anguita J, Akins DR, Pal U. Characterization of unique regions of Borrelia burgdorferi surface-located membrane protein 1. Infect Immun. 2010;78:4477–87. doi: 10.1128/IAI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol Microbiol. 2009;74:112–25. doi: 10.1111/j.1365-2958.2009.06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kariu T, Coleman AS, Anderson JF, Pal U. Methods for rapid transfer and localization of Lyme disease pathogens within the tick gut. J Vis Exp. 2011;48:e2544. doi: 10.3791/2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motaleb MA, Sal MS, Charon NW. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J Bacteriol. 2004;186:3703–11. doi: 10.1128/JB.186.12.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuels DS, Radolf JD. Borrelia: molecular biology, host interaction and pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. [Google Scholar]
- 33.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2012;192:4847–58. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llamas MA, Ramos JL, Rodriguez-Herva JJ. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol. 2000;182:4764–72. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barandun J, Delley CL, Weber-Ban E. The pupylation pathway and its role in mycobacteria. BMC Biol. 2012;10:95. doi: 10.1186/1741-7007-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong P, Houry WA. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J Struct Biol. 2004;146:79–89. doi: 10.1016/j.jsb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Ku CP, Lio JC, Wang SH, Lin CN, Syu WJ. Identification of a third EspA-binding protein that forms part of the type III secretion system of enterohemorrhagic Escherichia coli. J Biol Chem. 2009;284:1686–93. doi: 10.1074/jbc.M807478200. [DOI] [PubMed] [Google Scholar]
- 38.Lin CN, Sun WS, Lu HY, Ng SC, Liao YS, Syu WJ. Protein interactions and regulation of EscA in enterohemorrhagic E. coli. PLoS One. 2014;9:e85354. doi: 10.1371/journal.pone.0085354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creasey EA, Friedberg D, Shaw RK, et al. CesAB is an enteropathogenic Escherichia coli chaperone for the type-III translocator proteins EspA and EspB. Microbiology. 2003;149:3639–47. doi: 10.1099/mic.0.26735-0. [DOI] [PubMed] [Google Scholar]
- 40.Dixit PD, Maslov S. Evolutionary capacitance and control of protein stability in protein-protein interaction networks. PLoS Comput Biol. 2013;9:e1003023. doi: 10.1371/journal.pcbi.1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–62. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.