Abstract
Leber congenital amaurosis (LCA) and juvenile retinitis pigmentosa (RP) are severe hereditary diseases that causes visual impairment in infants and children. SPATA7 has recently been identified as the LCA3 and juvenile RP gene in humans, whose function in the retina remains elusive. Here, we show that SPATA7 localizes at the primary cilium of cells and at the connecting cilium (CC) of photoreceptor cells, indicating that SPATA7 is a ciliary protein. In addition, SPATA7 directly interacts with the retinitis pigmentosa GTPase regulator interacting protein 1 (RPGRIP1), a key connecting cilium protein that has also been linked to LCA. In the retina of Spata7 null mutant mice, a substantial reduction of RPGRIP1 levels at the CC of photoreceptor cells is observed, suggesting that SPATA7 is required for the stable assembly and localization of the ciliary RPGRIP1 protein complex. Furthermore, our results pinpoint a role of this complex in protein trafficking across the CC to the outer segments, as we identified that rhodopsin accumulates in the inner segments and around the nucleus of photoreceptors. This accumulation then likely triggers the apoptosis of rod photoreceptors that was observed. Loss of Spata7 function in mice indeed results in a juvenile RP-like phenotype, characterized by progressive degeneration of photoreceptor cells and a strongly decreased light response. Together, these results indicate that SPATA7 functions as a key member of a retinal ciliopathy-associated protein complex, and that apoptosis of rod photoreceptor cells triggered by protein mislocalization is likely the mechanism of disease progression in LCA3/ juvenile RP patients.
Introduction
Leber congenital amaurosis (LCA, MIM 204000) is the most severe early onset inherited retinal dystrophy. Patients suffer severe vision loss or blindness within their first year of life, typically accompanied by sensory nystagmus, amaurotic pupils with sluggish or no pupillary response and little or no response to light as measured by the electroretinogram (ERG). The molecular basis underlying LCA is highly heterogeneous; to date, 18 genes have been associated with recessive LCA (1–23). These genes are involved in various aspects of visual function, including the retinoid cycle, outer segment (OS) phagocytosis, protein transport and photoreceptor development. The genetic causes for LCA can overlap with milder, progressive retinal dystrophies, such as retinitis pigmentosa (RP). In addition, they can also overlap with syndromic phenotypes, often but not exclusively encompassing additional characteristics such as nephronophthisis (in Senior-Løken syndrome) and cerebellar vermis hypoplasia (in Joubert syndrome), or more extensive phenotypes affecting many additional organs as observed in Bardet-Biedl syndrome and Meckel-Gruber syndrome. These syndromic cases are often caused by a dysfunctional primary cilium, a microtubule-based sensory organelle that is present in most cell types. The photoreceptor cell also carries a sensory cilium, encompassing the highly specialized outer segment (OS) that contains the Opsin-filled membrane stacks, and the more prototypic basal body and adjacent transition zone. The latter is named the connecting cilium (CC) in photoreceptor cells, as it connects the protein biosynthetic inner segment with the phototransductive OS, acting as a conveyor belt for intraflagellar transport particles (IFTs) that translocate proteins and vesicles along the microtubule tracks of the CC. Strikingly, about a third of all retinal dystrophy genes encode proteins that localize to this structure (24), pinpointing it as the Achilles’ heel of the photoreceptor cell.
Previously, we have identified SPATA7 (spermatogenesis-associated protein 7) as the affected disease gene for the LCA3 locus (20). Follow-up studies from several groups estimate that mutations in SPATA7 account for about 1.7 and 4.6% of LCA patients in the British and Chinese populations, respectively (25–27). In the mouse, Spata7 is expressed in the retina (20) as well as in many other tissues, although it is most abundant in the testis where it was first identified (28). The SPATA7 protein is conserved from sea urchin to human. Sequence analysis of SPATA7 reveals no conserved protein domain except for a predicted coiled-coil domain at amino acids 81–109 (SMART). Despite its apparent importance in human eye disease, details of the molecular function of SPATA7 remain unknown.
To decipher SPATA7 function in the retina and understand the mechanism of disease in LCA3/ juvenile RP patients, we took an integrated approach involving accurate SPATA7 localization in cells and mouse retinas, protein interaction screening and scrutinizing the phenotype of a Spata7 knock-out model. Our data show that SPATA7 is a ciliary protein that localizes at the connecting cilium of photoreceptor cells. In addition, it directly interacts with RPGRIP1 in vitro and in vivo, a known LCA disease protein that co-localizes at the CC and facilitates protein trafficking between the inner segment (IS) and OS, suggesting a close functional interaction between SPATA7 and RPGRIP1. Indeed, in the homozygous Spata7 mutant retina, the level of RPRGIP1 at the connecting cilium is greatly reduced and the protein is mislocalized to the inner segment. In addition, in Spata7 mutant retinas, mislocalization of rhodopsin (RHO) is observed followed by apoptotic photoreceptor cell death. Taken together, we propose a model where SPATA7 plays a key role in RPGRIP1-mediated protein trafficking across the connecting cilium of photoreceptor cells and that apoptotic degeneration of these cells triggered by protein mislocalization is a likely mechanism of disease progression in LCA3/ juvenile RP patients.
Results
SPATA7 is a microtubule-associated protein and localizes to the connecting cilium of photoreceptor cells
To establish the cellular location of SPATA7, epitope-tagged SPATA7 was expressed in hTERT RPE-1 cells. Interestingly, overexpressed SPATA7 decorates the cellular microtubule network and localizes to the ciliary axoneme of ciliated cells (Fig. 1A–D, and Supplementary Material, Fig. S1A). Expression of different SPATA7 fragments in ciliated hTERT RPE-1 cells indicates that this microtubule association depends on the N-terminal part (amino acids 2–310) of SPATA7 which contains the predicted coiled-coil domain, but not the C-terminal region (Supplementary Material, Fig. S1B and C).
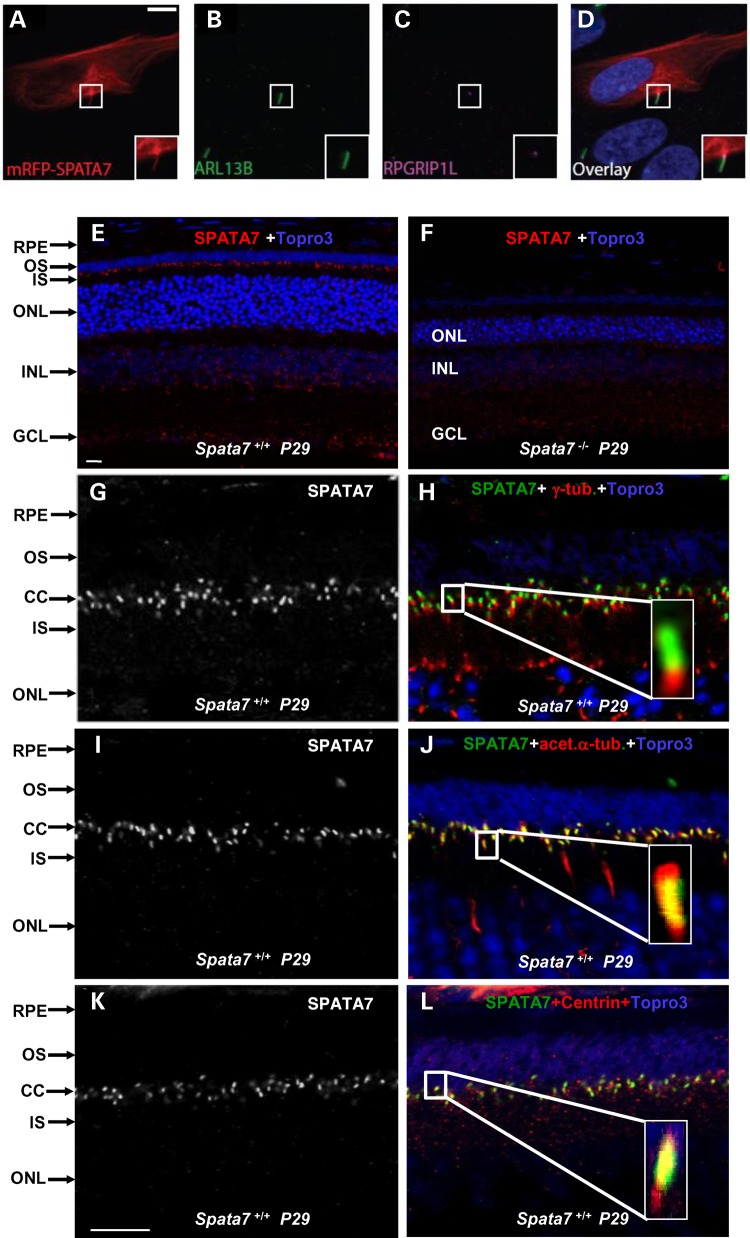
Figure 1.
SPATA7 is an MAP and localizes to connecting cilia in photoreceptors. (A–D) Localization of mRFP_SPATA7 (A, red) in hTERT RPE-1 cells, ARL13B (B, green) and RPGRIP1L (C, magenta) are used as markers for ciliary axonemes and basal bodies respectively. Scale bar: 10 µm. (E–L) SPATA7 localizes to connecting cilia in mouse photoreceptors. Immunofluorescence staining of SPATA7 (red) at P29 in wild-type (E) and Spata7 mutant (F) retinas; scale bar is 10 µm. Double labeling experiments were conducted to determine the subcellular localization of SPATA7 in photoreceptor cells. SPATA7 (green) co-stained with the basal body marker γ-tubulin (red) (G and H), the axonemal marker acetylated-α-tubulin (red) (I and J) and Centrin (red) (K and L). Topro3 (purple) is used to label nuclei, scale bar is 10 µm. Paraffin embedded mouse retinal sections were used for these immunofluorescence staining.
Given the results from the recombinant protein expression, we wanted to assess if SPATA7 also localizes to cilia in vivo. SPATA7 was shown previously to be expressed in the mouse retina using commercially available anti-SPATA7 antibodies (20). However, the sensitivity of these antibodies is low, resulting in low resolution. Furthermore, without Spata7 mutant mice as negative controls, it is difficult to evaluate the specificity of the antibodies. To further investigate the pattern of SPATA7 expression and gain a more precise subcellular localization in the retina, new polyclonal antibodies against SPATA7 were generated. Retinal sections from different time points during development were stained using the custom anti-SPATA7antibodies. Coinciding with developing photoreceptors, SPATA7 is first clearly detected at P4, and the immunoreactivity increases as time progresses (Supplementary Material, Fig. S2A, E and I). At P15, the strongest SPATA7 immunoreactivity is observed specifically in the photoreceptor cell layer (Supplementary Material, Fig. S2M and N). Furthermore, SPATA7 specifically localizes at the region between the inner segment (IS) and outer segment (OS), coinciding with the connecting cilium (Fig. 1E–L). The staining pattern is specific to SPATA7 since no signal in this region is observed in Spata7−/− mouse retinas stained with the same antibody (Fig. 1F). To verify the connecting cilium localization of SPATA7, double stainings with several cilium markers and anti-SPATA7 were conducted. The basal body of the connecting cilium can be marked by γ-tubulin (29). As shown in Figure 1G and H, SPATA7 localizes distal to γ- tubulin with little overlap. Consistent with this observation, SPATA7 staining overlaps with acetylated α-tubulin (30) and pan-Centrin (31,32), two markers of the axoneme that localizes distal to the basal body (Fig. 1J and L), and an enrichment of endogenous SPATA7 to the basal body of the cilium is also observed in hTERT RPE-1 cells (Supplementary Material, Fig. S3D). Similar to rod photoreceptor cells, SPATA7 is also found at the connecting cilium of cone photoreceptor cells in mouse retinas (Supplementary Material, Fig. S3B and C).
SPATA7/LCA3 interacts with RPGRIP1/LCA6
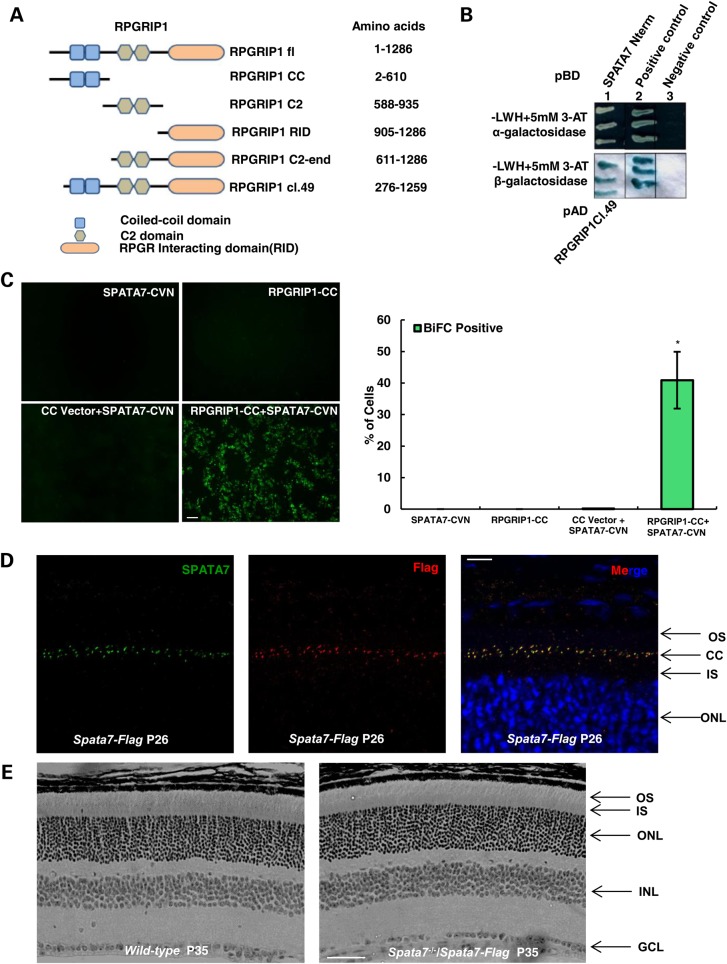
Given the predominant localization of SPATA7 at the connecting cilium of photoreceptor cells, where proteins often function in multi-subunit complexes, a GAL4-based yeast two hybrid system was used to screen a human retinal cDNA library to identify potential SPATA7 protein–protein interactions. Using an N-terminal fragment (amino acids 2–309) that contains the predicted coiled-coil region of SPATA7 as the bait, we identified one clone expressing RPGRIP1 (clone 49) as a potential binding partner from this screen (Fig. 2A and B). RPGRIP1 is known to be involved in establishing a retinal degeneration-associated protein complex in the connecting cilium, and the cognate gene is mutated in LCA6 (7). The specificity of this interaction was further confirmed using several complementary in vitro and in vivo methods (Fig. 2 and Fig. 3). First, while no fluorescent signals are detected in cells expressing either SPATA7 or RPGRIP1 alone, expression of both proteins induces fluorescence using the bimolecular fluorescence complementation (BiFC) assay, indicating that they are localizing in very close proximity of each other (Fig. 2C). Secondly, we validated the presence of SPATA7 and RPGRIP1 in vivo in the same retinal protein complex by co-immunoprecipitation (co-IP) from mouse retinal tissue. To improve efficiency of SPATA7 immunoprecipitation, we generated transgenic mice carrying a Flag-tagged Spata7 genomic construct (Spata7-Flag). As shown in Figure 2D, the fusion protein is located at the connecting cilium, comparable to the wild-type protein. Indeed, the genomic construct completely rescues the known mutant phenotypes of Spata7−/− mice (Fig. 2E). Using anti-Flag antibodies (Fig. 3C), a substantial amount of RPGRIP1 is pulled down by Flag-tagged SPATA7 (Fig. 3D), confirming the in vivo interaction of these two proteins in the retina.
Figure 2.
SPATA7 physically interacts with RPGRIP1 (A) RPGRIP1 fragments, identified in the Y2H screen (RPGRIP1 cl.49) and those used in downstream validation experiments. (B) Validation of interaction of SPATA7 N-terminal fragment and RPGRIP1 in Y2H assays. Protein–protein interactions induced yeast growth on selective media (−LWH + 3-AT), a positive (green) α-galactosidase assay (top panel) and a positive (blue) β-galactosidase assay (bottom panel). pBD depicts DNA binding domain fusions; pAD depicts transcription activation domain fusions. (C) The interaction between SPATA7 and RPGRIP1 was confirmed in cellulo by the BiFC assay. HEK293T cells were transfected with either RPGRIP1 alone or together with SPATA7 constructs. Strong fluorescence signals (green) were observed in cells expressing both SPATA7 and RPGRIP1, but not in cells that express only one protein. Bar = 50 µm. The percentage of BiFC positive cells was quantified by FACS (*P<0.001). (D) Paraffin-embedded retinal sections at P26 were co-stained for SPATA7 (green) and Flag (red) in the connecting cilium of photoreceptors of transgenic mice and show complete co-localization of SPATA7 with Flag at physiological levels of the proteins, scale bar is 10 um. (E) Hematoxylin and eosin (H and E) staining of paraffin-embedded retinal sections at P35. The Spata7-Flag transgene has fully rescued retinal phenotype of Spata7 knockout mouse. Scale bar is 50 um.
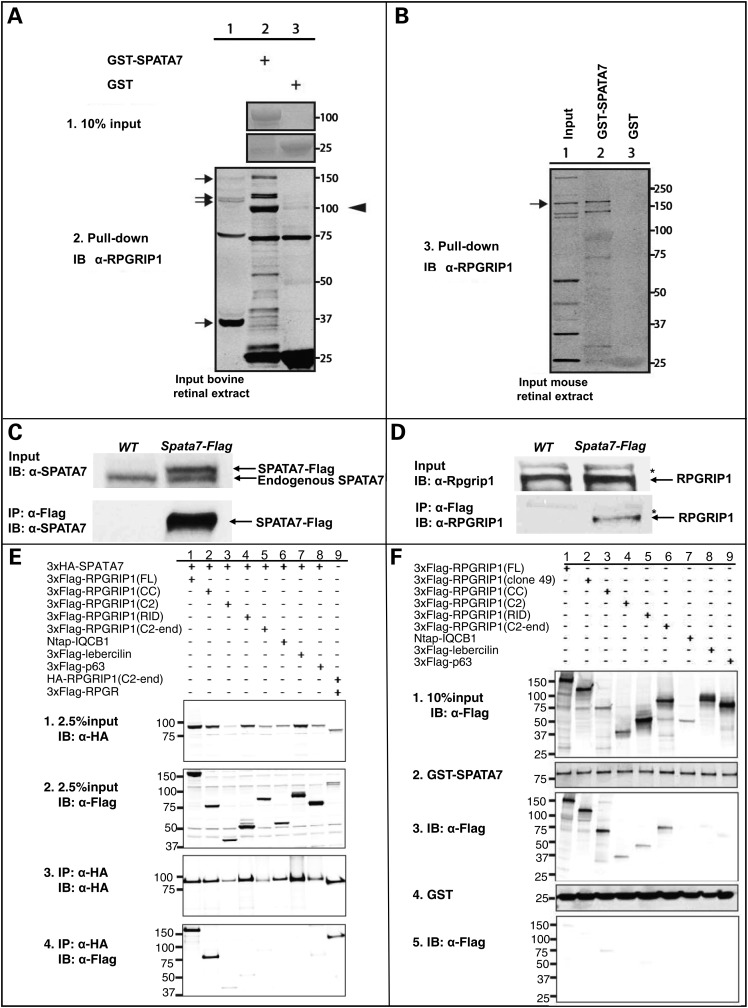
Figure 3.
Interaction of SPATA7 and RPGRIP1 validated by GST pull-down and Co-IP in bovine and mouse retinas. GST-SPATA7 pulls down endogenous RPGRIP1 isoforms of approximately 150 and 120 kDa, but not the smallest isoform of 37 kDa, from a bovine retinal lysate (A, lanes 1–2, arrows). The arrowhead points to a presumed degradation product of one of the larger isoforms as this was not detected in the input extract (lane 1). Two non-specific immunoreactive bands, which were also not recognized in the input (lane 1), migrated at 75 and 25 kDa in lanes 2 and 3, and are thus not relevant for this assay. (B) An endogenous murine RPGRIP1 isoform of 155 kDa can be pulled down from mouse retinal lysates by GST-SPATA7, but not by unfused GST. (C,D) Co-immunoprecipitation of RPGRIP1 with SPATA7 from mouse retinal extracts. Retinas from Spata7-Flag transgenic and wild-type mice were used for IP. RPGRIP1 is co-immunoprecipitated with SPATA7 from Spata7-Flag transgenic mouse retinas. *Non-specific bands. (E) 3xHA-SPATA7 co-precipitates efficiently with full-length RPGRIP1, as well as with the coiled-coil fragment of RPGRIP1 (2.5% input, panels 1, 2; co-IP assay in panels 3–4, lanes 1–2). In a weaker manner, 3xHA-SPATA7 co-precipitates with other RPGRIP1 fragments (panels 3–4, lanes 3–5). 3xHA-SPATA7 is unable to co-precipitate two other coiled-coil containing protein IQCB1 and lebercilin. RPGR-RPGRIP1 binding is used as a positive control (lane 9), a very weak binding of 3xHA-SPATA7 to 3xFlag-p63 (a negative control) is also observed (lane 8), at similar levels compared with the RPGRIP1 fragments not containing the CC domain, reiterating the weak affinity of these fragments to SPATA7 in this assay. (F) GST- SPATA7 (10% input, panel 2) pulls down all tested fragments of RPGRIP1 (10% input, panel 1; pull-down assay in panel 3, lanes 1–6), while unfused GST does not (panel 5, 10% input in panel 4). IQCB1, lebercilin and p63 are included as negative controls.
Thirdly, we performed a GST pull-down assay from retinal extracts. From bovine retinal extracts, four annotated isoforms of bovine RPGRIP1 are detected specifically at the approximate sizes of 150, 120 and 37 kDa using previously described anti-RPGRIP1 antibodies (Fig. 3A, lane 1, arrows) (33,34) (Uniprot). As shown in Figure 3A, GST-SPATA7 is able to pull down endogenous RPGRIP1. However, the smallest RPGRIP1 isoform, which lacks the predicted coiled-coil domain at amino acid 267–533 (Uniprot), is not detected in the pull-down fraction. Unfused GST shows no capacity to pull down endogenous RPGRIP1. Similarly, in a pull-down assay of GST-SPATA7 from mouse retinal extracts, endogenous RPGRIP1 was detected specifically at ∼155 kDa using mouse anti-RPGRIP1 antibodies (35), but not when using unfused GST (Fig. 3B, lanes 1–3).
SPATA7 binds to the coiled-coil domain of RPGRIP1
Previously, it has been shown that the C-terminal region of RPGRIP1 binds to RPGR, which is associated with X-linked RP (33), and the central C2 domain-containing region of RPGRIP1 binds specifically to NPHP4 (36), which is associated with the retinal renal ciliopathy Senior-Løken syndrome as well as isolated nephronophthisis (NPHP). To evaluate whether SPATA7 competes for any of these binding epitopes, the binding capacity of SPATA7 for different fragments of RPGRIP1 was assessed using co-immunoprecipitation and GST pull-down analyses. SPATA7 co-precipitated with full-length RPGRIP1, and the coiled-coil fragment of RPGRIP1 (Fig. 3E, lanes 1 and 2). However, the absence of this coiled-coil domain in RPGRIP1 (C2), RPGRIP1 (RID) and RPGRIP1 (C2-end) fragments resulted in a less efficient co-precipitation of RPGRIP1 (Figs. 2A and 3E, lanes 3–5). In addition, SPATA7 did not co-precipitate with two other coiled-coil domain containing proteins IQCB1 (IQ-calmodulin-binding motif-containing protein 1) and lebercilin (Fig. 3E, lanes 6 and 7, respectively), indicating that the SPATA7-RPGRIP1 interaction is specific. Consistent with co-immunoprecipitation data, GST-fused SPATA7 co-purified all different fragments of RPGRIP1. Also here, the strongest affinity in this assay was observed with full-length RPGRIP1, the RPGRIP1 clone 49 identified from the Y2H assay, and the coiled-coil domain containing fragment of RPGRIP1 (CC) (Fig. 3F, lanes 1–3) but not with RPGRIP1 fragments lacking the N-terminal coiled-coil domain (Fig. 3F, lanes 4–6). Taken together, these data indicate that although SPATA7 can interact with different domains of RPGRIP1, the vast majority of the affinity is contributed by the region containing the predicted coiled-coil domain of RPGRIP1.
To further delineate the interacting domains, different epitope-tagged fragments of RPGRIP1 and SPATA7 were expressed in ciliated hTERT RPE-1 cells. As previously shown (36), full-length RPGRIP1 diffusely localizes to the cytoplasm (Supplementary Material, Fig. S4A). Coexpression of full-length RPGRIP1 and SPATA7 yields a near complete recruitment of RPGRIP1 to the microtubule cytoskeleton, which is also decorated by and thus colocalizing with SPATA7 (Supplementary Material, Fig. S4B). Absence of the coiled-coil domain in both RPGRIP1(C2-end) and SPATA7 (fragment amino acids 311–568) results in two distinct localization patterns of these proteins upon expression in ciliated cells (data not shown). These data indicate that not only the coiled-coil domain of RPGRIP1 but also the N-terminal part of SPATA7, which contains a predicted coiled-coil domain, contributes to the full affinity and colocalization of SPATA7 and RPGRIP1.
Generation of Spata7 null mice
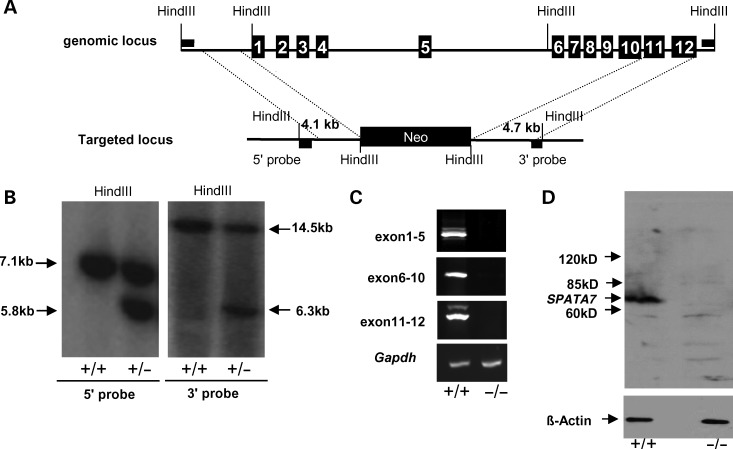
To further evaluate the role of SPATA7 at the CC and to elucidate mechanisms of LCA3/ juvenile RP pathogenesis, we set out to generate a targeted knock-out mouse model of Spata7. Mouse Spata7 contains 12 exons and encodes a protein of 582 amino acids, sharing 67% identity with its human ortholog. As shown in Figure 4A, the genomic region containing exons 1 through 10 of Spata7 was replaced by a Neo cassette. As a result, most of the coding region of Spata7 including the translation start site was removed, resulting in a null allele. Mice carrying the Spata7 null allele were confirmed by genomic Southern blot analysis (Fig. 4B). Homozygous mutant mice (Spata7−/−) are obtained at the expected frequency (25%) from mating of heterozygous mutant mice. Spata7−/− mice are viable, fertile and without gross morphological defects.
Figure 4.
Generation of Spata7 null mutant mice. (A) Spata7 targeting strategy. Numbered rectangles represent Spata7 exons. (B) Genomic Southern of Spata7 targeted allele. Fragments of 5.8 kb and 6.3 kb are detected in a heterozygous mouse using 5′ and 3′ probes, respectively. (C) No transcript is detected from retinas of Spata7 homozygous knockout mice. RT–PCR of wild-type and Spata7 knockout retinas was carried out using three different primer pairs spanning exons 1–5, 6–10 and 11–12, respectively. Gapdh was used as a positive control. (D) No SPATA7 protein is detected in mutant retina extracts by western blot.
To confirm the null allele, several methods, including RT–PCR and western blotting, were used to detect the Spata7 transcript and protein. As shown in Figure 4C, using three primer pairs covering the coding region of Spata7, positive bands can be amplified by RT–PCR from wild-type retinal mRNA. In contrast, corresponding bands are not detected from mRNA extracted from the adult Spata7−/− retina. Similarly, a protein of ∼64 kDa corresponding to the predicted size of SPATA7 is detected in the wild-type retina (Fig. 4D), while no corresponding band is detected in Spata7−/− retinas by western blotting (Fig. 4D). These data indicate that the mutant allele we constructed is indeed a molecular null.
Progressive degeneration of photoreceptors in the Spata7 mutant retina
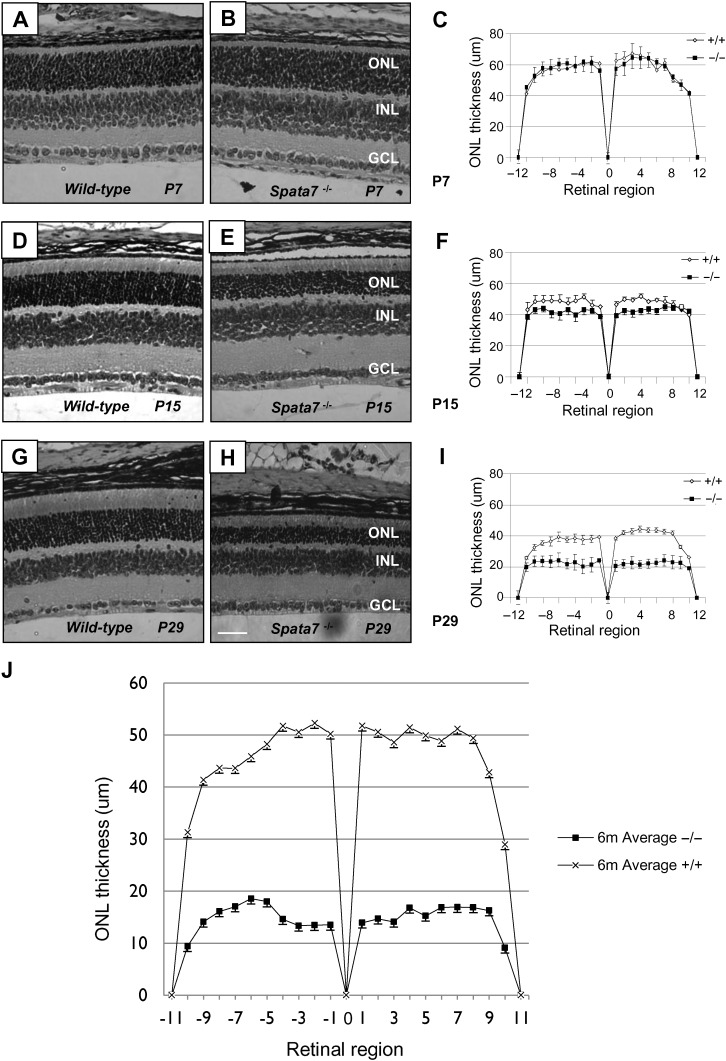
Consistent with the human phenotype, severe early-onset retinal defects are observed in Spata7 mutant mice. As shown in Figure 5, a marked reduction in the thickness of the outer nuclear layer (ONL) is observed in Spata7 mutant retinas compared with those of wild-type mice. Although no obvious differences are observed between wild-type and mutant retinas at P7 (Fig. 5A–C), by P15, a decrease in the thickness of the ONL is detected in Spata7 mutant retinas (Fig. 5D–F). A further reduction in the ONL, to about 50% of wild-type, is observed by P29 (Fig. 5G–I). At 6 months, the ONL is just 30–40% of wild-type (Fig. 5J). After 6 months, only the ONL still slowly continues to become thinner. These results suggest that photoreceptor cells degenerate in the absence of Spata7 function.
Figure 5.
Progressive photoreceptor degeneration in the Spata7 mutant retina. Hematoxylin and eosin (H&E) staining of paraffin-embedded retinal sections and retinal morphometry at P7 (A–C), P15 (D–F) and P29 (G–I). Retinal morphometry was also conducted at 6 months of age for wild-type and Spata7 mutants (J). For retinal morphometry, ONL thickness was measured at 20 equally spaced positions along the vertical meridian of the retina for three retinas of the same genotype. Three measurements were taken for each position, and an average value was calculated. Each point represents the mean ± SEM obtained for each group (n = 3). Position 0 corresponds to the optic nerve head. RPE, retinal pigment epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar: 50 µm.
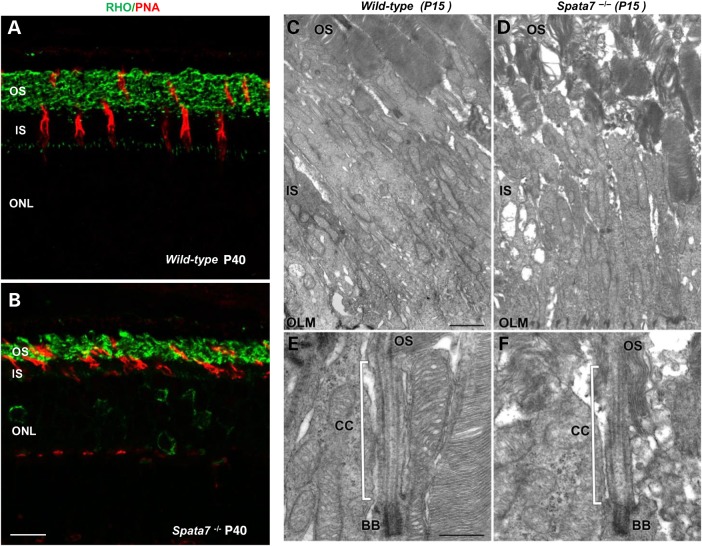
To determine if other cell types are affected in Spata7 mutant retinas, we performed immunolabeling with a panel of cell type-specific markers, including BRN3B, CHX10, PAX6 and Calbindin that mark ganglion cells, bipolar cells, amacrine cells and horizontal cells, respectively (37–40). As shown in Supplementary Material, Fig. S5, the staining patterns for all four antibodies appear normal in the Spata7 mutant retina, indicating that there are no gross defects in these cell types. In contrast, abnormal staining for photoreceptor cell markers is observed (Supplementary Material, Fig. S5B). As shown in Figure 6A and B, rods and cones, marked by RHO and Peanut agglutinin (PNA) staining, respectively (41), are disorganized and abnormal in Spata7 mutant retinas at Day P40.
Figure 6.
Photoreceptor cell defects in the Spata7 mutant retina. (A and B) Cones and rods were labeled with Peanut agglutinin lectin (PNA) (red) and rod Opsin (green), respectively, in paraffin sections of wild-type (A) and Spata7 mutant retinas (B) (scale bar is 10 µm). (C and D) Transmission electron microscopy was used to reveal detailed structure of photoreceptors at P15. The OSs of Spata7 mutant retinas (D) are disorganized compared with wild-type (C). Despite the defect in the OSs, the connecting cilia (CC) and basal bodies (BB) of Spata7 mutants (F) appear normal compared with wild-type (E). OLM-outer limiting membrane. Scale bar is 1 µm in C (D is the same magnification) and 0.5 µm in E (F is the same magnification).
In the mouse retina, over 90% of photoreceptor cells are rods (42). The extensive degeneration observed in the photoreceptor layer of Spata7 mutants must therefore involve rods. To assess if cone photoreceptors also degenerate in the Spata7 mutant retina, the number of cone cells in a unit area was counted in whole mount retinas. At the age of 7 months, only a small reduction in cone photoreceptor cells is observed across the Spata7 mutant retina compared with wild-type (data not shown), suggesting that cone photoreceptor degeneration proceeds at a substantially lower rate compared with rods in Spata7 mutant retinas.
The fine structure of photoreceptors was also examined using transmission electron microscopy (TEM). In wild-type retinas, membrane discs in OSs are well-organized into stacks (Fig. 6C). In contrast, in Spata7 mutant retinas, OSs are shortened and the disc membranes are disorganized (Fig. 6D). However, the ultrastructure of inner segments (IS) and the connecting cilium (CC) seem largely unaffected in Spata7−/− photoreceptors, with basal bodies and axonemes still present (Fig. 6E and F).
Defects in response to light in Spata7 mutants
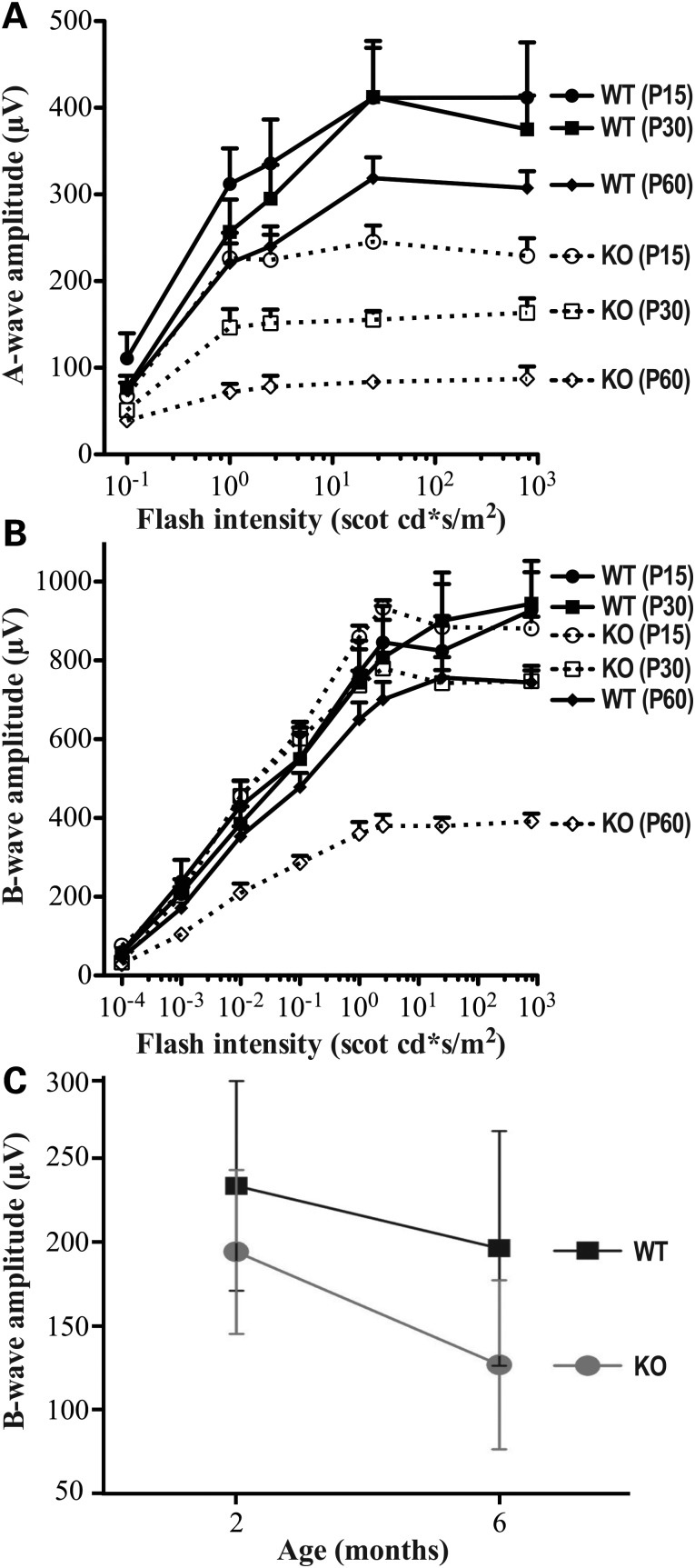
One of the hallmarks of LCA and juvenile RP is a dramatic reduction in response to light as determined by ERG measurements (20,43). To test whether a similar functional defect is observed in Spata7−/− retinas, both rod and cone ERGs were recorded. As shown in Figure 7, a decline in the scotopic (rod-mediated) A-wave amplitude is observed in Spata7 mutant mice by P15 and becomes more pronounced with age (Fig. 7A). By P60, the A-wave amplitude is further reduced to only 20% of that of wild-type retinas. In contrast, a substantial decline in the scotopic B-wave amplitude appears later in life and is first detected in Spata7−/− retinas at about 2 months of age (Fig. 7B). These data suggest that the primary defect in Spata7−/− mice is in rod photoreceptors. Consistent with the mild decrease in the number of cone cells, the double-flash B-wave, which measures cone photoreceptor response, shows only a slight age-dependent decline in Spata7−/− mutant mice. Specifically, the cone cell response is reduced by only 20 and 30% compared with wild-type retinas by 2 and 6 months, respectively (Fig. 7C). Retinal degeneration continues to progress as the A-wave amplitude is further reduced and is almost undetectable by the age of 12 months (Supplementary Material, Fig. S6).
Figure 7.
Spata7 mice exhibit defects in response to light. ERGs were performed on Spata7 mutant mice at different ages. Fractional scotopic a-wave (A) and b-wave amplitudes (B) of ERG recordings as a function of increasing light intensities at P15, 1 month and 2 months are shown. (C) Fractional b-wave amplitudes of double-flash cone b-waves for 2- and 6-month-old mice. n = 5 or more for each group. Each point represents the mean ± SEM obtained for each group.
Mislocalization of RPGRIP1 in the connecting cilia of Spata7 mutant mice
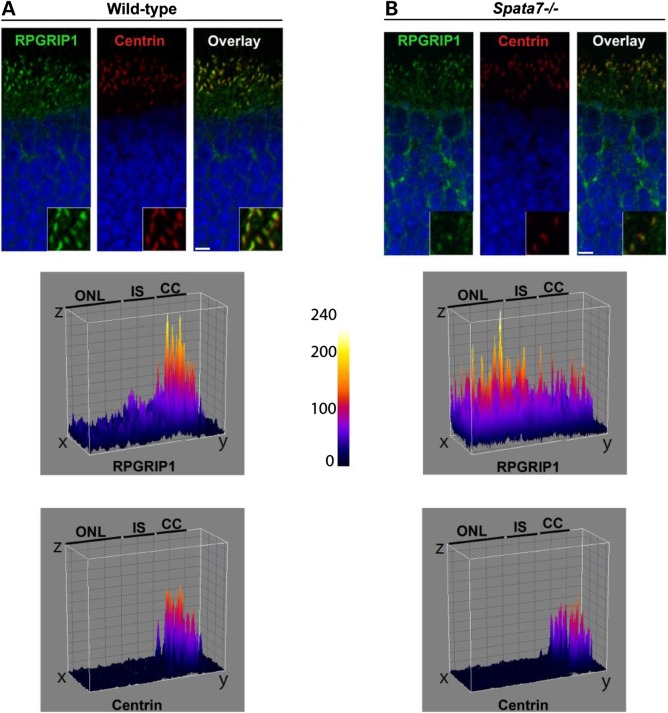
Given the interaction of SPATA7 and RPGRIP1 and photoreceptor cell defects observed in the absence of Spata7 function, the localization of RPGRIP1 in Spata7 mutant photoreceptor cells was examined. Normally, RPGRIP1 localizes to the connecting cilium (CC) of the photoreceptor cell and functions as a protein scaffold that is important for proper assembly and localization of several retinal ciliopathy-associated proteins, including RPGR and NPHP4 (44,45). Strikingly, a substantial reduction of RPGRIP1 protein levels at the CC and its mislocalization to the inner segment are observed in Spata7−/− mutant mice (Fig. 8B). In contrast, the ciliary marker centrin (Fig. 8A and B) as well as the photoreceptor basal body maker RPGRIP1L (data not shown) localize normally, suggesting that the RPGRIP1 mislocalization in the Spata7 mutant retina is specific. Taken together, our results strongly support a model for SPATA7 as a new member of a group of ciliary proteins that are essential for proper localization and/or assembly of CC protein complexes, which are important for proper protein trafficking in photoreceptor cells.
Figure 8.
RPGRIP1 is mislocalized in Spata7 mutant mouse photoreceptor cells. Compared with wild-type (A), partial mislocalization of RPGRIP1 (green) is detected in unfixed Spata7 mutant retina cryosections at P14 by IHC (B). In Spata7 mutants, RPGRIP1 (green) is detected in the ONL and its levels are significantly decreased in the CC while Centrin(red) showed almost no change . The surface plots (middle and bottom panels) quantitatively show the distribution of the RPGRIP1 and Centrin fluorescent signal in the corresponding images above. Scale bar is 10 µm.
Mislocalization of RHO in Spata7 mutant mice
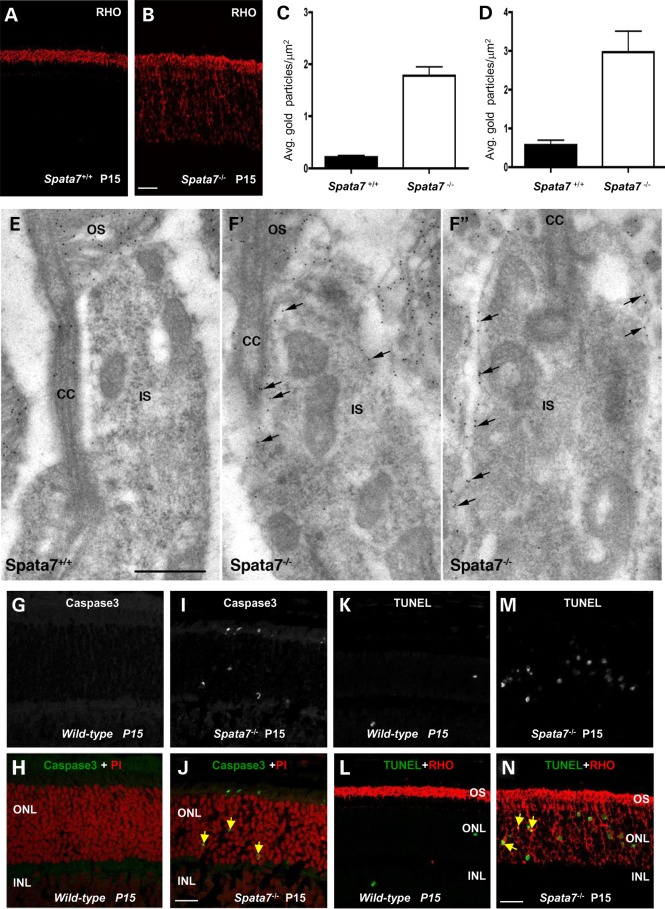
One of the most actively transported proteins between the IS and OS is RHO. Consistent with the model that SPATA7 is involved in protein trafficking, striking mislocalization of RHO in photoreceptor cells is observed in the Spata7−/− mutant retina. RHO is normally detected only in the OS due to active transport of the protein from the IS where it is synthesized (46). In contrast, in Spata7−/− retinas, substantial RHO immunoreactivity is detected in the inner segment (IS) as well as in the ONL at P15 (Fig. 9A and B). To pinpoint the location of mislocalized RHO in photoreceptor cells, immunogold labeling of RHO followed by electron microscopy was performed. Quantification of the RHO immunogold label revealed an 8-fold and 5-fold increase in the plasma membrane (Fig. 9C) and IS (Fig. 9D), respectively, of P11 Spata7−/− mutants compared with P11 wild-type. As shown in Figure 9E, F’ and F’’, mislocalized RHO is detected in the cytoplasm and plasma membrane throughout the IS.
Figure 9.
Rhodopsin is mislocalized in Spata7 mutant mouse photoreceptor cells. Mislocalized Rhodopsin is detected in paraffin-embedded Spata7 mutant retinas at P15 by IHC. Compared with wild-type (A), RHO is detected in the IS and ONL (B), scale bar is 20 µm. (C and D) Quantification of the density of RHO immunogold particles in the inner segment plasma membrane (PM) and IS. Two independent P11 retinas were labeled and gold particles of 20 inner segments from each genotype were counted. Significantly (t-test, P< = 0.0001) more particles are observed in both the PM and IS of Spata7 mutants compared with wild-type at P11. (E and F) Representative electron micrographs of photoreceptors in P11 wild-type and mutant (F’, F’’) retinal sections, immunogold labeled with RHO antibodies. Arrows in F’ and F’’ indicate immunogold labeling of mislocalized RHO. IS, rod inner segment; OS, rod outer segment; CC, connecting cilium. Scale bar = 500 nm (E, F’ and F’’ are the same magnification). (G–N) Several Caspase3 positive cells (green) are observed in P15 frozen sections (yellow arrows, J), and an increased number of TUNEL positive cells (green) are detected in P15 paraffin-embedded sections (N). The yellow arrow indicates a double-labeled nucleus in (N). Scale bar is 20 µm.
To determine if the protein mislocalization phenotype observed is a general phenomenon in the Spata7−/− mutant retina, the subcellular localizations of two other OS proteins, Peripherin(PRPH2) (also known as RDS) and ROM-1, were examined (47,48). Strong immunofluorescence staining for PRPH2 is detected exclusively in the OS in the Spata7−/− retina, indicating that PRPH2 localization is not substantially affected (Supplementary Material, Fig. S7A–G). Similarly, ROM-1 is properly restricted to the OS (Supplementary Material, Fig. S7I–O). It is well known that some of the proteins involved in the phototransduction pathway shuttle between the OS and IS depending on light conditions. One such protein is Transducin. Under normal light conditions, most Transducin is observed in the inner segment of photoreceptor cells while it is translocated to the outer segment in the dark adapted condition (49). As shown in Supplementary Material, Fig. S8, in the wild-type retina, Transducin is detected in the proper subcellular compartments of photoreceptors in light and dark conditions as expected; similarly, in Spata7−/− mutant retinas at P25, most Transducin is located in inner segments in light. In contrast, the vast majority of Transducin is properly translocated to the outer segments in dark adapted conditions, suggesting that Transducin trafficking is not substantially affected by the loss of Spata7 function. The small fraction of mislocalized Transducin observed is likely secondary to the ongoing photoreceptor cell degeneration at P25. Taken together, these results indicate that SPATA7 is required for the proper localization of only a subset of proteins in photoreceptor cells, such as RHO. To further test whether the photoreceptor degeneration observed in Spata7−/− mice is effected by light, retinas from mice raised in constant dark were examined by immunohistochemistry. These results show that photoreceptor degeneration in the Spata7−/− mutant retina is light independent (data not shown).
To assess if mislocalized RHO accumulates in the ER or Golgi, we stained Spata7−/− retinas using Concanavalin A (ConA), which detects mannose residues in N-glycans added to proteins in the ER and Golgi. In Spata7−/− mutant retinas, a substantial increase in ConA staining is observed in the inner segment (IS), coinciding with mislocalized RHO. This suggests that mislocalized RHO has processed through the ER and Golgi apparatus (Supplementary Material, Fig. S9E and G). Consistently, no obvious accumulation of RHO is observed in the ER or Golgi in the immunogold labeling experiment (data not shown), suggesting that mislocalized RHO is likely to be properly processed and folded.
RHO mislocalization precedes programmed cell death in Spata7−/− mutant retinas
RHO mislocalization has been a prevalent phenotype in both syndromic and nonsyndromic retinal degeneration (50,51). Studies of several mouse models, such as Bbs2, Ahi1, Rp1, Rpgrip1 and Cep290, have shown that mislocalization of RHO to the inner segment precedes photoreceptor cell degeneration (52–56). RHO mislocalization is also observed in mutations affecting protein transport components, such as Kinesin-2 and IFTs (46,57–61). It has been proposed that mislocalization of RHO itself can trigger cell death as opposed to being a secondary consequence of photoreceptor cell death (59).
To test if RHO mislocalization in Spata7−/− retinas is a likely cause for photoreceptor degeneration, the time course of RHO mislocalization and cell apoptosis was examined. RHO protein can be first detected around P2 before the initiation of outer segment morphogenesis and is localized to the OS by P7 (Supplementary Material, Fig. S10A, B, E and F). Interestingly, mislocalization of RHO in mutant retina is detected as early as P7 (Supplementary Material, Fig. S10C, D, G and H). Cell apoptosis in both wild-type and Spata7−/− retinas at these same time points was also examined. Previous work has indicated that there are many apoptotic nuclei in the wild-type mouse retina at early stages (P5), and then the number of apoptotic cells decreases markedly by P13 (62). Consistent with this report, using Caspase3 as a marker for cell apoptosis, apoptotic cells are observed in wild-type retinas at P7 and P11, but not at P15 (Fig. 9G and H and Supplementary Material, Fig. S10I, J, M and N). Moreover, most apoptotic cells observed at P7 and P11 are in the INL with few Caspase3 positive cells found in the ONL (Supplementary Material, Fig. S10I, J, M and N). In Spata7−/− retinas, substantially more Caspase3 positive cells are observed at P15 compared with wild-type (Fig. 9I and J). However, at Day P7 and P11, no substantial difference between wild-type and mutant retinas is observed and all apoptotic cells are found in the INL, but not in the ONL (Supplementary Material, Fig. S10I–P). These results indicate that photoreceptor apoptosis occurs later than RHO mislocalization. Similar results are obtained using the TUNEL assay. Specifically, more TUNEL positive cells are observed in Spata7−/− retinas compared with wild-type retinas by P15 but not at earlier time points (Fig. 9K and M).
Complete loss of Rho partially rescues photoreceptor degeneration in Spata7 mutants
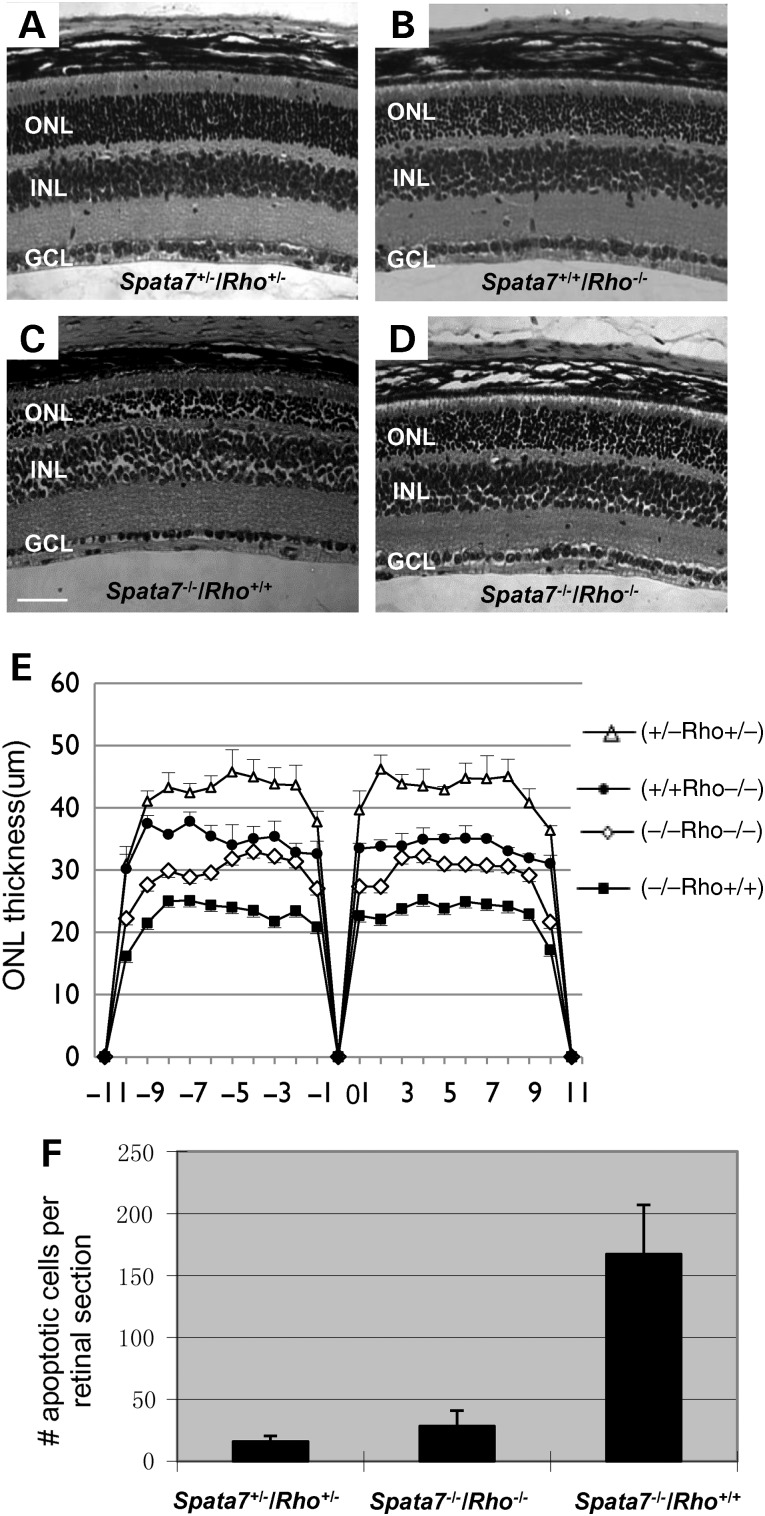
Given that RHO mislocalization precedes photoreceptor cell apoptosis, we hypothesized that mislocalization of RHO is one of the major defects in Spata7−/− retinas that triggers photoreceptor degeneration. If this is the case, we expect that reducing the dose of Rho might partially suppress the photoreceptor cell degeneration phenotype in Spata7−/− mutants. Mice that are homozygous mutant for both Spata7 and Rho were generated and their retinas were examined at 4 weeks of age. Retinas from mice that carry double heterozygous mutations for Spata7 and Rho (Spata7+/−/ Rho+/−) are histologically normal (Fig. 10A, Supplementary Material, Fig. S11A and B). Although Rho is required for the development and maintenance of proper OS structure and survival of photoreceptors, a relatively mild photoreceptor degeneration phenotype is observed in Rho−/− mice at 4 weeks of age (Fig. 10B and E and Supplementary Material, Fig. S11C) (63). In contrast, photoreceptors in Spata7−/− mutant retinas already show severe degeneration by P29, with only 4–5 rows of nuclei in the ONL at this age (Fig. 10C and Supplementary Material, Fig. S11D). Although no obvious rescue is observed by removing one copy of Rho in the Spata7−/− homozygous mutant background, when both copies of Rho are removed, the number of rows in the ONL in Spata7−/−/Rho−/− double mutants is comparable to that of the Rho−/− mutant alone (Fig. 10D and E and Supplementary Material, Fig. S11E). Consistent with this observation, substantially fewer TUNEL positive cells (P = 0.021) are observed in the Spata7−/−/Rho−/− double mutant ONL layer compared with Spata7−/− mutants, indicating rescue of the photoreceptor cell death phenotype by loss of Rho (Fig. 10F). Similarly, ER stress marked by CHOP staining is also suppressed in Spata7−/−/Rho−/− double mutant retinas (Supplementary Material, Fig. S9A–D). Together, these results support a model where mislocalization of RHO in Spata7−/− mutants is one of the major causes of photoreceptor degeneration. Indeed, no obvious increase in ConA staining is observed in the inner segment of photoreceptors in Spata7−/−/Rho−/− double knockout retinas, indicating that there is no substantial accumulation of glycosylated proteins in the inner segment (Supplementary Material, Fig. S9F).
Figure 10.
Loss of both copies of rod Opsin partially rescues the Spata7 mutant phenotype. H&E staining of paraffin embedded P29 retinal sections of Spata7+/−/Rho+/− (A), Spata7+/+/Rho−/− (B), Spata7−/−/Rho+/+ (C) and Spata7−/−/Rho−/− (D) mutant mice. Rescue of the Spata7 mutant phenotype is observed when both copies of Rho are removed as measured by retinal morphometry (E) and the number of apoptotic cells using the TUNEL assay (n = 3, Student's t-test, P = 0.021) (F). Each point represents the mean ± SEM obtained for each group in panel E (n = 3). Mice used in this experiment are littermates. Scale bar: 50 µm.
Discussion
In this study, we have combined both in vitro and in vivo molecular approaches to understand the molecular pathogenesis of LCA3 and juvenile RP, which is caused by mutations in SPATA7. We observe that the SPATA7 protein localizes to the connecting cilium of mouse photoreceptors. The connecting cilium is important in photoreceptor cells since it provides a structure through which proteins are transported from the site of protein biosynthesis (the inner segments) to the phototransductive outer segments and back (64,65). The OS contains large numbers of RHO molecules, efficiently stored in membrane stacks that continuously undergo photo-oxidative damage during the day. Therefore, the OS requires highly active renewal of about 10% each day, which is achieved through shedding at their proximal tips and subsequent phagocytosis by adjacent RPE cells. The same numbers of OS proteins therefore need to be replenished through the CC, thus requiring equally highly active and accurately organized mechanisms of ciliary protein transport. We here show that SPATA7 interacts with RPGRIP1, a protein encoded by another LCA associated gene (LCA6), pointing towards a specific LCA-associated ciliary protein module. Although an extensive effort has been invested in studying RPGRIP1 in the last decade, a mechanistic understanding of its function has remained elusive. RPGRIP1 can act as a protein scaffold that may assemble a CC-localized module of retinal ciliopathy-associated proteins, which was previously shown to also include RPGR, NPHP4, SSCCAG8, IQCB1 and CEP290 (66). SPATA7 predominantly binds to the N-terminal coiled-coil domain of RPGRIP1, potentially allowing a multimeric complex that includes binding of RPGR to the C-terminal RPGR interacting domain (RID) of RPGRIP1, and the binding of NPHP4 to the central C2 domains. Mouse models that lack RPGRIP1 or NPHP4 have suggested that this scaffold plays an important role in protein transport across the CC (45). The role of SPATA7 at the CC was investigated using a mouse model that mimics human juvenile RP disease by knocking out the Spata7 gene. Consistent with the clinical presentation of human patients, Spata7−/− mutant mice display progressive thinning of the photoreceptor cell layer and a dramatic reduction in response to light. Therefore, as a model system, Spata7−/− mutant mice provide us with an ideal opportunity for detailed characterization and analysis of LCA3/ juvenile RP pathogenesis.
Retinal defects in Spata7−/− mutant mice become obvious shortly after birth. Specifically, photoreceptor cell degeneration becomes evident at P15 as the thickness of the ONL begins to decrease. In addition, RHO staining and electron microscopy both show shortened and disorganized outer segments (OS) in photoreceptor cells. Although both rod and cone cells are affected, given that the majority of rod cells die by 6 months of age while 84% of cone cells still remain at this stage, cone cell loss may be secondary to defects in rod photoreceptor cells. Consistent with this idea, a dramatic reduction of the a-wave for scotopic ERGs is observed. In contrast, only a relatively mild reduction is observed in the double-flash cone b-wave, an assay that specifically tests cone cell function. Other than photoreceptor cell defects, all other cell layers in the retina appear normal based on both histology and staining for cell type-specific markers. Therefore, the primary defect in the Spata7 mutant retina appears to be specific to rod photoreceptor cells.
The reduction of photoreceptor cells in the Spata7 mutant retina is likely due to degeneration rather than developmental defects. First, at P0, before layer formation, the number of cells in the retina appears normal. Secondly, differentiation of photoreceptors and other retinal cell types appears to be normal as all layers are formed properly at P7. High magnification TEM shows that a substantial number of photoreceptor cells indeed develop with proper structure, including the inner segment (IS), OS and the connecting cilium (CC). Thirdly, photoreceptor degeneration progresses over time with an initial rapid drop of cell number between 2 and 4 weeks after birth, followed by a lower rate of degeneration between 4 weeks and 6 months. By the age of 6 months, only about 40% of cells in the ONL remain. Therefore, it is likely that Spata7 is not required for early cell differentiation but is required for the proper function and maintenance of photoreceptors.
Mislocalization of RHO in photoreceptor cells is likely to be the main defect that triggers degeneration in the Spata7−/− mutant retina. Importantly, accumulation of RHO within the IS precedes apoptosis. RHO accumulation in the inner segment can be detected as early as P7 while increased apoptosis is first observed at P15. These results raise the idea that the mechanism of photoreceptor cell degeneration in the Spata7−/− mutant retina is likely caused by RHO mislocalization. This hypothesis is further supported by the observation that photoreceptor degeneration in Spata7 mutants can be substantially rescued by loss of Rho function. In particular, Spata7−/−/Rho−/− double mutant retinas show much less cell apoptosis and a reduced rate of photoreceptor degeneration compared with loss of Spata7 function alone.
The mechanism of RHO mislocalization in Spata7−/− mutant mice is likely due to defects in protein trafficking through the connecting cilium mediated by CC complexes, such as the RPGRIP1 complex. First, RHO seems to be properly folded as mislocalized protein appears to be inserted into the plasma membrane. Secondly, the overall structure of photoreceptor cells is intact. Unlike many other known mutations that affect cilium formation in the retina, intact cilia are observed in Spata7−/− mutant photoreceptors. Thirdly, SPATA7 is observed to specifically localize to the connecting cilium of photoreceptor cells, consistent with a model where SPATA7 is involved in protein trafficking. This model is further strengthened by the direct interaction between SPATA7 and RPGRIP1. As SPATA7 was shown to decorate microtubules upon recombinant expression, it may be regarded as a novel microtubule-associated protein (MAP), although a direct interaction with microtubules has not been established. None of the other RPGRIP1 binding partners show microtubule binding capacity; therefore, SPATA7 may be required to firmly fix the scaffold to microtubules of the axoneme of the connecting cilium. Recruitment of full length RPGRIP1 by SPATA7 to colocalize to microtubules is consistent with this model. This could also be required to optimally align and thus structurally organize the scaffold at the CC, disruption of which could have major implications for CC function of all of the proteins connected to the RPGRIP1 scaffold. This is further supported by the partial mislocalization of RPGRIP1 to the IS and ONL of photoreceptors and decreased levels of RPGRIP1 at the CC in Spata7−/− mutant retinas. Therefore, we propose that SPATA7 mediates RHO transport as a member of the critical RPGRIP1-directed protein scaffold at the connecting cilium of photoreceptors. As no direct interaction between SPATA7 and RHO has been detected so far, the involvement of SPATA7 in RHO transportation might be indirect.
In summary, we propose that SPATA7 is a novel member of the retinal ciliopathy-associated RPGRIP1 scaffold at the connecting cilium of photoreceptors. Although not absolutely required, it plays a crucial role in protein trafficking across this cellular compartment. In the absence of SPATA7 in the connecting cilium, protein transport from the inner segment to the outer segment is defective, trigging photoreceptor apoptosis. Considering the high degree of conservation between humans and mice and the similar phenotypes observed in humans and mice carrying SPATA7 mutations, it is likely that a similar mechanism drives disease pathology in human LCA3/ juvenile RP patients.
Materials and Methods
Genotyping of Spata7 mutant mice
To genotype subsequent F2 and F3 generations of Spata7 knockout mice, we used a genomic PCR assay with a pair of primers from the deleted region. These primers are: P1 (5′-GTGGCAAGTACCTTTACTTGGTG-3), P2 (5′-TTCCACACCAGAATCAGTTCTTT-3′), P3 (5′-CTGACTAGGGGAGGAGTAGAAGG-3′) and P4 (5′-CCTCGACTGTGCCTTCTAGTTG-3′).
Immunoelectron microscopic analysis
For immunoelectron microscopy, eyecups were fixed in 0.2% glutaraldehyde, 4% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4. They were then processed for embedment in LR White. Ultrathin sections (70 nm) were etched with saturated sodium periodate for 15 min, blocked with 4% BSA in PBS for 1 h at room temperature and incubated with monoclonal rod Opsin antibody (1D4) in buffer overnight at 4°C. Sections were then washed and incubated with goat anti-mouse IgG conjugated to 12 nm gold (Jackson Lab) and stained with uranyl acetate. Immunolabeling density was determined by counting gold particles over the inner segment plasma membrane (defined as within 30 nm either side of the center of the membrane), and over the area enclosed by this membrane (designated as within the inner segment).
Dilutions and sources of antibodies used for immunohistochemistry
Rabbit anti-SPATA7 polyclonal antibody against the C-terminal 15 amino acids (1:2000, custom antibody made by Bethyl Laboratories), mouse anti γ-tubulin antibody (1:200, Sigma T6557), mouse anti-acetylated α-tubulin antibody (1:200, Sigma), mouse anti-pan Centrin (Millipore, 20H-5, 1:1000), mouse anti-rhodopsin (B6-30N) (1:200, a generous gift from W. Clay Smith), mouse anti-PRPH2 and mouse anti-ROM-1 (1:10, a generous gift from Dr R.S. Molday), mouse anti-PAX6 antibody (1:200, DSHB), goat anti-BRN3B (1:200, Santa Cruz), Sheep-CHX10 (1:200, Exalpha), mouse anti Calbindin (1:2000, Sigma), rabbit anti-Opsin R/G (1:500, Chemicon), Rabbit-Opsin blue (1:500, Chemicon), rabbit polyclonal anti-RPGRIP1 (1:2500, kindly provided by Dr Tiansen Li), Rod Transducin-α anti rabbit polyclonal (Santa-Cruz, 1:100), Fluorescence Concanavalin A (1:2000, Vector Laboratories, Burlingame, CA, USA). Cy3 anti-rabbit (1:500, Jackson Immunochemicals), Biotinylated Peanut Agglutinin (PNA,1:200, Vector Laboratories), Fluorescein Lectin Concanavalin A (1:2000, Vector Laboratories), rabbit polyclonal anti-GADD153 (F-168) (CHOP) (1:10, Santa-Cruz), mouse monoclonal anti-Flag (1:200, F3165, Sigma), Topro3 (dilution 1:2000), secondary antibodies were conjugated with Alexa fluor 488, 568 (1:500, Molecular Probes) and DAPI staining reagent (dilution 1:8000).
DNA constructs
The entry clone encoding full-length human SPATA7 isoform 2 with 568 amino acids [NCBI Reference Sequence: NM_001040428.3 (gene); NP_001035518.1 (protein) was kindly donated by Prof. N. Katsanis (center for Human Disease Modeling, Duke university, Durham, NC, USA)]. For SPATA7 fragment constructs, clones encoding the N-terminal amino acids 2–310 and the C-terminal amino acids 311–568 were generated by PCR using the SPATA7 full-length entry clone as a template. The entry clone encoding RPGRIP1 full length was kindly provided by Prof. Gerd Walz (University of Freiburg, Freiburg, Germany). Different RPGRIP1 fragments were generated by PCR using the RPGRIP1 full-length entry clone as a template. The RPGRIP1 C2-end constructs encoding a fragment spanning from the first C2 domain to the C-terminus of the protein (amino acids 611–1286) were generated as previously described (36), as was the RPGR-expressing construct containing exons 1–19 (33). Expression constructs were created from the entry clones using Gateway cloning procedures (Life Technologies) according to the manufacturer's instructions. Primers are available upon request. The sequence of all entry clones was verified by nucleotide sequencing.
Yeast two-hybrid assays
A GAL4-based yeast two-hybrid system was used for the identification of protein interactors of SPATA7 as previously described (67). Constructs encoding full-length or fragments of SPATA7 fused to a DNA-binding domain (GAL4-BD) were used as baits to screen a human oligo-dT primed retinal cDNA library, containing 4.5 × 106 independent recombinant clones (1.5 × 106 independent translated proteins) to saturation (68). The yeast strain PJ96-4A, which carries the HIS3 (histidine), ADE2 (adenine), MELI (α-galactosidase) and LacZ (β-galactosidase) reporter genes, was used as a host. Interactions were analyzed by assessment of reporter gene activation based on growth on selective media (HIS3 and ADE2 reporter genes), α-galactosidase colorimetric plate assays (MELI reporter gene) and β-galactosidase colorimetric filter lift assays (LacZ reporter gene).
For validation of yeast two hybrid assays, plasmids encoding DNA-binding domain (pBD vector, Gateway cloning, Life Technologies) fused SPATA7 N-terminal fragment was co-transformed with plasmids encoding GAL4-activation domain (pAD vector, Gateway) fused RPGRIP1 clone 49 (identified in the screen with retina library), in the PJ69-4 alpha yeast strain. GAL4-AD expressing plasmid was co-transformed with either GAL4-BD or Lamin C expressing plasmids as positive and negative controls, respectively. Co-transformed yeast was selected on media lacking leucine (-L) and tryptophan (-W) for the presence of both pAD and pBD. Activation of reporter genes was further detected on selection plates lacking histidine (-LWH) and adenine (-LWHA), as well as by α-galactosidase assays and β-galactosidase filter lift assays.
Co-immunoprecipitation
Plasmids expressing 3xHA-SPATA7 was co-transfected with plasmids expressing 3xFlag-RPGRIP1 full length, 3xFlag-RPGRIP (CC), 3xFlag-RPGRIP1 (C2), 3xFlag-RPGRIP1 (RID), 3xFlag-RPGRIP1 (C2-end), 3xFlag-lebercilin and Ntap-IQCB1 (IQ calmodulin-binding motif-containing protein1)in HEK293T cells using Effectene (Qiagen), respectively. As a positive control, the previously described interaction between RPGR and RPGRIP1 was used (33). Plasmid expressing functionally unrelated 3xFlag-p63 (Tumor protein 63) protein was used as a negative control. Forty-eight hours post-transfection, cells were lysed on ice in lysis buffer [50 mm Tris–HCl (pH7.5), 150 mm NaCl, 1% NP-40] supplemented with complete protease inhibitor cocktail (Roche Diagnostics). Lysates were incubated with anti-HA affinity matrix (Roche) for 2–3 h at 4°C. After incubation, beads with bound protein complexes were washed in ice-cold lysis buffer. Subsequently, 4x NuPAGE Sample Buffer was added to the beads and heated for 10 min at 70°C. Beads were precipitated by centrifugation, and supernatant was run on NuPAGE Novex 4–12% Bis–Tris SDS–PAGE gels. The interaction of target proteins was assessed by immunoblotting, followed by staining with either rabbit polyclonal α-Flag or rabbit polyclonal α-HA as primary antibodies and goat anti-rabbit IRDye800 as a secondary antibody. Fluorescence was analyzed on a Li-Cor Odyssey 2.1 infrared Scanner.
GST-pull down assays
Human SPATA7 was cloned into the pDEST15 vector (Gateway cloning system, Life Technologies). To generate GST-fusion proteins, BL21-DE3 bacteria were transformed with pDEST15 constructs. Cells were induced with 0.5 mm IPTG at 30°C for about 3 h until the culture OD600 reached 0.6–0.8 and harvested by centrifugation. Subsequently, cells were lysed in STE buffer [10 mm Tris–HCl (pH8.0), 1 mm EDTA, 150 mm NaCl] supplemented with 10 mg/ml Lysozyme, 0.5% Sarkosyl, 1% Triton X-100 and complete protease inhibitor cocktail (Roche Diagnostics). Lysates were incubated with Glutathione Sepharose 4B beads (Amersham Bioscience). After incubation, beads were washed with STE and TBSTD (TBS with 1% Triton X-100 and 2 mm DTT). The amount of GST-fusion proteins bound to beads was verified on a NuPAGE Novex 4%–12% Bis–Tris SDS–PAGE gel by staining with Simply Blue Safe stain (Life Technologies). Beads with bound GST-fusion proteins were incubated for 2 h at 4°C with lysates of HEK293T cells expressing different fragments of RPGRIP1, N-terminal tagged with 3xFlag epitope. After incubation, beads with bound protein complexes were washed in lysis buffer [50 mm Tris–HCl (pH7.5), 150 mm NaCl, 0.5% Triton X-100, freshly supplemented with complete protease inhibitor tablet (Roche)]. Beads were then taken up in 4X NuPAGE sample buffer and heated for 10 min at 70°C. Beads were precipitated afterward by centrifugation. The supernatants were run on NuPAGE Novex 4%–12% Bis–Tris SDS–PAGE gels, followed by immunoblotting and staining with mouse monoclonal α-Flag as primary antibody (Sigma-Aldrich) and goat anti-mouse IRDye800 as a secondary antibody. Recombinant proteins: 3xFlag-RPGRIP1 FL, 3xFlag-PGRIP1cl.49, 3xFlag-RPGRIP1 CC, 3xFlag-RPGRIP1 C2, 3xFlag-RPGRIP1 RID, 3xFlag-RPGRIP1 C2-end, Ntap-IQCB1, 3xFlag-lebercilin and 3xFlag-p63 are detected as bands of approximately 148, 114, 75, 45, 50, 80, 56, 83 and 75 kDa, respectively, on the immunoblot. Unfused GST is used as negative control. Fluorescence was analyzed on a Li-Cor Odyssey 2.1 infrared Scanner.
GST pull-down and co-immunoprecipitation from retinal extract
For GST pull-down experiments, retinas were dissected from fresh bovine eyes obtained from the local slaughter house or from wild-type mice at 8 months of age. Retinas were homogenated by sonication on ice for two times 30 s in extraction buffer [10 mm HEPES (pH 7.9), 10 mm NaCl, 3 mm MgCl2, freshly added 1 mm DTT, 1 mm Na3VO4], supplemented with complete protease inhibitor cocktail (Roche Diagnostic). Retinal extracts were incubated overnight at 4°C with the GST fusion proteins immobilized on glutathione sepharose 4B beads (previously described). Samples were washed three times with extraction buffer after overnight incubation. The beads with associated protein complexes were processed as described earlier. Immunoblotting with α-RPGRIP1 antibody (at 66 ng/ml, a gift from Paulo Ferrera; panel 3, 1:5000, a gift from Tiansen Li) was performed by standard western blotting procedures.
For co-immunoprecipitation experiments, retinas from adult Spata7-Flag transgenic and wild-type mice were lysed in the immunoprecipitation buffer [20 mm Tris, pH 7.5, 1 mm EDTA, 150 mm NaCl, 0.5% NP-40, protease inhibitor cocktail tablets (Roche, #11836170007)], pre-cleared with the Protein A/G-Sepharose (GE Healthcare, #17-5280-01,17-0618-01) and immunoprecipitated with anti-Flag M2 affinity gel (Sigma Aldrich, #A2220). After washes, the IP samples were eluted in Laemmli sample buffer and subjected to SDS–PAGE. Anti-SPATA7 antibodies (Proteintech, #12020-1-AP) and anti-RPGRIP1 antibodies (a gift from Tiansen Li) were used for western blotting.
Bimolecular fluorescence complementation assay
The BiFC assay was performed as previously described (69). Briefly, constructs expressing SPATA7-CVN (the N-terminus of Venus YFP) and RPGRIP1-CC (the C-terminus of Venus YFP) were transfected into 293T cells. After 48 h, fluorescence signals generated by the complementation between N- and C-terminal YFP as a result of SPATA7 and RPGRIP1 interaction was visualized using fluorescence microscopy and quantified by Fluorescence Activated Cell Sorting (FACS). Cells expressing only SPATA7-CVN, RPGRIP1-CC, or empty vectors were used as controls.
Additional details are provided in Supplementary material, Materials and Methods.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (EY004446, EY019908, EY02520), Retina Research Foundation & Research to Prevent Blindness to S.M.W., by a grant from the National Institutes of Health to D.S.W. (R01EY13408, D.S.W. is an RPB Jules and Doris Stein Professor), by the European Community's Seventh Framework Programmes FP7/2009 under grant agreement no: 241955 (SYSCILIA) to R.R., by the Netherlands Organization for Scientific Research (NWO Vici-865.12.005) to R.R., by the Foundation Fighting Blindness (C-CMM-0811-0546-RAD02) to R.R. and by grants from the Retina Research Foundation and the National Eye Institute (R01EY020540) to G.M. and R.C. Spata7 knockout mice were generated by the Mouse ES Cell Core at BCM that is partially supported by the BCM IDDRC (Grant Number 5P30HD024064-23) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. Confocal imaging was conducted on the microscope supported by grant from National Eye Institute (5P30EY002520) to S.M.W.
Supplementary Material
Acknowledgements
We thank Hanfang Tuan for critical reading and editing of the manuscript, Nichols Ralph for technical support and Dorus Mans for helpful discussions. We thank Dr W. Clay Smith for sharing mouse anti-RHO (B6-30N) antibody, Dr R.S. Molday for mouse anti-RDS and anti-ROM1 antibodies, Dr Paulo Ferreira for anti-RPGRIP1 antibodies and Dr Tiansen Li for anti-RPGRIP1 and anti-RPGR antibodies. We sincerely acknowledge support from Dr David Nelson for the mouse dark adaptation experiment.
Conflict of Interest statement. None declared.
References
- 1.Aldahmesh M.A., Al-Owain M., Alqahtani F., Hazzaa S., Alkuraya F.S. A null mutation in CABP4 causes Leber's congenital amaurosis-like phenotype. Mol. Vis. 2010;16:207–212. [PMC free article] [PubMed] [Google Scholar]
- 2.Bowne S.J., Sullivan L.S., Mortimer S.E., Hedstrom L., Zhu J., Spellicy C.J., Gire A.I., Hughbanks-Wheaton D., Birch D.G., Lewis R.A., et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2006;47:34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cremers F.P., van den Hurk J.A., den Hollander A.I. Molecular genetics of Leber congenital amaurosis. Hum. Mol .Genet. 2002;11:1169–1176. doi: 10.1093/hmg/11.10.1169. [DOI] [PubMed] [Google Scholar]
- 4.den Hollander A.I., Heckenlively J.R., van den Born L.I., de Kok Y.J., van der Velde-Visser S.D., Kellner U., Jurklies B., van Schooneveld M.J., Blankenagel A., Rohrschneider K., et al. Leber congenital amaurosis and retinitis pigmentosa with Coats-like exudative vasculopathy are associated with mutations in the crumbs homologue 1 (CRB1) gene. Am. J. Hum. Genet. 2001;69:198–203. doi: 10.1086/321263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Hollander A.I., Koenekoop R.K., Mohamed M.D., Arts H.H., Boldt K., Towns K.V., Sedmak T., Beer M., Nagel-Wolfrum K., McKibbin M., et al. Mutations in LCA5, encoding the ciliary protein lebercilin, cause Leber congenital amaurosis. Nat. Genet. 2007;39:889–895. doi: 10.1038/ng2066. [DOI] [PubMed] [Google Scholar]
- 6.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dryja T.P., Adams S.M., Grimsby J.L., McGee T.L., Hong D.H., Li T., Andreasson S., Berson E.L. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am. J. Hum. Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrada-Cuzcano A., Koenekoop R.K., Coppieters F., Kohl S., Lopez I., Collin R.W., De Baere E.B., Roeleveld D., Marek J., Bernd A., et al. IQCB1 mutations in patients with leber congenital amaurosis. Invest. Ophthalmol. Vis .Sci. 2010;52:834–839. doi: 10.1167/iovs.10-5221. [DOI] [PubMed] [Google Scholar]
- 9.Friedman J.S., Chang B., Kannabiran C., Chakarova C., Singh H.P., Jalali S., Hawes N.L., Branham K., Othman M., Filippova E., et al. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am. J. Hum. Genet. 2006;79:1059–1070. doi: 10.1086/510021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gal A., Li Y., Thompson D.A., Weir J., Orth U., Jacobson S.G., Apfelstedt-Sylla E., Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat. Genet. 2000;26:270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- 11.Hanein S., Perrault I., Gerber S., Tanguy G., Barbet F., Ducroq D., Calvas P., Dollfus H., Hamel C., Lopponen T., et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat. 2004;23:306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 12.Henderson R.H., Williamson K.A., Kennedy J.S., Webster A.R., Holder G.E., Robson A.G., FitzPatrick D.R., van Heyningen V., Moore A.T. A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Mol. Vis. 2009;15:2442–2447. [PMC free article] [PubMed] [Google Scholar]
- 13.Koenekoop R.K., Wang H., Majewski J., Wang X., Lopez I., Ren H., Chen Y., Li Y., Fishman G.A., Genead M., et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat. Genet. 2012;44:1035–1039. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlhens F., Bareil C., Griffoin J.M., Zrenner E., Amalric P., Eliaou C., Liu S.Y., Harris E., Redmond T.M., Arnaud B., et al. Mutations in RPE65 cause Leber's congenital amaurosis. Nat. Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 15.Perrault I., Hanein S., Gerber S., Barbet F., Ducroq D., Dollfus H., Hamel C., Dufier J.L., Munnich A., Kaplan J., et al. Retinal dehydrogenase 12 (RDH12) mutations in leber congenital amaurosis. Am. J. Hum. Genet. 2004;75:639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrault I., Rozet J.M., Calvas P., Gerber S., Camuzat A., Dollfus H., Chatelin S., Souied E., Ghazi I., Leowski C., et al. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nat. Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 17.Sohocki M.M., Bowne S.J., Sullivan L.S., Blackshaw S., Cepko C.L., Payne A.M., Bhattacharya S.S., Khaliq S., Qasim Mehdi S., Birch D.G., et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat. Genet. 2000;24:79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaroop A., Wang Q.L., Wu W., Cook J., Coats C., Xu S., Chen S., Zack D.J., Sieving P.A. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum. Mol. Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- 19.Thompson D.A., Li Y., McHenry C.L., Carlson T.J., Ding X., Sieving P.A., Apfelstedt-Sylla E., Gal A. Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nat .Genet. 2001;28:123–124. doi: 10.1038/88828. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., den Hollander A.I., Moayedi Y., Abulimiti A., Li Y., Collin R.W., Hoyng C.B., Lopez I., Abboud E.B., Al-Rajhi A.A., et al. Mutations in SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am. J. Hu . Genet. 2009;84:380–387. doi: 10.1016/j.ajhg.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang P.W., Wang J., Chen Y., Fu Q., Zhong J., Yi X., Wu R., Gan H., Shi Y., Barnett C., et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat.Genet. 2012;44:972–974. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 22.Falk M.J., Zhang Q., Nakamaru-Ogiso E., Kannabiran C., Fonseca-Kelly Z., Chakarova C., Audo I., Mackay D.S., Zeitz C., Borman A.D., et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat .Genet. 2012;44:1040–1045. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrault I., Hanein S., Zanlonghi X., Serre V., Nicouleau M., Defoort-Delhemmes S., Delphin N., Fares-Taie L., Gerber S., Xerri O., et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat. Genet. 2012;44:975–977. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 24.Estrada-Cuzcano A., Roepman R., Cremers F.P., den Hollander A.I., Mans D.A. Non-syndromic retinal ciliopathies: translating gene discovery into therapy. Hum. Mol. Genet. 2012;21:R111–R124. doi: 10.1093/hmg/dds298. [DOI] [PubMed] [Google Scholar]
- 25.Li L., Xiao X., Li S., Jia X., Wang P., Guo X., Jiao X., Zhang Q., Hejtmancik J.F. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PLoS One. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackay D.S., Ocaka L.A., Borman A.D., Sergouniotis P.I., Henderson R.H., Moradi P., Robson A.G., Thompson D.A., Webster A.R., Moore A.T. Screening of SPATA7 in patients with Leber congenital amaurosis and severe childhood-onset retinal dystrophy reveals disease-causing mutations. Invest. Ophthalmol.. Vis Sci. 2011;52:3032–3038. doi: 10.1167/iovs.10-7025. [DOI] [PubMed] [Google Scholar]
- 27.Perrault I., Hanein S., Gerard X., Delphin N., Fares-Taie L., Gerber S., Pelletier V., Merce E., Dollfus H., Puech B., et al. Spectrum of SPATA7 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum. Mutat. 2010;31:E1241–E1250. doi: 10.1002/humu.21203. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Liu H., Zhang Y., Qiao Y., Miao S., Wang L., Zhang J., Zong S., Koide S.S. A novel gene, RSD-3/HSD-3.1, encodes a meiotic-related protein expressed in rat and human testis. J. Mol. Med. (Berl.) 2003;81:380–387. doi: 10.1007/s00109-003-0434-y. [DOI] [PubMed] [Google Scholar]
- 29.Muresan V., Joshi H.C., Besharse J.C. Gamma-tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J. Cell. Sci. 1993;104(Pt 4):1229–1237. doi: 10.1242/jcs.104.4.1229. [DOI] [PubMed] [Google Scholar]
- 30.Arikawa K., Williams D.S. Acetylated alpha-tubulin in the connecting cilium of developing rat photoreceptors. Invest. Ophthalmol. Vis. Sci. 1993;34:2145–2149. [PubMed] [Google Scholar]
- 31.Giessl A., Pulvermuller A., Trojan P., Park J.H., Choe H.W., Ernst O.P., Hofmann K.P., Wolfrum U. Differential expression and interaction with the visual G-protein transducin of centrin isoforms in mammalian photoreceptor cells. J. Biol .Chem. 2004;279:51472–51481. doi: 10.1074/jbc.M406770200. [DOI] [PubMed] [Google Scholar]
- 32.Wolfrum U. Centrin in the photoreceptor cells of mammalian retinae. Cell. Motil. Cytoskeleton. 1995;32:55–64. doi: 10.1002/cm.970320107. [DOI] [PubMed] [Google Scholar]
- 33.Roepman R., Bernoud-Hubac N., Schick D.E., Maugeri A., Berger W., Ropers H.H., Cremers F.P., Ferreira P.A. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 34.Castagnet P., Mavlyutov T., Cai Y., Zhong F., Ferreira P. RPGRIP1s with distinct neuronal localization and biochemical properties associate selectively with RanBP2 in amacrine neurons. Hum. Mol. Genet. 2003;12:1847–1863. doi: 10.1093/hmg/ddg202. [DOI] [PubMed] [Google Scholar]
- 35.Hong D.H., Yue G., Adamian M., Li T. Retinitis pigmentosa GTPase regulator (RPGRr)- interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J. Biol. Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 36.Roepman R., Letteboer S.J., Arts H.H., van Beersum S.E., Lu X., Krieger E., Ferreira P.A., Cremers F.P. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc. Natl Acad. Sci .USA. 2005;102:18520–18525. doi: 10.1073/pnas.0505774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haverkamp S., Wassle H. Immunocytochemical analysis of the mouse retina. J. Comp. Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- 38.Liu I.S., Chen J.D., Ploder L., Vidgen D., van der Kooy D., Kalnins V.I., McInnes R.R. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 39.Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 40.Xiang M., Zhou L., Macke J.P., Yoshioka T., Hendry S.H., Eddy R.L., Shows T.B., Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanks J.C., Johnson L.V. Selective lectin binding of the developing mouse retina. J. Comp. Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- 42.Jeon C.J., Strettoi E., Masland R.H. The major cell populations of the mouse retina. J. Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dharmaraj S.R., Silva E.R., Pina A.L., Li Y.Y., Yang J.M., Carter C.R., Loyer M.K., El-Hilali H.K., Traboulsi E.K., Sundin O.K., et al. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic. Genet. 2000;21:135–150. [PubMed] [Google Scholar]
- 44.Arts H.H., Doherty D., van Beersum S.E., Parisi M.A., Letteboer S.J., Gorden N.T., Peters T.A., Marker T., Voesenek K., Kartono A., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 45.Patil H., Tserentsoodol N., Saha A., Hao Y., Webb M., Ferreira P.A. Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell. Death. Dis. 2012;3:e355. doi: 10.1038/cddis.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marszalek J.R., Liu X., Roberts E.A., Chui D., Marth J.D., Williams D.S., Goldstein L.S. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 47.Bascom R.A., Manara S., Collins L., Molday R.S., Kalnins V.I., McInnes R.R. Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron. 1992;8:1171–1184. doi: 10.1016/0896-6273(92)90137-3. [DOI] [PubMed] [Google Scholar]
- 48.Connell G., Bascom R., Molday L., Reid D., McInnes R.R., Molday R.S. Photoreceptor peripherin is the normal product of the gene responsible for retinal degeneration in the rds mouse. Proc. Natl Acad. Sci. USA. 1991;88:723–726. doi: 10.1073/pnas.88.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artemyev N.O. Light-dependent compartmentalization of transducin in rod photoreceptors. Mol. Neurobiol. 2008;37:44–51. doi: 10.1007/s12035-008-8015-2. [DOI] [PubMed] [Google Scholar]
- 50.Adamian M., Pawlyk B.S., Hong D.H., Berson E.L. Rod and cone opsin mislocalization in an autopsy eye from a carrier of X-linked retinitis pigmentosa with a Gly436Asp mutation in the RPGR gene. Am. J. Ophthalmol. 2006;142:515–518. doi: 10.1016/j.ajo.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 51.Concepcion F., Chen J. Q344ter mutation causes mislocalization of rhodopsin molecules that are catalytically active: a mouse model of Q344ter-induced retinal degeneration. PLoS One. 2010;5:e10904. doi: 10.1371/journal.pone.0010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol .Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J., Cheon K., Nusinowitz S., Liu Q., Bei D., Atkins K., Azimi A., Daiger S.P., Farber D.B., Heckenlively J.R., et al. Progressive photoreceptor degeneration, outer segment dysplasia, and rhodopsin mislocalization in mice with targeted disruption of the retinitis pigmentosa-1 (Rp1) gene. Proc. Natl Acad. Sci. USA. 2002;99:5698–5703. doi: 10.1073/pnas.042122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Louie C.M., Caridi G., Lopes V.S., Brancati F., Kispert A., Lancaster M.A., Schlossman A.M., Otto E.A., Leitges M., Grone H.J., et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet. 2010;42:175–180. doi: 10.1038/ng.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B., et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl Acad. Sci.USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y., Hong D.H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl Acad. Sci. USA. 2003;100:3965–3970. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jimeno D., Feiner L., Lillo C., Teofilo K., Goldstein L.S., Pierce E.A., Williams D.S. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest. Ophthalmol .Vis. Sci. 2006;47:5039–5046. doi: 10.1167/iovs.06-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krock B.L., Perkins B.D. The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT-particle-kinesin-II dissociation in vertebrate photoreceptors. J. Cell. Sci. 2008;121:1907–1915. doi: 10.1242/jcs.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopes V.S., Jimeno D., Khanobdee K., Song X., Chen B., Nusinowitz S., Williams D.S. Dysfunction of heterotrimeric kinesin-2 in rod photoreceptor cells and the role of opsin mislocalization in rapid cell death. Mol. Biol. Cell. 2010;21:4076–4088. doi: 10.1091/mbc.E10-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pazour G.J., Baker S.A., Deane J.A., Cole D.G., Dickert B.L., Rosenbaum J.L., Witman G.B., Besharse J.C. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell. Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sukumaran S., Perkins B.D. Early defects in photoreceptor outer segment morphogenesis in zebrafish ift57, ift88 and ift172 Intraflagellar Transport mutants. Vision. Res. 2009;49:479–489. doi: 10.1016/j.visres.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeiss C.J., Neal J., Johnson E.A. Caspase-3 in postnatal retinal development and degeneration. Invest. Ophthalmol. Vis. Sci. 2004;45:964–970. doi: 10.1167/iovs.03-0439. [DOI] [PubMed] [Google Scholar]
- 63.Lem J., Krasnoperova N.V., Calvert P.D., Kosaras B., Cameron D.A., Nicolo M., Makino C.L., Sidman R.L. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl Acad .Sci. USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams N.A., Awadein A., Toma H.S. The retinal ciliopathies. Ophthalmic. Genet. 2007;28:113–125. doi: 10.1080/13816810701537424. [DOI] [PubMed] [Google Scholar]
- 65.Liu Q., Tan G., Levenkova N., Li T., Pugh E.N., Jr, Rux J.J., Speicher D.W., Pierce E.A. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rachel R.A., Li T., Swaroop A. Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia. 2012;1:2046–2530. doi: 10.1186/2046-2530-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letteboer S.J., Roepman R. Versatile screening for binary protein-protein interactions by yeast two-hybrid mating. Methods Mol. Biol. 2008;484:145–159. doi: 10.1007/978-1-59745-398-1_10. [DOI] [PubMed] [Google Scholar]
- 68.Roepman R., Schick D., Ferreira P.A. Isolation of retinal proteins that interact with retinitis pigmentosa GTPase regulator by interaction trap screen in yeast. Methods Enzymol. 2000;316:688–704. doi: 10.1016/s0076-6879(00)16757-6. [DOI] [PubMed] [Google Scholar]
- 69.Chen L.Y., Liu D., Songyang Z. Telomere maintenance through spatial control of telomeric proteins. Mol. Cell. Biol. 2007;27:5898–5909. doi: 10.1128/MCB.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.