Abstract
Fever predicts clinical outcomes in sepsis, trauma and during cardiovascular stress, yet the genetic determinants are poorly understood. We used an integrative genomics approach to identify novel genomic determinants of the febrile response to experimental endotoxemia. We highlight multiple integrated lines of evidence establishing the clinical relevance of this novel fever locus. Through genome-wide association study (GWAS) of evoked endotoxemia (lipopolysaccharide (LPS) 1 ng/kg IV) in healthy subjects of European ancestry we discovered a locus on chr7p11.2 significantly associated with the peak febrile response to LPS (top single nucleotide polymorphism (SNP) rs7805622, P = 2.4 × 10−12), as well as with temperature fluctuation over time. We replicated this association in a smaller independent LPS study (rs7805622, P = 0.03). In clinical translation, this locus was also associated with temperature and mortality in critically ill patients with trauma or severe sepsis. The top GWAS SNPs are not located within protein-coding genes, but have significant cis-expression quantitative trait loci (eQTL) associations with expression of a cluster of genes ∼400 kb upstream, several of which (SUMF2, CCT6A, GBAS) are regulated by LPS in vivo in blood cells. LPS- and cold-treatment of adipose stromal cells in vitro suggest genotype-specific modulation of eQTL candidate genes (PSPH). Several eQTL genes were up-regulated in brown and white adipose following cold exposure in mice, highlighting a potential role in thermogenesis. Thus, through genomic interrogation of experimental endotoxemia, we identified and replicated a novel fever locus on chr7p11.2 that modulates clinical responses in trauma and sepsis, and highlight integrated in vivo and in vitro evidence for possible novel cis candidate genes conserved across human and mouse.
Introduction
Fever is a critical component of inflammation and host defense, and temperature homeostasis is tightly regulated in humans. Both hypo- and hyperthermia have been associated with adverse outcomes in acute inflammatory events including trauma, sepsis (1,2) and cardiovascular disease (3,4). Increased body temperature may slow the growth and reproduction of bacteria and viruses, and activate the heat-shock response, protecting against pathogenic infections (5), but an uncontrolled fever may itself be pathogenic. Mutations involved in rare familial fever disorders have been identified (6), however, common genetic variation underlying fever in the general population has not been described. Novel genetic determinants of fever may reveal biological processes of clinical importance particularly in the context of sepsis, trauma or myocardial infarction (MI).
The bacterial-derived endotoxin lipopolysaccharide (LPS) is a powerful pyrogen, and triggers fever through toll-like receptor 4 (TLR4) signaling, and downstream peripheral activation of inflammatory cytokines, and cyclooxygenase 2 (COX-2) mediated production of prostaglandin E2 (PGE2) (7,8). Multiple tissues and systems maintain temperature homeostasis through peripheral cues affecting the brain temperature set-point, and signaling through the sympathetic nervous system to effector tissues (muscle, brown adipose, vasculature and skin) to alter thermogenesis and heat loss (9). Thus, genetic modulation of fever-related signaling in several cell-types and tissues may affect the ability to mount an effective febrile response.
In the Genetics of Evoked Responses to Niacin and Endotoxemia (GENE) study, we applied experimental endotoxemia (LPS) in 294 healthy individuals of European ancestry (EA) and African ancestry (AA) as a controlled model of activation of innate immunity and evoked inflammation and fever (10,11). The response to LPS has been well described and characterized as a febrile illness (10,12). We examined the genomic determinants of the temperature response to LPS through genome-wide association study (GWAS) and identified a fever-associated intergenic region on chromosome 7p11.2 in EA individuals. We replicated this GWAS association in an independent human endotoxemia experiment (Fenofibrate and Omega-3 Fatty Acid Modulation of Endotoxemia, FFAME study). In clinical translation, we established preliminary clinical relevance of this ‘febrile-response’ region and mortality in established cohort studies of trauma and sepsis. Through bioinformatics as well as cell and rodent studies, we identified several candidate genes that may mediate the effect of this thermoregulatory locus. These candidate genes are LPS- and cold-modulated in vivo and in vitro, with evidence of genotype-specific expression. Thus, we establish the 7p11.2 genomic region as a novel fever locus of clinical relevance and uncover a cluster of potential candidate genes for thermoregulation in humans.
Results
GWAS of temperature response to endotoxin
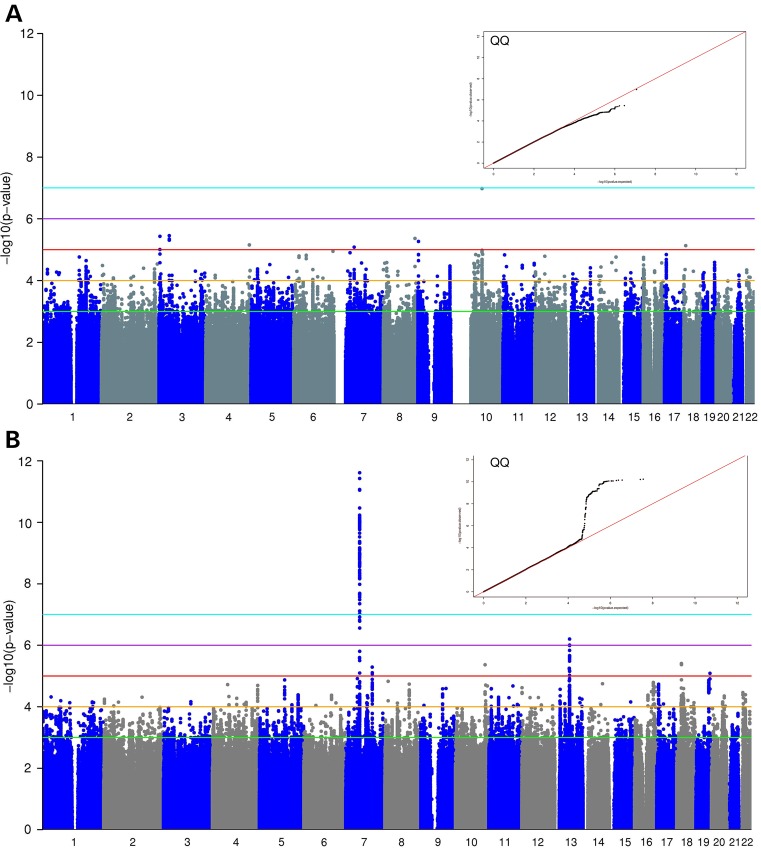
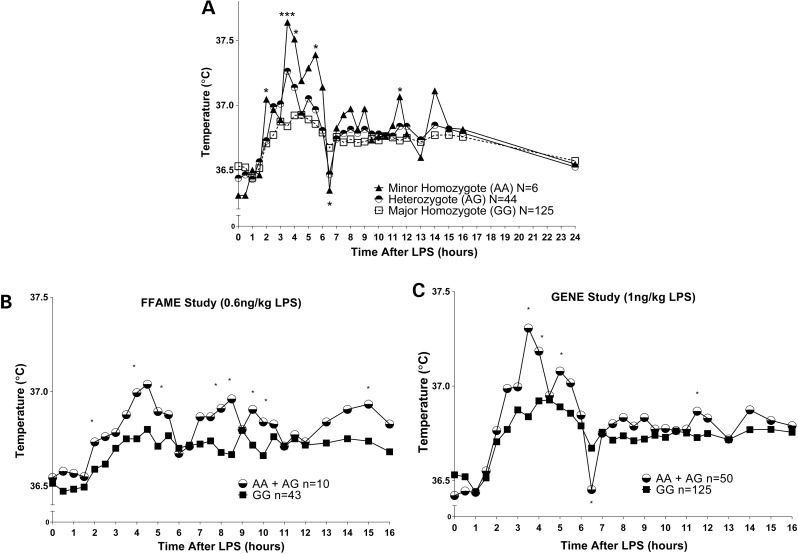
We performed GWAS for body temperature response following evoked endotoxemia in EA GENE study participants (n = 175, Table 1). We analyzed 574 042 genotyped single nucleotide polymorphisms (SNPs), and 7 043 028 imputed SNPs after quality control (QC) filtering (Affy 6.0 with 1000G-imputation, see Supplementary Material, Methods). While at baseline (pre-LPS) no SNPs were significantly associated with temperature (Fig. 1A), we detected a genome-wide significant peak on chromosome 7p11.2 for the change from baseline to peak temperature (3.5 h post-LPS) in response to evoked endotoxemia (the top SNP rs7805622 was directly genotyped, P = 2.4 × 10−12) (Fig. 1B). Minor allele carriers of rs7805622 had significantly higher peak temperature (by ∼0.5°C) than major allele homozygotes and also had greater temperature fluctuation over time (Fig. 2A). In post-hoc analysis, there were significant differences in temperature by rs7805622 genotypes at a number of times including a lower temperature for minor allele carriers at the nadir, 6.5 h post-LPS, (P = 0.008). Overall, 13 genotyped SNPs in the 7p11.2 region reached genome-wide significance (P < 5 × 10−8) for the change in temperature (Supplementary Material, Table S1). No imputed SNP reached a higher level of significance than the top genotyped SNP (best imputed SNP rs2329502 P = 5.85 × 10−11; 88 imputed SNPs reached genome-wide significance). There was no association between any SNPs and either baseline or change in temperature in AA participants; however, consistent with HapMap and 1000Genomes data, the minor allele frequency of rs7805622 in AA was lower (3% in AA versus 16% in EA). Coupled to smaller AA sample size (n = 96), which meant we were somewhat underpowered to detect an association (∼75% power at 5% significance level to detect a similar effect as that observed in EA individuals; using Genetic Power Calculator http://pngu.mgh.harvard.edu/~purcell/gpc/qtlassoc.html, last accessed on October 2014 (13,14)), and differences in linkage disequilibrium (LD) structure between EA and AA in this region (Supplementary Material, Figs S1 and S2), our ability to interrogate this temperature locus in AA was limited. No SNPs in any known familial fever genes (6) reached significance with baseline or change in temperature.
Table 1.
Characteristics of GENE study EA participants by rs7805622 genotypes at the chromosome 7p11.2 locus
| rs785622 | AA, N = 6, mean (SD) | AG, N = 44, Mean (SD) | GG, N = 125, Mean (SD) |
|---|---|---|---|
| Sex (male/female) | 5/1 | 28/16 | 63/60 |
| Age (years) | 24.7 (2.9) | 25.3 (7.1) | 24.5 (5.2) |
| BMI (kg/m2) | 23.9 (1.7) | 23.7 (2.5) | 23.5 (2.8) |
| Total cholesterol (mg/dl) | 173 (59) | 156 (36) | 147 (25) |
| Blood pressure (systolic/diastolic) (mmHg) | 110/63 (5/7) | 108/62 (11/9) | 112/64 (10/8) |
| Baseline temperature (°C) | 36.3 (0.2) | 36.4 (0.3) | 36.5 (0.3) |
| Peak temperature (˚C) | 37.6 (0.3) | 37.3 (1.1) | 36.8 (0.4) |
| C-reactive protein (mg/L) | 0.5 (0.3) | 1.7 (5) | 1.2 (2.5) |
| TNFα (pg/ml) | 1.2 (0.5) | 1.4 (0.8) | 1.3 (0.9) |
| IL-6 (pg/ml) | 1.4 (0.3) | 2.3 (1.6) | 3.0 (1.9) |
| IL-1RA (pg/ml) | 133 (52) | 136 (73) | 130 (77) |
BMI, body mass index; TNFα, tumor necrosis factor alpha; IL-6, interleukin 6; IL-1RA, interleukin 1 receptor agonist.
Figure 1.
Genome-wide association analysis of baseline temperature and change from baseline to peak in EA GENE study participants. As shown in the Manhattan and QQ plots, there were no genome-wide significant signals at baseline (A) but there was a highly significant signal on chromosome 7 for the peak-change in temperature following LPS (B).
Figure 2.
(A) Temperature responses in GENE study participants by rs7805622 genotypes at the chr 7p11.2 locus. (B) Temperature response to LPS by genotype at rs7805622 in FFAME replication sample, compared with equivalent presentation in original GENE discovery sample for comparison (C). Minor allele carriers (triangle and circle) had a higher peak temperature response than major allele homozygotes (square) with more dynamic fluctuations in temperature over time following endotoxin administration. (A) ***Genome-wide significance; *Significance (P < 0.01). (B and C) *Significance (one-sided P < 0.05).
There were modest association between 7p11.2 SNPs and plasma tumor necrosis factor alpha (TNFα), interleukin-6 (IL-6) and interleukin-1 receptor agonist (IL-1RA) responses to LPS in GENE study participants (strongest P = 0.001 for IL-6), although none of the SNP-associations reached genome-wide significance. While not excluding a role in local, cell-specific modulation of cytokine signaling, these data suggest that circulating cytokines are unlikely to mediate the stronger observed influence of this locus on fever.
Replication of association in an independent sample
We genotyped our top candidate SNP in an independent LPS study (FFAME, N = 80). Only one subject was homozygous for the minor allele, so this subject was grouped with the minor allele heterozygotes for analysis. The dose of LPS given in FFAME was slightly lower than that given in GENE (0.6 versus 1 ng/kg) and thus the peak fever response in FFAME was somewhat lower and delayed relative to GENE. In EA participants (n = 53), we observed significantly higher temperature 4 h post-LPS (one-sided P = 0.03) as well as at several other time-points post-LPS (e.g. 8.5 h post-LPS, one-sided P = 0.002) in minor allele carriers of rs7805622 (n = 10) in a pattern that was highly consistent with that observed in GENE participants (Fig. 2B and C). As this trial involved therapeutic interventions, we controlled for the treatment group in the analysis.
Clinical relevance of the 7p11.2 temperature locus in trauma and sepsis patients
Temperature regulation is an important homeostatic component of the host response in numerous pathologies, including trauma and sepsis, where extremes of hypothermia and fever influence outcomes (1). Hypothesizing that the 7p11.2 temperature locus identified experimentally in the GENE study would impact clinical responses during trauma and sepsis, we examined the influence of this locus on temperature responses and death in EA individuals enrolled in both the University of Pennsylvania (UPenn) Trauma cohort (n = 210) and the UPenn Molecular Epidemiology of Severe Sepsis in the ICU (MESSI) sepsis cohort (n = 151). Our focus was on EA patients because our discovery in GENE was observed only in EA individuals. The top genotyped SNP at the locus, rs7805622, was associated with differences in the peak temperature following trauma. Although the directionality was in the opposite direction to the peak temperature following LPS in the GENE study, this finding supports the pattern of dynamic bidirectional association between this allele and temperature observed in both the GENE and FFAME discovery and replication samples (Supplementary Material, Results). In EA participants of both the UPenn trauma study and the sepsis study, there were trends toward a higher rate of death with the rs7805622 variant. In both trauma and sepsis, the overall rate of death in rs7805622AA individuals was approximately double that of major allele carriers (43 versus 12% in trauma, and 38 versus 23% in sepsis) (Table 2). Although the number of deaths was small and the association only reached statistical significance in the trauma study (P = 0.047), the trend was consistent across both UPenn studies, and was significant also in meta-analysis of both studies (P = 0.046; using Fisher's combined probability trend test). These data provide supportive evidence for the clinical significance of the 7p11.2 temperature locus.
Table 2.
Characteristics of the UPenn Major Trauma Cohort and MESSI study EA subjects
| UPenn Major Trauma Cohort study, N =210, mean (SD) | MESSI study, N =151, mean (SD) | |
|---|---|---|
| Age (years) | 46 (20) | 59 (15) |
| Sex (M/F) | 138/72 | 94/57 |
| High temperature (°C) | 38.2 (0.7) | 38.0 (1.1) |
| Low temperature (°C) | 36.0 (0.9) | 36.5 (0.7) |
| Apache III | 39.1 (16) | 66.7 (25) |
| Mortality by genotype, % AA/AG/GG | 43/13/11% | 38/19/26% |
| *P value mortality by genotype | 0.047 | 0.3 |
High and low temperatures recorded during the first 24 h after admission to ICU. APACHE = Acute Physiology and Chronic Health Evaluation Score III. Relative to carriers of major allele (G) at rs7805622, minor allele homozygotes (AA) tended to have higher rates of death following trauma or sepsis. This effect only reached statistical significance in trauma, but was consistent across both studies, and significant in meta-analysis on trauma and sepsis (P = 0.046).
*P value from Fisher's Exact test.
Expression quantitative trait loci for the 7p11.2 temperature SNPs
The top fever SNPs at 7p11.2 do not map to any protein-coding genes; indeed the closest gene is ∼400 kb away. Furthermore, there was little evidence, through ENCODE bioinformatic interrogation, for functional effects at the 7p11.2 locus itself (see Supplementary Material, Results). Thus, to explore potential candidate genes mediating the downstream effect of the 7p11.2 locus on temperature, we interrogated expression quantitative trait loci (eQTL) datasets for the 7p11.2 SNP region in a variety of tissues of potential direct relevance to inflammation and thermoregulation. We screened eQTL associations for all 13 genome-wide significant 7p11.2 fever SNPs. While multiple of these SNPs had significant eQTL associations, consistent with moderate-strong LD between these 13 SNPs, we present findings (Table 3) for the top GWAS fever SNP (rs7805622) as well as a genome-wide significant SNP (rs7781869, P = 8.98 × 10−12 for fever; r2 = 0.36 with rs7805622 in 1000G Phase 2 EUR) with the strongest eQTL association signals (Table 3 and below).
Table 3.
(A) Tissue patterns of expression quantitative trait loci (eQTLs) for the top genome-wide significant 7p11.2 fever SNP (rs7805622), the genome-wide significant 7p11.2 fever SNP (rs7781869) with the strongest eQTL associations and the strongest eQTL SNP for each of the upstream candidate genes; (B) summary of experimental evidence for modulation by LPS and cold exposure of these eQTL candidate genes
| A |
B |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Lymphoblastoid cells (N = 109)a | Liver (N = 956)b | Subcutaneous fat (N = 610)b | Omental fat (N = 742)b | Monocytes (N = 1490)c | LPS-modulated | Cold-modulated | ||
| P-value | P-value | P-value | P-value | P-value | In vivo (human) | In vitro ASCs (by genotype) | In vivo (mouse) | In vitro ASCs (by genotype) | ||
| CCT6A | rs7805622 | NS | 0.014 | NS | NS | NS | *** | *** | ||
| rs7781869 | 0.002 | 0.019 | NS | 0.01 | ND | |||||
| Strongest eQTL SNP | rs7788786 P = 9.9 × 10−4 | rs1722282 P = 0.0001 | rs4591982d P = 0.02 | rs4591982d P = 0.004 | NS | |||||
| CHCHD2 | rs7805622 | 0.04 | NS | NS | NS | NS | *** | |||
| rs7781869 | 5.0 × 10−5 | NS | NS | NS | ND | |||||
| Strongest eQTL SNP | rs816407 P = 1.1 × 10−14 | rs7791829 P = 0.0004 | NS | NS | rs6593296 P = 7.7 × 10−87 | |||||
| GBAS | rs7805622 | NS | NS | NS | NS | NS | * | *** | ||
| rs7781869 | 0.004 | NS | NS | NS | ND | |||||
| Strongest eQTL SNP | rs7788786 P = 6.0 × 10−6 | NS | rs2270244 P = 0.007 | rs12719017 P = 0.02 | NS | |||||
| MRPS17 | rs7805622 | 0.07 | NS | NS | NS | NS | *** | |||
| rs7781869 | 5.0 × 10−4 | NS | NS | NS | ND | |||||
| Strongest eQTL SNP | rs2634080 P = 3.6 × 10−4 | NS | NS | rs4591982d P = 0.008 | NS | |||||
| PSPH | rs7805622 | 0.05 | NS | 0.006 | NS | NS | ** | *** | ** | |
| rs7781869 | 9.8 × 10−5 | NS | 4.7 × 10−5 | 0.0003 | ND | |||||
| Strongest eQTL SNP | rs7781869 (as above) | rs7791829, P = 2.5 × 10−5 | rs4591982d, P = 1.4 × 10−5 | rs4591982d, P = 3.1 × 10−6 | rs11772761, P = 4.7 × 10−6 | |||||
| SUMF2 | rs7805622 | 0.005 | NS | NS | 0.03 | 2.5 × 10−7 | *** | * | *** | |
| rs7781869 | 7.0 × 10−4 | NS | NS | NS | ND | |||||
| Strongest eQTL SNP | rs12155441 P = 1.2 × 10−4 | rs7793536 P = 0.008 | rs10800 P = 0.002 | rs1994518 P = 0.004 | rs12668532 P = 3.5 × 10−21 | |||||
| ZNF713 | rs7805622 | 0.01 | NS | NS | NS | NS | * | |||
| rs7781869 | 6.0 × 10−4 | NS | NS | 0.001 | ND | |||||
| Strongest eQTL SNP | rs1113765, P = 2.7 × 10−6 | NS | rs7804578, P = 9.4 × 10−5 | rs3813505, P = 4.5 × 10−6 | NS | |||||
NS, no significance; ND, no data.
aSNP genotypes obtained from HapMap version 3, release version 2; gene expression from Illumina Sentix Human-6 Expression BeadChip v2.
bSNPs genotyped using Illumina 650Y BeadChip array; gene expression from Agilent 44 K Custom array.
cSNPs genotyped using Affymetrix 6.0; gene expression from Illumina HT-12 BeadChip v3.
dThe best SNP has high LD (>0.8) with either of the top GWAS SNPs.
*P < 0.05; **P < 0.01; ***P < 0.05.
In HapMap3 lymphoblastoid cells (15), a number of clustered genes were differentially expressed by genotype at SNPs in our 7p11.2 fever locus. This cluster included SUMF2, CCT6A, CHCHD2, GBAS, MRPS17, PSPH and ZNF713 ∼400 kb upstream of the 7p11.2 locus (see Supplement Material, Fig. S1). In monocytes of 1490 individuals (16), we again observed significant cis-eQTL between 7p11.2 SNPs and SUMF2 gene expression. In large datasets for liver (n = 956), omental adipose (n = 742) and subcutaneous adipose (n = 610) (17,18) we confirmed the eQTLs for CCT6A, CHCHD2, GBAS, MRPS17, PSPH, SUMF2 and ZNF713. While none of the 7p11.2 locus fever SNPs were significant eQTLs in GTEX datasets (19) (not shown), GTEX dataset sample sizes for each tissue are substantially smaller (n < 200) than the lymphoblastoid, monocyte, liver and adipose resources that we interrogated above.
We used the same eQTL datasets to query the strongest eSNP for each of the seven candidate genes (Table 3). Notably, the strongest eQTL SNP for PSPH in lymphoblastoid cells (rs7781869; P = 9.8 × 10−5) was itself one of the 13 genome-wide significant 7p11.2 fever SNPs (P = 8.98 × 10−12 for fever). The strongest eQTL SNP for several genes in subcutaneous and omental adipose (rs4591982, CCT6A, MRPS17, PSPH) is located in the top fever SNP region, was also genome-wide significant for fever (imputed SNP, P = 6.97 × 10−10) and is in high LD with rs7781869 (r2 = 0.82). While SNPs located within the gene cluster had stronger effects on expression levels than the 7p11.2 fever locus SNPs for some genes, e.g. CHCHD2 (Table 3), several of these top eQTL SNPs located in the gene cluster had nominally significant associations with fever (Supplementary Material, Table S2) (e.g. rs10800 located in the 3′ UTR of SUMF2, the strongest eQTL for SUMF2 in subcutaneous fat; P = 8.8 × 10−5 for peak fever in GENE). There is moderate LD (all r2's < 0.4) between the top 13 fever 7p11.2 SNPs and the top eQTL SNPs in the gene cluster region (e.g. Supplementary Material, Fig. S1, rs7805622 regional LD using 1000G Phase 2 EA data; this is representative for all 13 fever 7p11.2 locus SNPs).
We performed conditional analyses between our lead GWAS SNP (rs7805622) and each of several eQTL SNPs (for genes in the upstream cluster) in order to explore independence of their associations with LPS-evoked fever (Supplementary Material, Results and Table S4). First, we conditioned the lead GWAS SNP (rs7805622) on the top eQTL SNP (rs7781869) for PSPH (in lymphoblastoid cells). Inclusion of rs7781869 greatly attenuated the association of rs7805622 with fever (2.4 × 10−12 to 0.001) while the rs7781869 association with fever was lost in this conditional analysis (8.9 × 10−12 to 0.09). This might suggest that the association signals are not independent of each other perhaps reducing the possibility of coincidental overlap. However, given the LD between these SNPs (r2 = 0.36), which could introduce multicolinearity into the model, we interpret these data with caution. We also conditioned our lead GWAS SNP (rs7805622) on two other top eQTL SNPs, not in LD with the top GWAS SNP (rs12668532, top eQTL SNP for SUMF2 in monocytes; and rs6593296, top eQTL SNP for CHCHD2 in monocytes). Inclusion of rs12668532 (the top eQTL SNP for SUMF2 in monocytes) with our lead GWAS SNP (rs7805622) led to slight attenuation of association of rs7805622 with fever (2.4 × 10−12 to 5.2 × 10−8) while a modest association of rs12668532 with fever was completely attenuated (0.001–0.8) when analyzed with rs7805622. Thus, these two SNPs may not be independent of each other in their relation to fever and in this case are not in LD eliminating concerns regarding model multicolinearity. Inclusion of rs6593296 (the top eQTL SNP for CHCHD2 in monocytes) with rs7805622 had no impact on the association of rs7805622 with fever while rs6593296 itself had no association with fever. Thus, rs7805622 appears to act on fever independently of the top CHCHD2 eQTL SNP (rs6593296). Because rs6593296 itself has no association with fever, it appears that some genes (e.g. CHCHD2) in this eQTL gene cluster are less likely to be involved in the GWAS signal for fever.
While we cannot exclude a coincidental overlap between eQTL SNPs and the genome-wide significant 7p11.2 fever locus SNPs, the observed eQTL combined with the LD and eQTL SNP fever-association patterns in this regions are suggestive of a regulatory function (e.g. enhancer) at the 7p11.2 locus acting through expression of one or more of the upstream gene cluster in mediating the association with fever.
Expression of SUMF2, CCT6A and GBAS is regulated by experimental endotoxemia
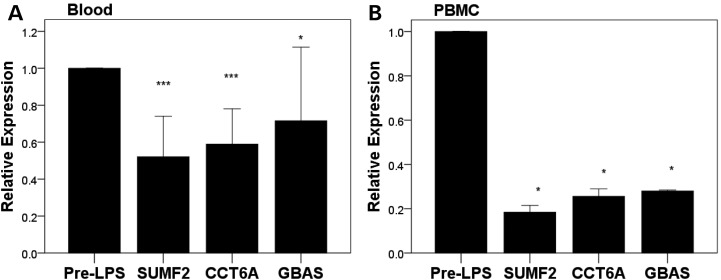
In order to prioritize specific candidate genes, we examined, in vivo in GENE participants, LPS-modulation of expression of each eQTL gene identified for the 7p11.2 temperature locus. RNASeq data from GENE study subjects were analyzed, before and after LPS, from whole blood (n = 10) and peripheral blood mononuclear cells (PBMCs) (n = 2). In blood (Fig. 3A), SUMF2 expression was reduced by ∼50% (P < 0.0001), CCT6A by ∼60% (P < 0.0001) and GBAS by ∼18% (P = 0.05). In PBMCs (Fig. 3B), SUMF2 was reduced by ∼18% (P = 0.02), CCT6A by ∼26% (P = 0.02) and GBAS by ∼70% (P = 0.003). In independent support of the GENE study findings, SUMF2 expression was reduced in whole blood (by ∼80%, P = 0.0007; n = 6) in a separate moderate-dose LPS study (20). These data suggest tissue-specific LPS-regulation of several of these candidate genes for the 7p11.2 fever locus.
Figure 3.
Amongst the candidate genes from eQTL analysis, SUMF2, CCT6A and GBAS are LPS-regulated in selective GENE study participants at RNASeq. (A) Blood RNASeq data were generated for 10 GENE study individuals and (B) PBMC RNASeq data in two participants. SUMF2 was significantly down-regulated by LPS in blood (A, P < 0.0001) and PBMC (B, P = 0.02). CCT6A was similarly down-regulated in blood (A, P < 0.0001) and PBMCs (B, P = 0.02). GBAS expression was reduced slightly in blood (A, P = 0.05) and was considerably down-regulated in PBMCs (B, P = 0.003). ***P < 0.001; *P < 0.05.
Expression of candidate genes is regulated by LPS and cold exposure in human genotype-specific adipose stromal cells
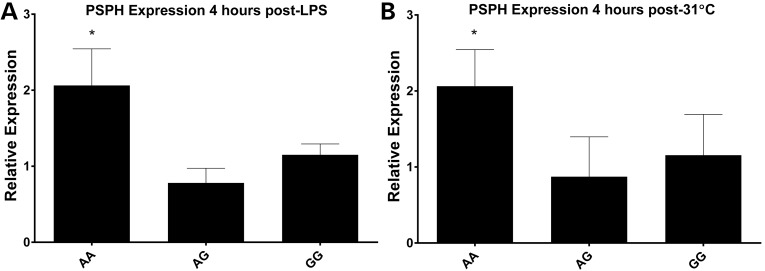
Using human adipose stromal cells (ASCs) obtained from GENE study subjects, we explored differences in gene expression by genotype at rs7805622 in response to 24-h LPS treatment (100 ng/ml) or cold exposure (31°C), an established peripheral thermoregulatory probe (21). In four cultured ASC lines (1 AA, 1 AG and 2 GG at rs7805622), expression of several genes, particularly PSPH, increased with LPS and cold exposure, and appeared to differ by genotype, with rs7805622AA cells having a significant increase in expression 4-h post-LPS compared with G allele carriers (P < 0.01) (Fig. 4A and B). There were also significant LPS-induced changes by genotype in ZNF713 (4-h post-LPS, P < 0.0001) and SUMF2 (1-hour post-LPS, P < 0.01), with increased expression in AA but not AG or GG (P values derived from unpaired T-test). Although there is only moderate LD between rs7805622 and the top eQTL SNP (rs7781869) in the overall population, the genotypes for rs7805622 and rs7781869 were identical within these four subjects.
Figure 4.
Differences in PSPH expression in human ASCs by rs7805622 genotypes at chromosome 7p11.2. PSPH expression increased 4-hours after LPS (100 ng/ml) (A, *P < 0.01) and cold exposure (31°C) (B, *P < 0.01) in ASCs of the minor allele homozygote (AA) but not in major allele carriers (AG and GG) for rs7805622.
Expression of candidate genes is regulated by cold exposure in mice
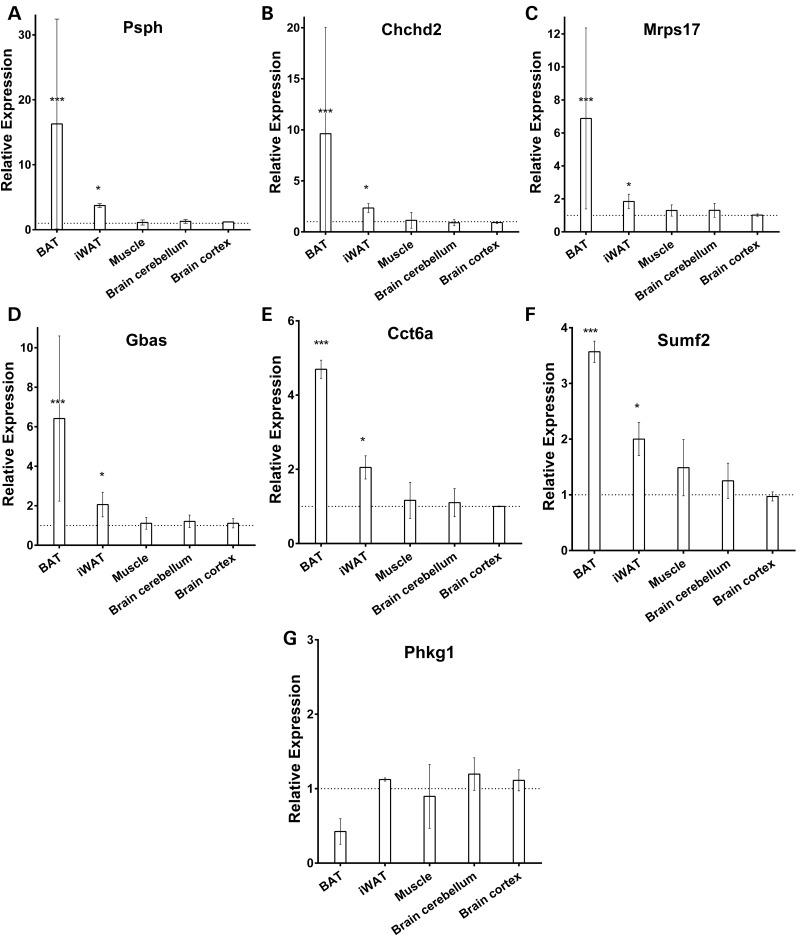
We examined expression of the eQTL-identified candidate genes in multiple tissues from mice exposed to cold (5°C) for 6 days versus mice maintained at 28°C. Several genes, including Psph, Chchd2, Gbas, Mrps17, Cct6a and Sumf2 (located on a syntenic block on mouse Chr5qG1.3) were markedly up-regulated in subscapular brown adipose tissue (BAT), slightly up-regulated in inguinal white adipose tissue (iWAT) but not in any other tissue in response to cold (all P < 0.0001 for BAT; <0.05 for WAT, P values from unpaired T-test) indicating potential roles in the peripheral thermogenesis arm of thermoregulation. As a hypothesized null control, there was no change in expression of Phkg1, also located in the human 7p11.2 and mouse 5qG1.3 region but not a significant eQTL gene for the temperature locus (Fig. 5A–G). As expected, several genes with established roles in BAT thermogenesis (e.g. Ucp1, Cidea, Cox8b) were modulated by cold stress in BAT (data not shown).
Figure 5.
Induction of candidate eQTL genes in BAT of male mice (n = 4) exposed to cold (5C versus 28C) for 6 days. (A) Psph, (B) Chchd2, (C) Mrps17, (D) Gbas, (E) Cct6a and (F) Sumf2 mRNA levels were increased (***all P < 0.0001) in BAT, as well as in iWAT (*all P < 0.05) during cold stimulation. (G) Phkg1 (a non-eQTL gene) was not differentially expressed in any tissue by cold exposure. All mRNA expression values were compared with control condition (28°C) within the same tissue.
Discussion
Through genomics of experimental endotoxemia in healthy EA individuals, we identified a genome-wide significant association between intergenic SNPs on chromosome 7p11.2 and the LPS-induced febrile response. This locus had no association with basal temperature, highlighting the importance of studying the evoked phenotype, as has been suggested for inflammatory pathophysiologies (22,23). Further, SNPs at the locus were only weakly related to LPS-induced plasma cytokine responses in the same individuals, suggesting that the 7p11.2 genomic region may influence thermoregulation independent of the systemic cytokine response. We replicated the association in an independent endotoxemia experiment even at a lower dose of LPS. We extended the association to clinically important settings by demonstrating associations with temperature change and death in preliminary clinical translation in cohorts of patients with trauma and sepsis. Through LD interrogation and eQTL analyses in multiple relevant tissues we identified a number of genes in cis ∼400 kb from the GWAS locus with significant expression differences by genotype at our top GWAS SNPs. Three of these genes, SUMF2, CCT6A and GBAS were LPS-regulated in vivo in the GENE study sample. Further, several genes, including PSPH, were regulated by LPS or cold exposure in vitro in ASCs in a genotype-specific manner, as well as being regulated in BAT by cold exposure in vivo in mice, thus providing candidates that require further functional and clinical focus in follow-up.
The febrile response is a crucial component of host adaptation, clinical course and survival during acute pathogenic stress and physical trauma. Body temperature is tightly regulated, in part via inflammatory prostaglandin signaling. Fever can be induced by peripheral circulating PGE2 as well as PGE2 produced within the brain in response to cytokine signaling (24,25). The relative importance of PGE2 production in the brain versus the periphery is not fully understood and may relate to different phases of the febrile response (8). Neuronal signaling mediates temperature changes via increased thermogenesis in BAT, shivering thermogenesis in skeletal muscle, and cutaneous vasoconstriction to reduce heat loss (26). Individual variation in the ability to mount a febrile response may have major consequences in the context of acute illnesses such as sepsis or trauma, yet common genetic variation impacting febrile responses in the general population has not been described. There may also be consequences in the context of cardiometabolic disease, e.g. fever during or following MI is associated with worse outcomes (27).
The human endotoxemia model allows for the precise interrogation of inflammatory mediators and responses in a controlled setting. Despite the modest sample size, we identified and replicated a striking association of LPS-induced temperature response with a locus on 7p11.2. While a difference by genotype at this locus was completely absent for resting temperature, interrogation of dynamic responses during the LPS provocation revealed the association; arguably, this evoked physiology is more informative for the relevant clinical pathophysiologies and disease than resting temperature. The minor allele at the top 7p11.2 SNP (rs7805622A) was related to peak temperature following LPS in the GENE study but also appeared to modulate dynamic temperature fluxes over time including the nadir temperature post-LPS. Remarkably, both the direction and temporal pattern of association of the rs7805622A allele with temperature were replicated in the independent FFAME study. Thus, genomic variation in the 7p11.2 region may modulate dynamic temperature fluctuations during infection and inflammatory stress.
The top 7p11.2 SNPs are located in and close to novel lincRNAs (RP11-760D2.1, RP11-760D2.5) of unknown function. While lincRNAs are currently receiving substantial attention for their potential functional roles in regulation of gene expression and cellular processes (28), little is known yet of the biological roles of this class of non-coding RNAs. We were unable to determine whether the 7p11.2 lincRNAs have any functional relevance. Indeed, they lack conservation or synteny with lincRNAs in mice and appear to be expressed in testes in human but no other tissue of relevance to fever and thermoregulation. Furthermore, it is possible that the SNPs at 7p11.2 exert enhancer and regulatory effects on surrounding genes, independent of any potential functions of the lincRNAs. Indeed, the eQTL signals (in multiple fever-relevant tissues) for the upstream gene cluster (SUMF2, CCT6A, GBAS, CHCHD2, ZNF713, MRPS17 and PSPH) suggest that the 7p11.2 locus (29) might act on fever via one or more of the eQTL genes. In particular, the top eQTL SNP for PSPH in lymphoblastoid cells (rs7781869) was amongst the genome-wide significant 7p11.2 SNPs for fever (P = 8.98 × 10−12). Yet, which of the genes in this cluster is important and in what capacity, i.e. the pre-febrile response to pyrogens in leukocytes, or alternatively in an effector role in neurons, BAT, skeletal muscle or skin remains to be elucidated. As the eQTL candidate genes are clustered together, and may be co-regulated, it is also possible that more than one of these genes have functional relevance, consistent with non-random functionally related gene clustering in eukaryotes (30). However, we highlight PSPH as a gene prioritized in the eQTL findings, as well as in the in vitro and in vivo analysis. Furthermore, it appears that BAT may be a promising target tissue for further functional studies.
PSPH (phosphoserine phosphatase), highlighted through eQTL analyses, and which had differential expression by 7p11.2 genotypes in human ASCs in response to LPS and cold (the rs7805622 minor allele associated with greater temperature fluctuations had higher PSPH expression in response to cold and LPS), also was the most up-regulated of the eQTL genes in mouse BAT in response to cold. PSPH, which is linked to the neurodevelopmental disorder Williams syndrome (31), is rate-limiting for l-serine production, which together with glutamate is a key signaling molecule in the brain (32). Serine is linked to fever inhibition in rabbits (33), making this gene an attractive biological candidate for further study in both central and peripheral aspects of thermoregulation. At deep RNASeq in GENE study samples, expression of sulfatase modifying factor 2 (SUMF2) was regulated by LPS in blood and PBMCs and was identified in the eQTL analysis in multiple tissues. SUMF2 interacts with interleukin-13 (IL-13), a cytokine associated with allergic inflammation, and its expression in lymphocytes was shown to inhibit secretion of IL-13 (34). IL-13 promotes activation of alternative M2 monocyte/macrophages, thereby inhibiting pro-inflammatory cytokines and chemokines (35). Thus, 7p11.2 locus modulation of SUMF2 expression and downstream IL-13 secretion might impact the fever response during endotoxemia, sepsis and trauma. Of the other candidate genes, several are localized to mitochondria, e.g. GBAS, CHCHD2 and MRPS17 (36). Given the importance of mitochondria in oxidative phosphorylation and thermogenesis, these genes are also candidates for the effector phase of the febrile response in mitochondria-rich tissue such as skeletal muscle and brown adipose. We acknowledge, however, that we cannot exclude a coincidental overlap between eQTL SNPs and the genome-wide significant 7p11.2 fever locus SNPs. These regional eQTL findings are correlative and do not infer causality for gene(s) in the upstream cluster.
Our study had several strengths. Despite a relatively small sample size, we detected a strong signal for LPS-induced fever that was not apparent at rest, highlighting the power of the evoked model and analysis of dynamic data as reported also by others (22,23). We replicated this finding in a smaller endotoxemia experiment even at a lower dose of LPS. Importantly, we demonstrated potential clinical importance through our finding that the 7p11.2 locus had nominal associations with both temperature and death in patients with trauma and sepsis. Several large tissue expression datasets were interrogated to uncover eQTLs for our top SNPs and to identify genes with differential expression in tissues of biological relevance to temperature homeostasis. In support of specific eQTL genes, we demonstrated their in vivo LPS-regulation (RNASeq and microarray) in two independent endotoxemia studies. Furthermore, several eQTL candidate genes displayed in vitro genotype-specific regulation by LPS and cold and were modulated in BAT in vivo during cold stress in mice suggesting a potential role for the locus and these genes in modulating peripheral thermogenesis. While we highlight a cluster of novel candidate genes with supporting evidence and potential mechanisms for a subset, we have yet to definitively implicate a single gene specifically or define the exact cell or tissue mediating the fever phenotype at the 7p11.2 locus. Larger studies and further elucidation of underlying mechanisms will define clinical utility and therapeutic potential in the treatment of trauma, sepsis, resuscitation and possibly chronic inflammatory diseases.
Materials and Methods
Clinical studies
The GENE study: human low-dose endotoxemia
The GENE study recruited healthy individuals (N = 294, 52% female, 65% EA, 35% AA) ancestry to a UPenn inpatient Clinical and Translational Research Center (CTRC) protocol as previously described (11). Participants completed an endotoxin challenge (1 ng/kg E coli-derived LPS; US standard reference, lot no. CCRE-LOT-1+2, Clinical Center, Pharmacy Department at the National Institutes of Health, Bethesda, MD) (11,37). Multiple clinical variables were assessed during the visit, with temperature recorded every 30 min for the first 8-h post-LPS and hourly thereafter. The GENE study was approved by UPenn's Institutional Review Board (IRB), with regulatory oversight by the FDA (LPS: IND# 5984) and an NIH-appointed data-safety and monitoring board. All subjects provided written informed consent.
The FFAME study: human very low-dose endotoxemia
The FFAME study recruited healthy individuals (N = 80, 65% EA, 20% AA, 15% Asian ancestry) to a UPenn CTRC evoked endotoxemia protocol (0.6 ng/kg LPS) following a 6–8 week double-blind placebo-controlled intervention with fenofibrate or long-chain omega-3 polyunsaturated fatty acids (n− 3 PUFA) as described (38). Temperature was recorded every 30 min for the first 8-h post-LPS and hourly thereafter. This study was approved by the UPenn IRB and all participants provided written informed consent.
The UPenn LPS study: human moderate-dose endotoxemia
Healthy volunteers were studied as reported (20,37,39). Subjects completed an endotoxin challenge (3 ng/kg LPS), with serial blood samples collected. Microarray analysis was carried out in a subset of participants in blood (n = 6). This study was approved by the UPenn IRB, and written informed consent was provided by all participants.
The UPenn Major Trauma Cohort study and MESSI study
Trauma and sepsis subjects were recruited after admission to the Hospital of the University of Pennsylvania (HUP) intensive care unit (ICU), as previously described (40,41) (and Supplementary Material, Methods). Clinical and demographic data were abstracted from the electronic medical record, and hospital mortality determined for all subjects. Peak and nadir temperatures for the first 24 h of hospitalization were recorded. Subject characteristics are summarized in Table 2. Informed consent was obtained from patients or their proxies by trained study personnel.
Genotype-specific adipose stromal cell studies
Fresh adipose tissue was obtained by needle aspiration from GENE study subjects and ASCs generated as described in the Supplementary Material, Methods. ASC lines from young healthy male volunteers, selected by genotype at rs7805622 (AA n = 1, AG n = 1, GG n = 2), were cultured to confluence, and either treated with 100 ng/ml LPS, subjected to cold (incubator set at 31°C) or maintained at 37°C for 24 h. All conditions were performed in triplicate. Cells were harvested for RNA extraction using Trizol reagent (Life Technologies, Foster City, CA).
Mouse studies
Wild-type male mice (N = 4, C57BL/6 COX-2 Flox/Flox) were maintained at 28°C for 1 month, and then challenged at 5°C for 6 days (n = 2), or maintained at 28°C for another 6 days (n = 2) as per an IACUC-approved protocol. Mice were sacrificed and tissues harvested (subscapular brown adipose, inguinal white adipose, skeletal muscle, brain cerebellum and brain cortex). Tissues were frozen immediately and maintained at −80°C prior to RNA extraction.
Laboratory methods
Cytokine measurement
Plasma levels of TNFα, IL-6 and IL-1RA were measured in GENE study subjects by high-sensitivity enzyme-linked immunosorbent assay (Quantikine; R&D Systems, Minneapolis, MN) according to manufacturer's instructions, as described (11).
Nucleic acid extraction and analysis
DNA was extracted from buffy coat samples using a QIASymphony SP instrument (GENE and FFAME Studies) or QIamp mini kit (Trauma and MESSI studies) (Qiagen, Valencia, CA). GENE study subjects were genotyped using the Affymetrix 6.0 SNP chip (Affymetrix, Santa Clara, CA) at the Center for Applied Genomics at the Children's Hospital of Philadelphia (e.g. (42)). Genotyping QC and filtering were applied as described in the Supplementary Material, Methods. After filtering, there were n = 175 EA and n = 96 AA individuals for GWAS analysis. Selected SNPs (rs7805622, rs10238214, rs10248128) were genotyped in the FFAME, Trauma and MESSI studies using Taqman primers and probes on an ABI7900 real-time PCR system (Applied Biosystems, Foster City, CA). RNA was extracted and gene expression analyzed using RNASeq and qPCR as described in the Supplementary Material, Methods.
Statistical and bioinformatic analyses
GWAS of Affy 6.0 SNP genotyped and 1000 genomes-imputed data were analyzed by linear regression using Plink v1.06 (after filtering, n = 574 042 genotyped SNPs) and Mach2qtl v.1.1.0 (43,44) (after filtering, n = 7 043 028 imputed SNPs). QC filtering was applied as described in detail in the Supplementary Material, Methods. Briefly, we filtered for MAF > 0.1, call rate >95%, HWE P > 1 × 10−8 and imputation r2 > 0.3. The change in temperature to the average peak time (3.5 h post-LPS, inverse normal transformed) and temperature at baseline (Pre-LPS) were analyzed as our primary and secondary outcome variables. Analysis was adjusted for age and sex (e.g. Model 1, Supplementary Material, Table S3). Inclusion of principal components (Model 2, Supplementary Material, Table S3) did not impact findings, e.g. rs7805622, P = 5.1 × 10−12 and 2.4 × 10−12 in PC-adjusted and non PC-adjusted models, respectively. Association analyses in FFAME applied race-specific linear regression models adjusting for the age, sex and treatment group. Association analyses between SNPs and temperature in the Trauma and MESSI study samples applied race-specific linear regression models, adjusting for age, sex and, for Trauma only, crystalloid resuscitation. Association analyses between SNPs and death were carried out by Fisher’s Exact Test. Data were analyzed using SPSS 21 (IBM, Armonk, NY). Expression eQTL interrogation was carried out in Genevar 3.2.0 (http://www.sanger.ac.uk/resources/software/genevar) (15) using lymphoblastoid cell data from HapMap3 individuals (n = 109 CEU); monocyte eQTL data from 1490 individuals were queried using GHSExpress (http://genecanvas.ecgene.net/uploads/ForReview/, last accessed on October 2014) (16). Gene expression (Agilent custom microarray) and SNP genotype (Illumina 650Y BeadChip array) data from liver (n = 956), omental adipose (n = 742) and subcutaneous adipose (n = 610) were tested for association between SNP genotypes and mean log ratio expression measures of genes in each tissue located within 1 Mb of the SNPs of interest, using the kruskal.test procedure in R as previously described (17,18). We obtained LD estimates using 1000G Phase 1 version 2 (March 2012, EUR or AFR) (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G.2012-02-14.html, last accessed on September 2014) and visualized these using LocusZoom (1000G EUR or AFR March 2012) (45). RNASeq data were analyzed using Cufflinks v1.3.0 (46) for transcript assembly, and gene expression pre- and post-LPS was compared using the cuffdiff option. To ensure reliable expression estimates, we required the fragments per kilobase of exon per million fragments mapped value to be >10 for at least one of the two samples under comparison. Gene expression differences in LPS- or cold-treated cells and in mouse tissues were assessed by unpaired T tests (SPSS 21, IBM Armonk NY).
Supplementary Material
Funding
The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000003, NIH-NHLBI SCCOR Project grant (P50-HL-083799), R01-HL-113147 and by the Penn Genome Frontiers Institute under a grant with the Pennsylvania Department of Health (which disclaims responsibility for any analyses, interpretations or conclusions) to M.P.R.; NIH-NHLBI SCOR Project grant (P50-HL-60290) and HL079063 to J.D.C., and K23-HL102254 to N.J.M. J.F.F. was supported by a postdoctoral fellowship grant from the American Heart Association (12POST11840017). J.D.C. is also supported by R01-HL081619, R01-HL087115, R01-HL096845 and K24-HL-115354. M.P.R. is also supported by R01-HL-111694, R01-DK-090505, U01-HL-108636 and K24-HL-107643. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Data and Materials Availability
Summary GWAS results for the association of genotyped SNPs with evoked fever are available in the Supplementary Data file. RNASeq data have been deposited in the Gene Expression Omnibus (GEO) database (accession numbers GSE46323 and GSE50792). The GENE study is assigned FDA clinicaltrials.gov registration number NCT00953667.
Supplementary Material
Acknowledgements
We acknowledge the time and commitment of the participants, investigators and staff of the GENE study.
Conflict of Interest statement. The authors have no relevant disclosures or conflicts of interest.
References
- 1.Mizushima Y., Ueno M., Idoguchi K., Ishikawa K., Matsuoka T. Fever in trauma patients: friend or foe? J. Trauma. 2009;67:1062–1065. doi: 10.1097/TA.0b013e3181b848fc. [DOI] [PubMed] [Google Scholar]
- 2.Schortgen F. Fever in sepsis. Minerva Anestesiol. 2012;78:1254–1264. [PubMed] [Google Scholar]
- 3.Nallamothu B.K., Payvar S., Wang Y., Kosiborod M., Masoudi F.A., Havranek E.P., Foody J.M., Casscells S.W., Krumholz H.M. Admission body temperature and mortality in elderly patients hospitalized for heart failure. J. Am. Coll. Cardiol. 2006;47:2563–2564. doi: 10.1016/j.jacc.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Guinea G.V., Atienza J.M., Fantidis P., Rojo F.J., Ortega A., Torres M., Gonzalez P., Elices M.L., Hayashi K., Elices M. Increases of corporal temperature as a risk factor of atherosclerotic plaque instability. Ann. Biomed. Eng. 2008;36:66–76. doi: 10.1007/s10439-007-9397-4. [DOI] [PubMed] [Google Scholar]
- 5.Hasday J.D., Singh I.S. Fever and the heat shock response: distinct, partially overlapping processes. Cell Stress Chaperones. 2000;5:471–480. doi: 10.1379/1466-1268(2000)005<0471:fathsr>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aksentijevich I., Kastner D.L. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat. Rev. Rheumatol. 2011;7:469–478. doi: 10.1038/nrrheum.2011.94. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Ballou L.R., Morham S.G., Blatteis C.M. Cyclooxygenase-2 mediates the febrile response of mice to interleukin-1beta. Brain Res. 2001;910:163–173. doi: 10.1016/s0006-8993(01)02707-x. [DOI] [PubMed] [Google Scholar]
- 8.Blatteis C.M., Li S., Li Z., Feleder C., Perlik V. Cytokines, PGE2 and endotoxic fever: a re-assessment. Prostaglandins Other Lipid. Mediat. 2005;76:1–18. doi: 10.1016/j.prostaglandins.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Solinas G. Molecular pathways linking metabolic inflammation and thermogenesis. Obes. Rev. 2012;13(Suppl 2):69–82. doi: 10.1111/j.1467-789X.2012.01047.x. [DOI] [PubMed] [Google Scholar]
- 10.Suffredini A.F., Fromm R.E., Parker M.M., Brenner M., Kovacs J.A., Wesley R.A., Parrillo J.E. The cardiovascular response of normal humans to the administration of endotoxin. N. Engl. J. Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson J.F., Patel P.N., Shah R.Y., Mulvey C.K., Gadi R., Nijjar P.S., Usman H.M., Mehta N.N., Shah R., Master S.R., et al. Race and gender variation in response to evoked inflammation. J. Transl. Med. 2013;11:63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martich G.D., Boujoukos A.J., Suffredini A.F. Response of man to endotoxin. Immunobiology. 1993;187:403–416. doi: 10.1016/S0171-2985(11)80353-0. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S., Cherny S.S., Sham P.C. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 14.He J., Li H., Edmondson A.C., Rader D.J., Li M. A Gaussian copula approach for the analysis of secondary phenotypes in case–control genetic association studies. Biostatistics. 2012;13:497–508. doi: 10.1093/biostatistics/kxr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics (Oxford, England) 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenawalt D.M., Dobrin R., Chudin E., Hatoum I.J., Suver C., Beaulaurier J., Zhang B., Castro V., Zhu J., Sieberts S.K., et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium G.T. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrke M., Reilly M.P., Millington S.C., Iqbal N., Rader D.J., Lazar M.A. An inflammatory cascade leading to hyperresistinemia in humans. PLoS Med. 2004;1:e45. doi: 10.1371/journal.pmed.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L., Wu J., Cohen P., Kazak L., Khandekar M.J., Jedrychowski M.P., Zeng X., Gygi S.P., Spiegelman B.M. Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:12480–12485. doi: 10.1073/pnas.1310261110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco L.M., Bucasas K.L., Wells J.M., Nino D., Wang X., Zapata G.E., Arden N., Renwick A., Yu P., Quarles J.M., et al. Integrative genomic analysis of the human immune response to influenza vaccination. Elife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R., Mias G.I., Li-Pook-Than J., Jiang L., Lam H.Y., Miriami E., Karczewski K.J., Hariharan M., Dewey F.E., Cheng Y., et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazarus M., Yoshida K., Coppari R., Bass C.E., Mochizuki T., Lowell B.B., Saper C.B. EP3 prostaglandin receptors in the median preoptic nucleus are critical for fever responses. Nat. Neurosci. 2007;10:1131–1133. doi: 10.1038/nn1949. [DOI] [PubMed] [Google Scholar]
- 25.Ek M., Engblom D., Saha S., Blomqvist A., Jakobsson P.J., Ericsson-Dahlstrand A. Inflammatory response: pathway across the blood-brain barrier. Nature. 2001;410:430–431. doi: 10.1038/35068632. [DOI] [PubMed] [Google Scholar]
- 26.Morrison S.F., Nakamura K. Central neural pathways for thermoregulation. Front. Biosci. 2011;16:74–104. doi: 10.2741/3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito K., Anzai T., Yoshikawa T., Maekawa Y., Sugano Y., Kohno T., Mahara K., Okabe T., Asakura Y., Ogawa S. Increased body temperature after reperfused acute myocardial infarction is associated with adverse left ventricular remodeling. J. Card. Fail. 2007;13:25–33. doi: 10.1016/j.cardfail.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D.R., Hendrickson D.G., et al. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.M., Sonnhammer E.L. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaeken J., Detheux M., Fryns J.P., Collet J.F., Alliet P., Van Schaftingen E. Phosphoserine phosphatase deficiency in a patient with Williams syndrome. J. Med. Genet. 1997;34:594–596. doi: 10.1136/jmg.34.7.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabatabaie L., Klomp L.W., Berger R., de Koning T.J. l-serine synthesis in the central nervous system: a review on serine deficiency disorders. Mol. Genet. Metab. 2010;99:256–262. doi: 10.1016/j.ymgme.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Glyn J.R., Lipton J.M. Central administration of serine causes hypothermia and inhibits fever in rabbits. Brain Res. Bull. 1980;5:653–660. doi: 10.1016/0361-9230(80)90201-4. [DOI] [PubMed] [Google Scholar]
- 34.Liang H., Li Z., Xue L., Jiang X., Liu F. SUMF2 interacts with interleukin-13 and inhibits interleukin-13 secretion in bronchial smooth muscle cells. J. Cell Biochem. 2009;108:1076–1083. doi: 10.1002/jcb.22336. [DOI] [PubMed] [Google Scholar]
- 35.Yakubenko V.P., Hsi L.C., Cathcart M.K., Bhattacharjee A. From macrophage interleukin-13 receptor to foam cell formation: mechanisms for alphaMbeta2 integrin interference. J. Biol. Chem. 2013;288:2778–2788. doi: 10.1074/jbc.M112.381343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittag J., Lyons D.J., Sallstrom J., Vujovic M., Dudazy-Gralla S., Warner A., Wallis K., Alkemade A., Nordstrom K., Monyer H., et al. Thyroid hormone is required for hypothalamic neurons regulating cardiovascular functions. J. Clin. Invest. 2013;123:509–516. doi: 10.1172/JCI65252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson P.D., Mehta N.N., Wolfe M.L., Hinkle C.C., Pruscino L., Comiskey L.L., Tabita-Martinez J., Sellers K.F., Rickels M.R., Ahima R.S., et al. Innate immunity modulates adipokines in humans. J. Clin. Endocrinol. Metab. 2007;92:2272–2279. doi: 10.1210/jc.2006-2545. [DOI] [PubMed] [Google Scholar]
- 38.Ferguson J.F., Mulvey C.K., Patel P.N., Shah R.Y., Doveikis J., Zhang W., Tabita-Martinez J., Terembula K., Eiden M., Koulman A., et al. Omega-3 PUFA supplementation and the response to evoked endotoxemia in healthy volunteers. Mol. Nutr. Food Res. 2014;58:601–613. doi: 10.1002/mnfr.201300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta N.N., McGillicuddy F.C., Anderson P.D., Hinkle C.C., Shah R., Pruscino L., Tabita-Martinez J., Sellers K.F., Rickels M.R., Reilly M.P. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes. 2010;59:172–181. doi: 10.2337/db09-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah C.V., Lanken P.N., Localio A.R., Gallop R., Bellamy S., Ma S.F., Flores C., Kahn J.M., Finkel B., Fuchs B.D., et al. An alternative method of acute lung injury classification for use in observational studies. Chest. 2010;138:1054–1061. doi: 10.1378/chest.09-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah C.V., Localio A.R., Lanken P.N., Kahn J.M., Bellamy S., Gallop R., Finkel B., Gracias V.H., Fuchs B.D., Christie J.D. The impact of development of acute lung injury on hospital mortality in critically ill trauma patients. Crit. Care Med. 2008;36:2309–2315. doi: 10.1097/CCM.0b013e318180dc74. [DOI] [PubMed] [Google Scholar]
- 42.Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R., et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.