Abstract
Analysis of the T-cell receptor (TCR) repertoire of innate CD4+ T cells selected by major histocompatibility complex (MHC) class II-dependent thymocyte–thymocyte (T-T) interaction (T-T CD4+ T cells) is essential for predicting the characteristics of the antigens that bind to these T cells and for distinguishing T-T CD4+ T cells from other types of innate T cells. Using the TCRmini Tg mouse model, we show that the repertoire of TCRα chains in T-T CD4+ T cells was extremely diverse, in contrast to the repertoires previously described for other types of innate T cells. The TCRα chain sequences significantly overlapped between T-T CD4+ T cells and conventional CD4+ T cells in the thymus and spleen. However, the diversity of the TCRα repertoire of T-T CD4+ T cells seemed to be restricted compared with that of conventional CD4+ T cells. Interestingly, the frequency of the parental OT-II TCRα chains was significantly reduced in the process of T-T interaction. This diverse and shifted repertoire in T-T CD4+ T cells has biological relevance in terms of defense against diverse pathogens and a possible regulatory role during peripheral T-T interaction.
Introduction
Previously, we have shown that thymocyte–thymocyte (T-T) interactions that occur in a major histocompatibility complex (MHC) class II-restricted manner produce functionally competent CD4+ T cells with innate properties (that is, T-T CD4+ T cells).1, 2, 3, 4 Another group has provided support for this idea using a transgenic mouse system.5, 6 Most importantly, a recent report from our group demonstrated that T-T CD4+ T cells are present and have innate functions in humans, similar to what has been observed in transgenic mouse models.4
T-T CD4+ T cells differ from other types of innate T cells in several key aspects. As reported previously, T-T CD4+ T cells are restricted by classic MHC class II molecules,3, 5 and their T-cell receptor (TCR) repertoire is likely to be diverse.3 Innate T cells, such as NKT cells and γδ T cells, express restricted TCRs that bind conserved pathogen- or stress-related molecules. iNKT cells, which develop from CD4+CD8+ double-positive thymocytes through homotypic interactions, in a manner similar to that of T-T CD4+ T cells, interact through the invariant TCR. This TCR recognizes only non-classic MHC class Ib (CD1d) loaded with glycosphingolipid, thereby creating a very limited TCR repertoire.7, 8, 9, 10 In general, the various types of innate T cells use a single, fixed TCRα chain specific to each population and oligoclonal TCRβ chains with additional junctional variations.9, 11, 12 Therefore, determination of the TCRα repertoire of T-T CD4+ T cells is important in terms of distinguishing this T-cell population from other innate T cells and predicting the population's functional role in the immune defense mechanism.
Attempts have been made to analyze the TCR repertoire using transgenic mouse models with very limited TCR variability.13, 14, 15, 16 The opportunities for recombination were sharply reduced to limit TCR diversity, so that only imprecise joining, nucleotide insertion or deletion between the fixed Vα and Jα gene segments was permitted. Using a similar strategy, we generated an OT-II TCR-derived transgenic mouse model with limited TCR diversity: both the TCRα and the TCRβ transgenes were derived from an OT-II TCR-transgenic mouse, and the original OT-II TCRα transgene was further manipulated to generate a mini-construct. The established model, a TCRmini Tg mouse, was consequently used as the bone marrow (BM) donor to build a chimera in which CD4+ T cells were selected exclusively by MHC class II-expressing thymocytes. Using this model, we were able to analyze the TCR repertoire of T-T CD4+ T cells and compare it with that of conventional CD4+ T cells, in terms of both clonal diversity and degree of skewing.
Materials and methods
DNA constructs and transgenesis
The mini-transgene construct, which was designed to encode TCRα chains with CDR3 variability and recombination between the Vα2.3 and Jα31, and the Vα2.3 and Jα2 gene segments, was derived from the OT-II monoclonal transgenic TCR. The Vα2.3 and Jα31 gene segments were amplified from RNA that was extracted from OT-II TCR-transgenic splenocytes17 and cloned into the pGEMT-Easy vector (Promega, Madison, WI USA), and a rearrangement substrate composed of an RSS downstream of Vα2.3, a 481-bp spacer and the natural RSS from the Jα31 segment was inserted in between the Vα2.3 and the Jα31 fragments by PCR. The complete rearrangement machinery was then subcloned into the pTα expression cassette, as described previously.18
To obtain the desired transgenic mice, the complete construct was injected into fertilized (B6xSJL)F2 eggs. Flow cytometry (FACSCalibur; Becton Dickinson, Mountain View, CA, USA) using the anti-Vα2 (Vα2.3)-PE antibody (Ab) (BD Biosciences, San Jose, CA, USA) and anti-CD3-FITC was used to screen transgenic founders, and PCR using primers that detect the Vα2.3 and Jα31 segments confirmed that the mini-transgene was well introduced.
To eliminate any effect of the endogenous TCRα chain, the mice bearing the mini-transgene were bred with TCRCα−/− mice, which have the normal TCRα chain knocked out.19 The generated mice, which carried whole Vβ elements (referred to as TCRmini Tg Op), were crossed with OT-II TCRβ-transgenic mice, which are deficient with respect to normal TCRα chain expression (TCRVβ5.2/Cα−/−), to generate the TCRmini Tg mice.
Mice
C57BL/6 mice were obtained from the animal facility at the Biomedical Center for Animal Resource Development, Seoul National University College of Medicine. Plck-CIITATgCIITA−/− and Plck-CIITATgPIV−/− mice were previously generated.3, 20 TCRCα−/−, Rag1−/− and I-Ab−/− mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). The Plck-CIITATgCIITA−/− mice were bred with TCRCα−/− mice to obtain Plck-CIITATg/CIITA−/−/Cα−/− mice. All mice were maintained under specific pathogen-free conditions at the Biomedical Center for Animal Resource Development, Seoul National University College of Medicine, and were between 6 and 10 weeks of age when analyzed. Experiments were performed after approval received from the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Resources, Seoul National University.
BM chimeras
Rag1−/− or Rag1−/−/I-Ab−/− mice at 6–10 weeks of age were irradiated with two split doses of 400 cGy from a 137Cs source with a 4-h interval, and BM cells were transferred within 24 h of the second irradiation. The BM cells were flushed from the femurs and tibiae of TCRmini Tg or TCRmini Tg Op, Plck-CIITATg/CIITA−/−/Cα−/− and TCRα−/− mice. A single-cell suspension of BM cells was filtered through a sterile nylon mesh, incubated with CD4 and CD8 magnetic beads (Miltenyi Biotec, Auburn, CA, USA), and subjected to MACS depletion according to the manufacturer's protocols (Miltenyi Biotec). The T-cell-depleted BM cells (3.0 × 106) were mixed at a ratio of 1:1 and prepared in a volume of 300 μl of phosphate-buffered saline. Subsequently, these cells were injected intravenously into the lateral tail vein of the irradiated recipient mice. At 6–8 weeks post transplantation, the chimeras were killed for flow cytometric analysis of the thymocytes and splenocytes and subjected to single-cell sorting.
Single-cell sorting and RT-PCR
Single-cell suspensions of thymocytes or lymphocytes from the BM chimeras were prepared and stained with appropriate Ab sets. Using a FACSAria sorter (BD Biosciences), CD4+CD8−CD24−CD25−TCRVα2+ thymocytes or CD4+CD8−CD25−TCRVα2+ lymphocytes were directly sorted into 96-well PCR plates that contained 10 μl of RT buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 2% Triton X-100, 1 μg of bovine serum albumin, 125 μM dNTPs, 50 ng of oligo(dT)12–18, a specific primer (5′-CTGAACTGGGGTAGGTGGCGTT-3′) for detecting the constant region of TCRα (COSMOGENETECH, Seoul, Korea), 8 U of RNase inhibitors and 30 U of Moloney murine leukemia virus reverse transcriptase (KOSCHEMCO, Seongnam, Korea) in each well. The plates were incubated for 90 min at 37 °C and cooled to 8 °C. Amplification of the resulting cDNA was performed by adding 40 μl of Taq buffer (50 mM KCl, 10 mM Tris-HCl (pH 9.3), and 2.5 mM MgCl2) containing 2.5 U of Taq polymerase, 250 μM dNTPs, and 10 pmol of the sense and antisense primers and involved annealing at 50 °C and 35 cycles of 93 °C for 3 min. In the second round of PCR, 2 μl of the first PCR products was then used under the same conditions in 48 μl of Taq buffer that contained 1 U of Taq polymerase, 125 μM dNTPs and 5 pmol of the nested primers (24 cycles). The following primer sets were used for the amplification of the CDR3α region of the TCRα chain: Vα2F1 (5′-TCCATACGTTCAGTGTCCGATAAA-3′) and CαR1 (5′-TGGCGTTGGTCTCTTTGAAG-3′) for the first round of PCR and Vα2F2 (5′-AAAGGGAGAAAAAGCTCTCC-3′) and CαR2 (5′-GGCCCCATTGCTCTTGGAATC-3′) for the second PCR. The PCR products were loaded onto an agarose gel, and samples that yielded a single band were selected for PCR gel purification. The purified Vα2 PCR products were sequenced directly using CαR2 (5′-GGCCCCATTGCTCTTGGAATC-3′) (COSMOGENETECH).
Flow cytometry
The following Abs were purchased from BD Pharmingen (San Diego, CA, USA): APC-conjugated anti-CD4 (GK1.5); PE-conjugated anti-Vα2 (B20.1) and anti-CD62L (MEL-14); and FITC-conjugated anti-CD8 (53-6.7), anti-CD24 (M1/69), anti-CD25 (2A3), anti-CD44 (IM7), anti-CD69 (H1.2F3), and anti-Vβ5.1&5.2 (MR9-4). Fresh suspensions of thymocytes and splenocytes were resuspended in FACS buffer (1 × phosphate-buffered saline with 0.1% bovine serum albumin and 0.1% sodium azide). After staining with fluorescence-conjugated Abs for 30 min at 4 °C, the live cells, which were gated as the population negative for propidium iodide (Sigma Chemical Co., St. Louis, MO, USA) staining, were analyzed using a FACSCalibur that was equipped with CellQuest Pro software (Becton Dickinson).
ELISPOT assay
The frequencies of IFN-γ-secreting ovalbumin-specific CD4 T cells in the spleens of wild-type (WT) and CIITATgPIV−/− mice were measured using an ELISPOT kit (BD Biosciences; 552569). The WT and CIITATgPIV−/− mice were primarily immunized by subcutaneous injection with 100 μg of ovalbumin emulsified in complete Freund's adjuvant. Two and 4 weeks after the primary injection, second and third injections were performed with 100 μg of ovalbumin in incomplete Freund's adjuvant. One week after the last challenge, 1.0 × 105 CD4+ T cells isolated from the spleens of the immunized mice were cultured with 2.0 × 105 T-cell-depleted WT splenocytes pulsed with ovalbumin (100 μg ml−1) in complete RPMI 1640 media for 20 h at 37 °C in a 5% CO2 incubator. As a positive control, WT CD4 T cells were cultured with T-cell-depleted splenocytes in the presence of anti-CD3 Ab (2C11, 1 μg ml−1), and as a negative control, CD4 T cells from WT or CIITATgPIV−/− mice were cultured with T-cell-depleted splenocytes and without any antigen. The experimental procedures were performed exactly according to the manufacturer's instructions. The resulting spots were counted using a computer-assisted ELISPOT Reader System (AID, Strassberg, Germany).
Statistical analysis
The Jaccard classic (Cj) and Morisita-Horn (MH) indices21 were used as measures of similarity. These indices range from 0 (no similarity) to 1 (complete similarity). To exclude the impact of the sample size on the similarity level, the total number of individual sequences in each repertoire was standardized in terms of the smallest sample size prior to calculating the Cj and MH indices. We iterated these steps 10 000 times and obtained average index values. To estimate repertoire diversity, we employed two different indices: the species accumulation curve22 and the abundance-based coverage estimator (ACE).14, 23 In this paper, species accumulation curve is occasionally referred to as Obs (standing for ‘unique sequence observed'). We evaluated the significance of the diversity difference between two different TCR repertoires using the species accumulation curve and the ACE-based accumulation curve by calculating the empirical P-values using a permutation test.24 We first pooled two repertoires and then randomly partitioned the pooled repertoire into two groups of the same sizes as the original repertoires. For each group, the species accumulation curve was plotted, and the diversity difference between the two groups was calculated by summing the differences between the numbers of distinct species at given sample sizes. We iterated this process 1000 times and obtained the empirical P-value. We used the same method to estimate the empirical P-values using the ACE-based accumulation curves.
Results
T-cell development and the TCR repertoire of OT-II-based TCRmini transgenic mice
To generate a manageable set of TCR repertoires for the comparison of T-T CD4+ T cells with conventional CD4+ T cells, we engineered a transgenic mouse model in which the T cells express a limited TCR repertoire.13, 14 The TCRβ chain was fixed (Vβ5.2) using a transgene, and only the TCRα chain was allowed to undergo genetic rearrangement of a single Vα2.3 segment to either the Jα31 or the Jα2 segment (Mini1.1 construct), the combination of which was based on the OVA323-339-specific H2-Ab-restricted OT-II TCR. The mouse was backcrossed with a TCRCα−/− mouse to eliminate the influence of the endogenous TCRα chain, and the resulting mouse was designated as TCRmini Tg (Mini1.1/Cα−/−/Vβ5.2).
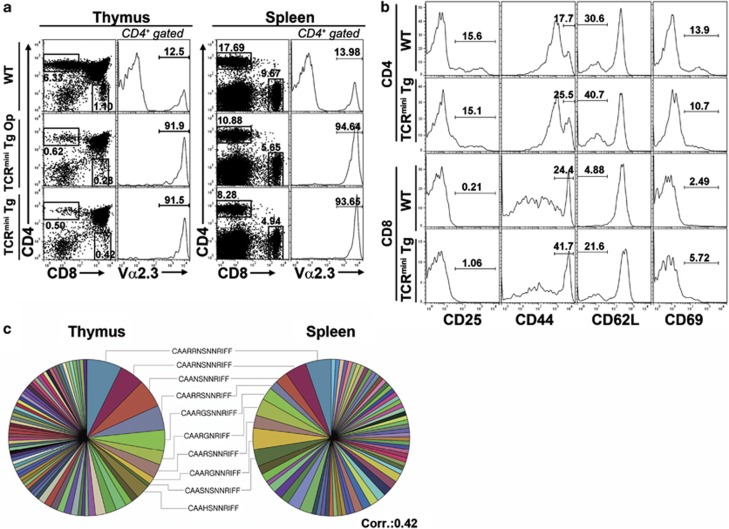
The TCRmini Tg mice seemed to carry out all the thymic ontogenic processes, and most of the CD4 single-positive (SP) and CD8 SP thymocytes and splenocytes expressed Vα2.3 (Figure 1a). The total numbers of CD4 SP and CD8 SP thymocytes generated in the TCRmini Tg mice were approximately 10% of those generated in the WT mice, and the ratio of CD4+ T cells to CD8+ T cells was 2:1 in the periphery. Although the TCRmini Tg mouse model was engineered based on the H2-Ab-restricted OT-II TCR, the generation of mature CD4 SP and CD8 SP thymocytes suggested that the TCRmini Tg CD4 SP and CD8 SP thymocytes recognized both MHC class I and II molecules, as observed for the OT-I TCR-based system.13 The peripheral mature T cells did not acquire a memory phenotype, excluding the possibility of homeostatic expansion of oligoclonal T cells (Figure 1b). Therefore, it seems that the TCRmini Tg mice in our model closely recapitulate normal T cell ontogeny.
Figure 1.
Thymic selection and maturation in TCRmini Tg mice. (a) Development of thymic and splenic T-cell populations in TCRmini Tg mice. The profiles of the CD4+ and CD8+ T cells were obtained from three mice each of the following types: WT, Mini1.1/Cα−/− (TCRmini Tg Op), and Mini1.1/Cα−/−Vβ5.2 (TCRmini Tg). The histograms represent gated CD4 SP thymocytes or CD4+ splenocytes (number of total thymocytes in WT mice: 10.99±2.37 × 107, TCRmini Tg Op: 11.49±3.32 × 107, TCRmini Tg: 10.95±4.43 × 107; number of total splenocytes in WT mice: 8.27±0.98 × 107, TCRmini Tg Op: 5.96±0.66 × 107, TCRmini Tg: 5.53±2.29 × 107). (b) Activated phenotype of splenic CD4+ and CD8+ T cells of C57BL/6 and TCRmini Tg mice, respectively: these cells were stained with PE-conjugated anti-CD25, anti-CD44, anti-CD62L and anti-CD69. (c) Four cell populations (CD4+CD8−CD24lowTCRβ5.2hi and CD4−CD8+CD24lowTCRβ5.2hi SP thymocytes and CD4+CD8−TCRβ5.2hi and CD4−CD8+TCRβ5.2hi splenocytes) pooled from two TCRmini Tg mice were sorted into 96-well plates (one cell per well). Each well was checked for Vα2 expression and CDR3α sequences between Vα2.3-Jα31 by RT-PCR and nested PCR. Pie charts were generated based on the CDR3α sequences of the CD4+CD8−CD24lowTCRβ5.2hi SP thymocytes (total cell number=137; number of different sequences=79) and CD4+CD8−TCRβ5.2hi splenocytes (total cell number=117; number of different sequences=71). Each color in the pie chart represents a different CDR3α sequence; several of the colors are used repeatedly for different sequences because of the high level of diversity. The area of each pie segment reflects the respective frequency of each TCRα sequence. The data are representative of two independent experiments.
The potential of the TCRmini Tg mouse to produce a diverse TCR repertoire was a prerequisite for the comparative analysis of the target TCR repertoires (T-T CD4+ T cells versus conventional CD4+ T cells). The TCR repertoire of the TCRmini Tg mouse was analyzed by single-cell sorting into microtiter plates, PCR amplification of the joining region and sequencing. The length of CDR3α in the TCRmini Tg mouse displayed a Gaussian-like distribution for the diverse TCR repertoires of four different subpopulations: CD4 SP and CD8 SP thymocytes and CD4+ and CD8+ peripheral T cells (Supplementary Figure 1a). Visualization of the clonotypic distributions of the TCRs between the CD4 SP thymocytes and the CD4+ peripheral T cells in a pie chart indicated that the thymic clones were not significantly altered in the periphery (Figure 1c). The CDR3α sequences of the CD4 SP and CD8 SP thymocytes showed certain subsets of distinctive TCR sequences according to their lineages (Supplementary Figure 1b). The total CDR3α sequences obtained from the thymuses and spleens of the TCRmini Tg mice are shown in Supplementary Table 1.
T-cell development through T-T interactions using the TCRmini Tg mouse system
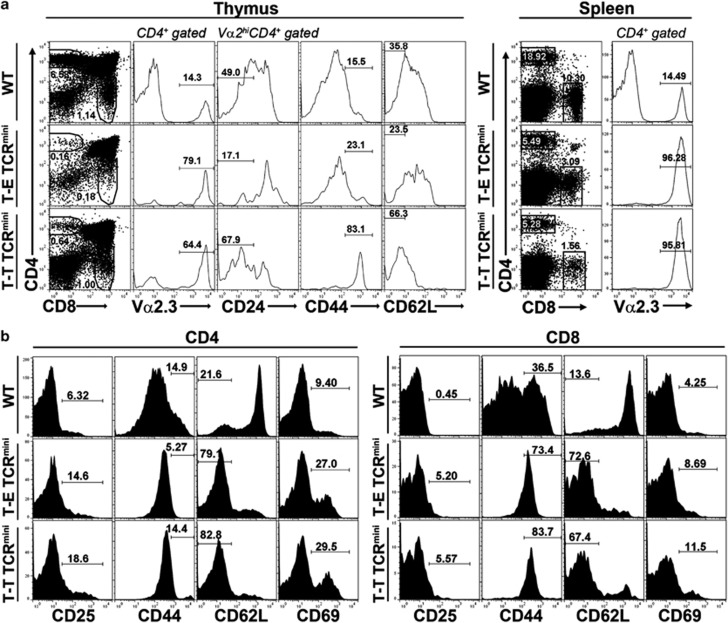
To examine the TCR repertoire created by the T-T interaction in detail, we established a BM chimera in which TCRmini Tg immature thymocytes were positively selected by MHC class II-expressing thymocytes (T-T TCRmini). Thus, a previously developed murine line (Plck-CIITATg)3 that expresses the MHC class II molecule only on its thymocytes was backcrossed with a TCRCα−/− mouse19 (Plck-CIITATg/Cα−/−, designated as the Tg/Cα−/− mouse). Subsequently, a BM chimera was produced by transferring mixed BM cells from the TCRmini Tg mice and Tg/Cα−/− mice (1:1) into irradiated (800 cGy) Rag1−/−/I-Ab−/− hosts (TCRmini Tg+Tg/Cα−/− → Rag1−/−/I-Ab−/−; designated as the T-T TCRmini mouse). In this chimera, T cells that express a limited TCR repertoire (TCRmini Tg) are destined to be positively selected exclusively by MHC class II-expressing thymocytes. In comparison, in the control chimera, TCRmini Tg thymocytes are selected exclusively by cortical thymic epithelial cells (cTECs) (TCRmini Tg+Cα−/− → Rag1−/−; designated as T-E TCRmini). Six weeks after engraftment, the TCRmini Tg BM cells successfully reconstituted the CD4+ T-cell populations in the thymuses and spleens of the T-T TCRmini and T-E TCRmini mice (Figures 2a and b). The CD4 SP thymocytes and mature peripheral CD4+ T cells from the T-T TCRmini and T-E TCRmini mice showed substantial expression levels of Vα2.3 (Figure 2a) and Vβ5.2 (Figure 3a). Both the CD4 SP thymocytes and the peripheral CD4+ T cells in the T-T TCRmini mice had the memory-like phenotype of CD44hi and CD62Llow (Figure 2a), whereas the levels of CD25 and CD69 expressed by mature CD4+ T cells remained low (data not shown). In T-E TCRmini mice, the phenotype of CD4 SP thymocytes was comparable with the WT phenotype; however, the peripheral CD4+ T cells were CD44hi and CD62Llow. In contrast to the thymic compartment, the memory-like phenotype of the peripheral CD4+ T cells in T-E TCRmini mice might be due to the homeostatic expansion of T cells with high-affinity TCRs. We presumed that both types of mice had increased chances of relatively higher-affinity TCR–MHC interactions because of their extremely restricted TCR diversity.
Figure 2.
Normal thymic development and maturation in T-T TCRmini and T-E TCRmini mice. (a) Thymic developmental profiles of mixed BM chimeras (TCRmini Tg+Tg/Cα−/− → Rag−/−I-Ab−/− for T-T TCRmini; TCRmini Tg+Cα−/− → Rag−/− for T-E TCRmini). These chimeras were killed 6 weeks after BM cell engraftment. Usage of TCRVα2.3 chain was checked in CD4 SP thymocytes of WT, T-E TCRmini and T-T TCRmini mice, and surface expression of CD24, CD44, and CD62L on Vα2.3hi-gated CD4 SP thymocytes was shown as histograms. The data are representative of two independent experiments (numbers of total thymocytes in WT mice: 5.83 × 107 and 6.1 × 107, T-T TCRmini mice: 1.28 × 107 and 6.73 × 107, T-E TCRmini mice: 1.54 × 108 and 1.12 × 108; numbers of CD4 SP thymocytes in T-T TCRmini mice: 8.16 × 104 and 38.3 × 104, T-E TCRmini mice: 24.6 × 104 and 19.1 × 104). (b) Representative developmental profiles of splenic CD4+ and CD8+ T cells from WT, T-E TCRmini, and T-T TCRmini mice. The data are representative of two independent experiments (numbers of total splenocytes in WT mice: 8.0 × 107 and 4.73 × 107, T-T TCRmini mice: 3.6 × 107 and 4.93 × 107, T-E TCRmini mice: 5.75 × 107 and 5.98 × 107; numbers of CD4 T cells in the spleen in T-T TCRmini mice: 19.8 × 104 and 28.2 × 104, T-E TCRmini mice: 30.3 × 104 and 28.9 × 104).
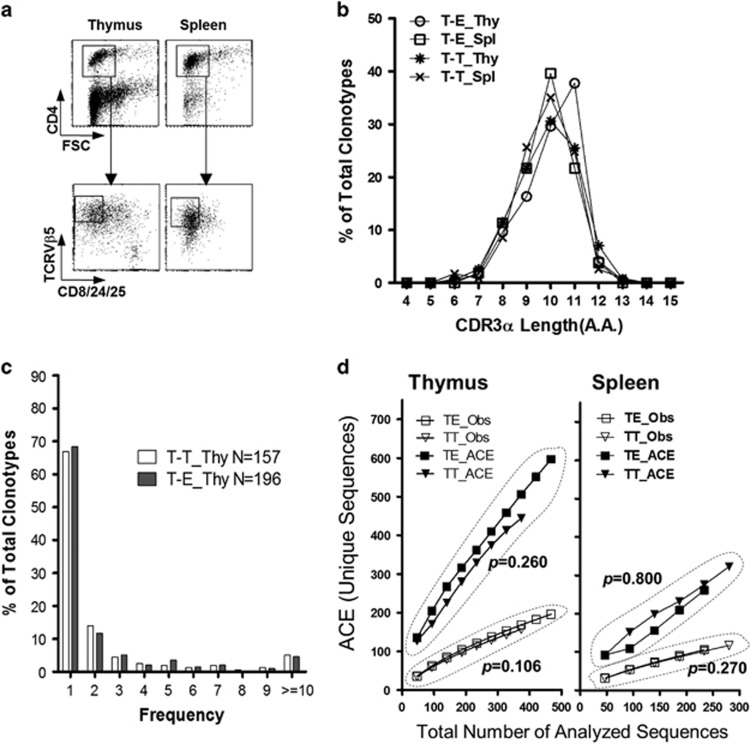
Figure 3.
Extensive TCR diversity determined by thymocyte–thymocyte interactions. (a) Thymocytes and splenocytes were extracted from the T-T TCRmini and T-E TCRmini mice, and CD8+ cells were depleted by magnetic cell sorting. The remaining cells were stained with anti-CD4-APC, anti-CD8/24/25-FITC and anti-TCRVβ5.1&5.2-PE Abs, and the gated populations were subjected to single-cell sorting into 96-well PCR plates using a FACSAria. (b) CDR3α length distributions for TCR sequences obtained from pooled CD4 SP thymocytes or splenic CD4+ T cells from the T-T TCRmini and T-E TCRmini mice (n=2 each). The y axis indicates the proportion that the number of clonotypes found with each CDR3α length represents among all clonotypes. (c) A power-law distribution for the TCR CDR3α frequencies in the CD4 SP thymocytes in both mouse systems is shown, based on the number of occurrences in each population (n=157 and n=199 for T-E TCRmini and T-T TCRmini, respectively). (d) The total number of unique protein sequences obtained from CD4 SP thymocytes and splenic CD4+ T cells from T-T TCRmini and T-E TCRmini mice was estimated on the basis of the number of unique sequences observed (Obs) and the ACE (upper row). The ACE estimates the percentage of individuals with rare sequences (that is, those sequences found 10 times or less) based on abundance data. The ACE was calculated following a published method.44 The empirical P-values were estimated from a one-tailed permutation test.24 Spl, spleen; T-E, T-E TCRmini; Thy, thymus; T-T, T-T TCRmini.
T-T interactions shape a diverse TCR repertoire in the T-T TCRmini mouse model
For comparison of the TCRα repertoires of T-T TCRmini T cells and T-E TCRmini T cells, CD4+CD8−CD25−CD24lowVβ5hi thymocytes and splenocytes were strictly sorted out (Figure 3a), and the amino acid (AA) sequences of their CDR3α regions were determined (Supplementary Table 2a and 2b). A CDR3α length of 10-11 AAs predominated, with a recurrent Gaussian-like distribution in the T-T TCRmini CD4+ T cells of the thymus and spleen as well as in the T-E TCRmini CD4+ T cells (Figure 3b). We sequenced a total of 376 clones from the T-T TCRmini CD4 SP thymocytes and 467 clones from T-E TCRmini CD4 SP thymocytes and found that the number of distinct TCR clonotypes was similar between the two groups, that is, 157 and 199 clonotypes, respectively (respective totals of 261 and 218 clones and 117 and 106 clonotypes from the T-T TCRmini and T-E TCRmini splenic CD4+ T cells). In both groups, we confirmed that the TCR CDR3α regions with high occurrence were encoded by several different nucleotide sequences, which excluded the possibility of selective amplification of specific clonotypes or PCR contamination (Supplementary Table 3).16
In the CD4 SP thymocytes of both the T-T TCRmini and the T-E TCRmini mice, a few clonotypes were found at high frequency (>10 occurrences), whereas most of the clonotypes were found only once in each system (68.4% and 66.9% for T-T TCRmini and T-E TCRmini, respectively) (Figure 3c,Supplementary Table 4). This power-law distribution was also observed for spleen cells (data not shown), which indicates that the CD4+ T cells of the T-T TCRmini and T-E TCRmini mice have diverse TCR repertoires with a few dominant sequences. To compare the overall diversity and estimated total number of unique TCR sequences between the two populations, we used the ACE, which represents a statistical measure of diversity based on the estimated number of distinct CDR3α sequences.14, 23 As shown in Figure 3d, the estimator curves indicated that the possibility of the T-T TCRmini T cell clonotypes being found in the mother population was similar to that of the T-E TCRmini CD4+ T cell clonotypes in both the thymus and the periphery (P>0.05) (Supplementary Figure 2a). These statistical approaches support the notion that T-T TCRmini T cells are diverse, with comparable diversity to T-E TCRmini T cells.
Clonal distribution of the TCR repertoires shaped by T-T or T-E interactions
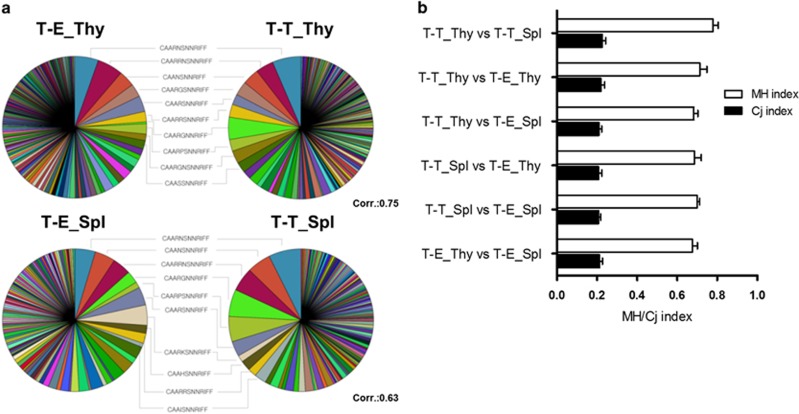
To compare the CDR3α repertoires of the T-T TCRmini and T-E TCRmini mice, we plotted their clonal distributions in pie charts that show the total CDR3α sequences of both groups (Figure 4a). Interestingly, the clonotypes that were highly ranked in terms of abundance among the total clones substantially overlapped across the two repertoires. As shown in Table 1, 8/10 of the most abundant thymic CDR3α sequences were shared between the T-T TCRmini and the T-E TCRmini CD4+ T cells, which indicates that these two systems prefer the same TCR clonotypes. Overlapping clones were detected to a similar degree in the spleen, and the overall clonal distribution of thymic CD4+ T cells in each group was not altered significantly in the periphery (Figure 4a). This finding was confirmed at the population level using the MH and Cj similarity indices15, 16, 25 (Figure 4b). In the comparison of the T-T TCRmini and T-E TCRmini CD4+ T cells, the MH index after normalization ranged from 0.683 to 0.713, and the Cj index ranged from 0.208 to 0.219.
Figure 4.
Clonal similarities between the TCR repertoires derived from T-T and T-E interactions. (a) Pie charts for the CDR3α sequences obtained from the single-cell RT-PCR products of CD4 SP thymocytes (upper row) and CD4+ splenocytes (lower row) from the T-T TCRmini and T-E TCRmini mice (N=376 and N=467 for T-T_Thy and T-E_Thy, respectively; N=261 and N=218 for T-T_Spl and T-E_Spl, respectively). The 10 most abundant and shared sequences between the T-T TCRmini and the T-E TCRmini mice are listed between the pie charts. (b) The Cj and MH similarity indices were calculated using normalized samples (10 000 iterative samplings). The value of these indices ranges from 0 (no similarity) to 1 (complete agreement). The Cj index is defined as the number of common CDR3α sequences divided by the total number of CDR3α sequences in two repertoires, and the MH index addresses the frequency of each CDR3 sequence. The results are shown as the mean±s.d. from 10 000 iterations.
Table 1. Ten most abundant sequences in T-T and T-E TCRmini groups.
|
T-T CD4 SP Thymocytes |
T-E CD4 SP Thymocytes |
||||
|---|---|---|---|---|---|
| CDR3α Seq. | T-T TCRmini | T-T TCRminiOp | CDR3α Seq. | T-E TCRmini | T-E TCRmini Op |
| CAARNSNNRIFF | 24 | 24 | CAARRNSNNRIFF | 27 | 13 |
| CAARGNNRIFF | 19 | 5 | CAARNSNNRIFF | 25 | 13 |
| CAARRNSNNRIFF | 15 | 26 | CAANSNNRIFF | 17 | 8 |
| CAANSNNRIFF | 14 | 18 | CAARSNNRIFF | 17 | 2 |
| CAARGSNNRIFF | 12 | 10 | CAARGSNNRIFF | 14 | 6 |
| CAARGNSNNRIFF | 11 | 3 | CAARPSNNRIFF | 11 | 8 |
| CAARRSNNRIFF | 11 | 10 | CAARRSNNRIFF | 11 | 5 |
| CAARPSNNRIFF | 10 | 10 | CAARGNRIFF | 10 | 0 |
| CAARSNNRIFF | 9 | 10 | CAASRNSNNRIFF | 10 | 3 |
| CAASSNNRIFF | 9 | 7 | CAASSNNRIFF | 9 | 3 |
| % of all sequences | 35.54% | 33.06% | % of all sequences | 32.33% | 22.93% |
| No. of total sequences | 376 | 372 | No. of total sequences | 467 | 266 |
Abbreviations: SP, single positive; TCR, T cell receptor; T–T, thymocyte–thymocyte.
The shadowed sequences in gray color represent shared TCR CDR3α sequences.
TCR repertoires derived from T-T or T-E interactions without Vβ fixation
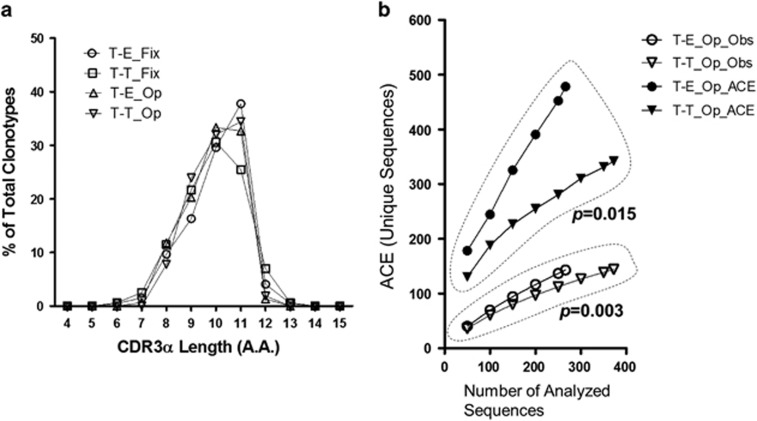
To exclude the possibility of skewing of the overall TCR repertoire distribution by TCRβ chain fixation,26 we analyzed the TCRα chain repertoire of the T-T TCRmini mice in the absence of the OT-II-derived TCRVβ5.2 transgene (T-T TCRmini Op). These TCRs use the same limited TCRα chains as the TCRmini Tg, but they are able to pair with randomly variable TCRβ chains. As was the case for T-T TCRmini and T-E TCRmini mice, the CDR3α length of 10-11 AAs predominated in the CD4 SP thymocytes of the T-T TCRmini Op and T-E TCRmini Op mice (Figure 5a). The number of CDR3α clonotypes observed (Obs) was higher in the T-E TCRmini Op mice than in the T-T TCRmini Op mice. Similarly, the ACE curves indicated a more diverse TCR repertoire in the T-E TCRmini Op mice than in the T-T TCRmini Op mice (P<0.05) (Figure 5b; Supplementary Figure 2b). This was also the case when the ACE was estimated between T-E TCRmini and T-T TCRmini Op mice (Supplementary Figure 3b). To further investigate whether TCRβ chain fixation itself could alter the diversity of CDR3α sequences in both the T-T and the T-E conditions, the significance of the difference in the diversity of the CDR3α sequences was evaluated between TCRβ chain-fixed and the non-fixed TCRmini mice. Although the difference did not reach statistical significance (Thymus_T-E_Fixed vs Open: P=0.196; Thymus_T-T_Fixed vs Open: P=0.106; Supplementary Figure 3a), the ACE curve of T-T TCRmini Op mice showed that the calculated unique sequences (y axis) had a tendency to be less increased than in T-T TCRmini mice as the total number of analyzed sequences (x axis) increased. However, this tendency was not found when T-E TCRmini and T-E TCRmini Op mice were compared, indicating that TCRβ chain fixation could affect the shaping of the TCRα repertoire, at least in the T-T condition. In conclusion, the TCRα repertoire of the T-T TCRmini CD4+ T cells was more restricted than that of the T-E TCRmini CD4+ T cells when the TCRβ chain was open.
Figure 5.
TCR repertoires of CD4 SP thymocytes derived from T-T or T-E interactions without Vβ fixation. (a) CDR3α length distribution of the CD4 SP thymocytes pooled from two T-T TCRmini mice and two T-E TCRmini mice with a wide range of Vβ elements (referred to as T-T_Op and T-E_Op, respectively) compared with T-T TCRmini and T-E TCRmini mice with fixation (referred to as T-T_Fix and T-E_Fix, respectively) (numbers of total thymocytes in T-T_Op: 6.28 × 107 and 7.03 × 107, T-E_Op: 3.88 × 107 and 7.58 × 107; numbers of CD4 SP thymocytes in the thymus in T-T_Op mice: 8.2 × 104 and 38.3 × 104, T-E_Op mice: 24.6 × 104 and 19.1 × 104). (b) The total number of unique sequences obtained from the CD4 SP thymocytes of the T-T TCRmini and T-E TCRmini mice was assessed based on the number of unique sequences observed (Obs) and the ACE. The empirical P-values were estimated from a one-tailed permutation test.
Loss of parental TCRα chain pairing in the CD4 SP thymocytes in OT-II-based T-T TCRmini mice
It has been reported that in a mouse transgenic for the TCRβ chain, selected TCRα chains tend to be biased toward the parental sequences that originally paired with the given TCRβ chain,27 which strongly suggests a role for self-ligands in TCRα selection during the positive selection process. This phenomenon has been observed for conventional CD4+ T cells as well as for the TCRα repertoire of NKT cells13, 27, 28, 29 (Table 2). The tendency to seek out the parental TCRα chains was also found in the CD4 SP thymocytes of the T-E TCRmini mice, as parental TCRα sequences appeared in 10/467 clones from the T-E TCRmini mice. This finding is in contrast to the TCRα chain repertoire of the T-T TCRmini mice, which included only 1/376 clones containing parental sequences. The absence of a bias toward the parental sequences in the T-T TCRmini mice may have particular importance in the sense that the peptides recognized by T-T TCRmini CD4+ T cells during the positive selection process may be different from those recognized by T-E TCRmini T cells.27
Table 2. Frequency of parental TCRα chain in Vβ transgenic mouse models.
| Vβ transgenic mouse models: parental TCRα sequence | Reference | ||
|---|---|---|---|
| OT-II TCR-derived TCRβ5.2 transgene: CAARGNRIFF (Vα2.3-Jα31) | Present data | ||
| CD4 SP Thymocytes | Incidence | % of Pop. | |
| T-T TCRmini | 1/376 | 0.27% | |
| T-E TCRmini | 10/467 | 2.14% | |
| TCRmini Tg | 4/137 | 2.92% | |
| OT-I TCR-derived TCRβ5.2 transgene: CAASDNYQL (Vα2.3-Jα26) | 13 | ||
| CD8 SP Thymocytes | Incidence | % of Pop. | |
| Limited mice | 6/100 | 6.00% | |
| MBP-specific TCR-derived TCRβ8.2 transgene: CAASANSG (Vα2.3-Jα11) | 28 | ||
| MBP-TCRβ Transgenic | Incidence | % of Pop. | |
| CD4 SP Thymocytes | 19/29 | 65.52% | |
| CD8 SP Thymocytes | 0/21 | 0.00% | |
| DN32H6 (NK1.1+ T cells)-derived TCRβ8.2 transgene: VVGDRGSA (Vα14-Jα281) | 29 | ||
| CD4+Vβ8+ thymocytes | Incidence | % of Pop. | |
| DN32H6β transgenic #1 | 15/17 | 88.24% | |
| DN32H6β transgenic #2 | 14/18 | 77.78% | |
| Transgene negative littermate | 15/23 | 65.22% | |
Abbreviation: MBP, myelin basic protein; TCR, T cell receptor.
Discussion
The present analyses of the CDR3α region demonstrate that CD4+ T cells selected by T-T interaction possess a diverse TCR repertoire, challenging the notion that only cTECs are specialized in presenting positively selecting ligands.30, 31, 32 The wide variety of TCRα chains on T-T CD4+ T cells suggests that this population plays critical roles in immune defense compared with previously established innate T cells, which are strictly limited in terms of their TCRα sequences. Moreover, the bias toward pairing of parental TCRα sequences, which is an intrinsic property of monoclonal TCRs (Table 2), was barely detected in the T-T TCRmini mice, in contrast to the situation in the T-E TCRmini mice. This finding suggests that something is set in motion during the positive selection process.
To elucidate the details of the TCR repertoire, murine models with artificial gene constructs have been developed previously.9, 13, 15 The OT-II-based TCRmini Tg mouse in the present study developed a CD4:CD8 ratio (≈2:1) and activation/memory marker profiles that were comparable with those of the WT mouse. Our model also showed distinctive TCR clones in both the CD4+ and the CD8+ lineages (Supplementary Figure 1b), as was the case for previous transgenic mice.13, 15 On the basis of this system, we generated CD4+ T cells with limited TCR diversity that were restricted by the MHC class II molecule using a mixed BM chimera (TCRmini Tg+Tg/Cα−/− → Rag1−/−/I-Ab−/−, T-T TCRmini).
Given that T-T CD4+ T cells have an innate phenotype, it was intriguing to discover high clonal similarity between T-T CD4+ T cells and conventional CD4+ T cells in TCRmini Tg mice. The frequency of clonal overlap between T-T CD4+ T cells and conventional CD4+ T cells in TCRmini Tg mice was higher than previous estimates between two functionally distinct T cell subsets (regulatory T cells and conventional T cells).14, 15, 16 This result suggests that antigen-presenting thymocytes and cTECs may share abundant and immunodominant epitopes, contributing to determination of the dominant TCR sequences. The alternative possibility is that ‘germline-encoded' AA residues in the TCRβ CDR2 region participate in the generation of the dominant TCRs during positive selection.33, 34 When specific residues of the TCRβ chain (CDR2) were mutated, the TCR failed to be positively selected, suggesting that the overall shaping of the TCR repertoire is determined by this ‘built-in' specificity.33, 35 Given that these genetically determined AA residues of the TCR exist in both the T-T TCRmini and the T-E TCRmini mice, they may account for the sharing of dominant TCR sequences between the two mouse models.
However, it is easily assumed that thymocytes and cTECs have a different antigenic hierarchy, that is, dissimilar repertoires of positively selecting peptide ligands. Antigens that would be uniquely loaded onto MHC class II molecules in thymocytes are unlikely to be presented by cTECs. Representative peptides specifically related to immature thymocytes are their own TCR fragments (idiotopes) and activation molecules such as CD25 (ergotopes).36, 37 The CD4+ T cells selected by ergotopes on MHC II+ thymocytes would generate different TCR clonotypes from those selected by cTECs. Similarly, because MHC II+ immature thymocytes that share identical TCR sequences would be relatively few in number, CD4+ T cells selected by idiotopes may express very unique and rare TCR clonotypes.
The putative difference in the positively selecting peptides is further supported by the discovery of peptide-presentation apparatuses that are used exclusively by cTECs. cTECs express distinct proteases, including cathepsin L 30, 31, 32 and lysosomal thymus-specific serine protease,38 highly and specifically, which indicates that cTECs present a different antigenic repertoire than thymocytes do. Additionally, the near-complete loss of the tendency to revert to the parental OT-II TCRα sequences in T-T TCRmini mice reflects the presence of different selecting peptides between the two mouse models (Table 2). In TCRβ chain-transgenic mice, it is known that the biased TCRα chain repertoire of the ‘parental' TCRα chains is frequently expressed on mature T cells.27 This phenomenon has been demonstrated not to be due to the simple clonal expansion of a few T cell clones; the role of intrathymic self-peptides was found to be critical for expressing the biased TCRα chains.28, 39 Although too few clones were sequenced to determine the overall TCR repertoire in detail in the current study, a portion of clonotypes that were mutually exclusive to the T-T TCRmini or T-E TCRmini mice (59% and 67%, respectively, of the thymic clones) can be explained in this regard.
Considering the ACE curves of T-T TCRmini Op (without TCRβ chain fixation) and T-E TCRmini Op, when the two curves were not in parallel, the actual TCR diversity of T-T CD4+ cells in vivo might have been restricted compared with that of conventional CD4+ cells. Furthermore, as reflected by the reduced number of mature CD4+ T cells in CIITATgpIV−/− mice,4 in which only T-T interaction is possible, without thymocyte–cTEC interaction, the restricted TCR diversity might have been due to an increased chance of negative selection for T-T CD4+ cells. As both dendritic cells and thymocytes are derived from hematopoietic progenitor cells, it is likely that the negatively selecting ligands of dendritic cells overlap more frequently with the ligands of MHC II+ thymocytes than with the ligands of cTECs. Therefore, when CD4+ T cells that survive positive selection by thymocytes are subsequently recognized by dendritic cells in the thymic medulla, a more substantial proportion of these T cells would be eliminated40 than in WT mice. This phenomenon probably would not be observed in T-T CD4+ cells from TCRmini Tg mice because of their very limited TCR diversity. Nevertheless, reduced CD4+ T cell numbers as well as the biased TCR repertoire in CIITATgpIV−/− mice do not seem to cause defective T cell responses against antigenic stimuli. When CIITATgpIV−/− and WT B6 mice were immunized with ovalbumin (Supplementary Figure 4), there was no significant difference in their frequency of ovalbumin-reactive CD4+ T cells. These results suggested that T-T CD4+ T cells might have a sufficiently diverse TCR repertoire to respond to the diverse epitopes of ovalbumin compared with T-E CD4+ T cells, even though OT-II parental clones were poorly selected by T-T interactions.
The restricted but diverse TCR repertoire and the significant overlap of TCR sequences with conventional CD4+ T cells led us to assume that T-T CD4+ T cells are able to participate in the immune response against highly variable pathogens, such as viruses. This assumption is supported by the result that T-T CD4+ T cells, and particularly those expressing promyelocytic leukemia zinc finger (PLZF) protein, induce the development of CD8+ T cells with an innate phenotype. Moreover, T-T CD4+ T cells are likely to recognize peptide antigens, in contrast to other types of innate T cells that respond to non-peptide antigens. On a theoretical basis, we further speculate that MHC class II-dependent T-cell regulation is likely to exist in the periphery, which recapitulates thymic T cell–T cell interaction. If CD4 SP thymocytes are selected by thymocytes and then migrate to the periphery, they may easily recognize peripheral activated T cells expressing MHC class II molecules. When the activated T cells present idiotopes or ergotopes on their MHC molecules, the T cells selected by thymocytes are likely to interact with them and may provide certain signals through the activated T cells' MHC class II molecules for regulatory purposes, resulting in the activated T cells becoming either anergic or apoptotic.41, 42, 43
In summary, CD4+ T cells that are selected by thymocytes are found to use far more diverse TCRs than previously characterized innate T cells.11, 12 The diversity of the TCR repertoire may support the prompt protective role of T-T CD4+ T cells against highly variable pathogens, particularly in the human perinatal period. Moreover, it is likely that immature thymocytes present distinctive positively selecting ligands during T-T interaction, which leads to possible peripheral T cell–T cell interaction for the purpose of immune regulation. This dual role of T-T CD4+ T cells has implications for the evolutionary pathway of the sophisticated and complex immune system that exists in humans.
Acknowledgments
This work was sponsored by Support for Creative-Pioneering Researchers through Seoul National University in 2013 (to S.H.P.) and supported by the National Research Foundation of Korea (NRF) grants funded by the Korea Government, the Ministry of Science, ICT & Future Planning (NRF-2012M3A9A8053249 to S.H.P.; NRF-2013M3A9A7046303 to K.-H.C.; NRF-2012R1A1A2007188 to J.-R.K).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Park SH, Bae YM, Kim TJ, Ha IS, Kim S, Chi JG, et al. HLA-DR expression in human fetal thymocytes. Hum Immunol. 1992;33:294–298. doi: 10.1016/0198-8859(92)90338-n. [DOI] [PubMed] [Google Scholar]
- Choi EY, Park WS, Jung KC, Chung DH, Bae YM, Kim TJ, et al. Thymocytes positively select thymocytes in human system. Hum Immunol. 1997;54:15–20. doi: 10.1016/s0198-8859(97)00012-8. [DOI] [PubMed] [Google Scholar]
- Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, et al. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, et al. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–364. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27:698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia-Neves M, Waltzinger C, Mathis D, Benoist C. The shaping of the T cell repertoire. Immunity. 2001;14:21–32. doi: 10.1016/s1074-7613(01)00086-3. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunology. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Fehling HJ, Lemeur M, Benoist C, Mathis D. A vector driving the expression of foreign cDNAs in the MHC class II-positive cells of transgenic mice. J Immunol Methods. 1993;166:287–291. doi: 10.1016/0022-1759(93)90370-m. [DOI] [PubMed] [Google Scholar]
- Philpott KL, Viney JL, Kay G, Rastan S, Gardiner EM, Chae S, et al. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 1992;256:1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- Min HS, Lee YJ, Jeon YK, Kim EJ, Kang BH, Jung KC, et al. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunology. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE, Magurran AE. Ecological diversity and its measurement. Princeton University Press: Princeton, NJ, USA; 1998. [Google Scholar]
- Gotelli NJ, Colwell RK. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett. 2001;4:379–391. [Google Scholar]
- Chao A, Lee S-M. Estimating the number of classes via sample coverage. J Am Statist Assoc. 1992;87:210–217. [Google Scholar]
- Edgington E, Onghena P. Randomization tests. 4th edCRC Press: Boca Raton, FL, USA; 2007. [Google Scholar]
- Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol. 2007;44:1057–1064. doi: 10.1016/j.molimm.2006.06.026. [DOI] [PubMed] [Google Scholar]
- He X, Viret C, Janeway CA., Jr Self-recognition and the biased mature repertoire in TCR beta transgenic mice: the exception that supports the rule. Trends Immunol. 2002;23:467–469. doi: 10.1016/s1471-4906(02)02306-2. [DOI] [PubMed] [Google Scholar]
- Sant'Angelo DB, Lucas B, Waterbury PG, Cohen B, Brabb T, Goverman J, et al. A molecular map of T cell development. Immunity. 1998;9:179–186. doi: 10.1016/s1074-7613(00)80600-7. [DOI] [PubMed] [Google Scholar]
- Viret C, Lantz O, He X, Bendelac A, Janeway CA., Jr A NK1.1+ thymocyte-derived TCR beta-chain transgene promotes positive selection of thymic NK1.1+ alpha beta T cells. J Immunology. 2000;165:3004–3014. doi: 10.4049/jimmunol.165.6.3004. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunology. 2005;174:1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- Anderson G, Lane PJ, Jenkinson EJ. Generating intrathymic microenvironments to establish T-cell tolerance. Nat Rev Immunol. 2007;7:954–963. doi: 10.1038/nri2187. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourilsky P, Chaouat G, Rabourdin-Combe C, Claverie JM. Working principles in the immune system implied by the "peptidic self" model. Proc Natl Acad Sci U S A. 1987;84:3400–3404. doi: 10.1073/pnas.84.10.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jung KC, Park SH. MHC class II-dependent T-T interactions create a diverse, functional and immunoregulatory reaction circle. Immunol Cell Biol. 2009;87:65–71. doi: 10.1038/icb.2008.85. [DOI] [PubMed] [Google Scholar]
- Gommeaux J, Gregoire C, Nguessan P, Richelme M, Malissen M, Guerder S, et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. 2009;39:956–964. doi: 10.1002/eji.200839175. [DOI] [PubMed] [Google Scholar]
- Sant'Angelo DB, Waterbury PG, Cohen BE, Martin WD, Van Kaer L, Hayday AC, et al. The imprint of intrathymic self-peptides on the mature T cell receptor repertoire. Immunity. 1997;7:517–524. doi: 10.1016/s1074-7613(00)80373-8. [DOI] [PubMed] [Google Scholar]
- Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- Drenou B, Blancheteau V, Burgess DH, Fauchet R, Charron DJ, Mooney NA. A caspase-independent pathway of MHC class II antigen-mediated apoptosis of human B lymphocytes. J Immunology. 1999;163:4115–4124. [PubMed] [Google Scholar]
- Kudo H, Matsuoka T, Mitsuya H, Nishimura Y, Matsushita S. Cross-linking HLA-DR molecules on Th1 cells induces anergy in association with increased level of cyclin-dependent kinase inhibitor p27(Kip1) Immunol Lett. 2002;81:149–155. doi: 10.1016/s0165-2478(01)00341-8. [DOI] [PubMed] [Google Scholar]
- Holling TM, Schooten E, van Den Elsen PJ. Function and regulation of MHC class II molecules in T-lymphocytes: Of mice and men. Hum Immunol. 2004;65:282–290. doi: 10.1016/j.humimm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- RK C.EstimateS: Statistical estimate of species richness and shared species from samples. Version 8.2. User's Guide and application. . http://viceroy.eeb.uconn.edu/estimates1997
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.