Abstract
Peripheral nerve injury is a common clinical problem. Nerve growth factor (NGF) promotes peripheral nerve regeneration, but its clinical applications are limited by several constraints. In this study, we found that the time-dependent expression profiles of eight let-7 family members in the injured nerve after sciatic nerve injury were roughly similar to each other. Let-7 microRNAs (miRNAs) significantly reduced cell proliferation and migration of primary Schwann cells (SCs) by directly targeting NGF and suppressing its protein translation. Following sciatic nerve injury, the temporal change in let-7 miRNA expression was negatively correlated with that in NGF expression. Inhibition of let-7 miRNAs increased NGF secretion by primary cultured SCs and enhanced axonal outgrowth from a coculture of primary SCs and dorsal root gangalion neurons. In vivo tests indicated that let-7 inhibition promoted SCs migration and axon outgrowth within a regenerative microenvironment. In addition, the inhibitory effect of let-7 miRNAs on SCs apoptosis might serve as an early stress response to nerve injury, but this effect seemed to be not mediated through a NGF-dependent pathway. Collectively, our results provide a new insight into let-7 miRNA regulation of peripheral nerve regeneration and suggest a potential therapy for repair of peripheral nerve injury.
Introduction
Peripheral nerve injury is a common global clinical problem, which significantly affects the patients' quality of life and causes enormous financial burden.1 The peripheral nervous system (PNS) is capable of spontaneous regeneration in response to traumatic injury, but functional recovery is often unsatisfactory especially in the case of severe injury, where clinical intervention is thereby necessary. Nerve growth factor (NGF), the first discovered member of neurotrophin family, contributes to the development and phenotype maintenance of the PNS and ensures the functional integrity of cholinergic neurons in the central nervous system (CNS).2 Accordingly, many experimental studies have examined the beneficial effects of NGF on peripheral nerve regeneration, but clinical applications of NGF are still limited by several constraints, including the deleterious side effects of NGF and the complexity in NGF delivery.3
MicroRNAs (miRNAs) are endogenously encoded, evolutionarily conserved small RNAs (~22 base pairs), and they regulate gene expression predominantly by promoting degradation or inhibiting protein translation of target mRNAs.4,5 The regulatory role of miRNAs in neural development, degeneration, and regeneration is starting to be recognized.6,7,8 The therapeutic potential of miRNAs has also been explored for several diseases.9,10,11 As is well known, Schwann cells (SCs) are the principal glial cells in the PNS, and play a pivotal role in peripheral nerve regeneration by virtue of their interactions with re-growing axons.12 Based on this premise, many recent studies have shown that some miRNAs may induce phenotype modulation of SCs during peripheral nerve regeneration,13,14,15 and our group also reports on the impacts of several miRNAs on SCs behavior at an early stage after peripheral nerve injury.16,17,18,19
The lethal-7 (let-7) gene is a founding member of miRNA family. Let-7 miRNAs, originally identified in Caenorhabditis elegans, are conserved in vertebrates and invertebrates.20 The involvement of let-7 miRNAs in carcinogenesis has attracted considerable attention. Moreover, it has been known that let-7 miRNAs regulate neuronal cell fate, and affect neurodegeneration and neuronal regeneration.21,22,23,24 Until now, however, few studies have identified the expression profile of let-7 miRNAs after peripheral nerve injury, and further discussed their significance for peripheral nerve regeneration.
Deciphering the putative targets of miRNAs is usually a key to elucidating the function of miRNAs in different cell processes, and let-7 miRNAs are certainly no exception. Interestingly, some members of let-7 family are predicted to target NGF25 that is a crucial neurotrophin molecule (as we mentioned above). This study was aimed to investigate the expression changes of let-7 miRNAs and the possible effects of let-7 miRNAs on cellular behaviors of neural cells (SCs and axons) following peripheral nerve injury. Our findings from in vitro and in vivo studies will contribute to determining let-7 miRNAs regulation of peripheral nerve regeneration, and suggest a potential therapeutic target for peripheral nerve injury.
Results
Expression profiling of let-7 miRNAs in injured sciatic nerves
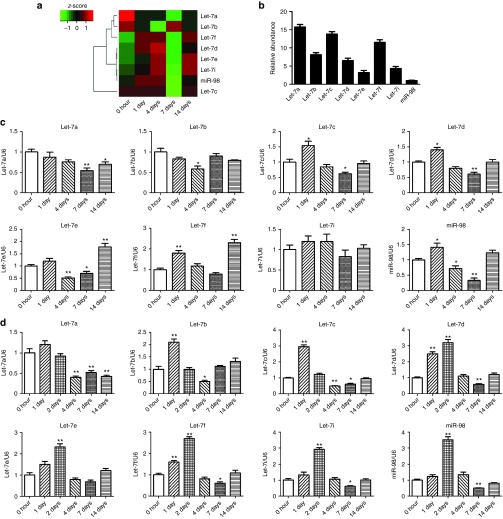
Solexa sequencing was used to investigate the expression profile of miRNAs in the proximal nerve segment after sciatic nerve transection. The time-dependent expression profiles of the eight members of let-7 family, including let-7a, 7b, 7c, 7d, 7e, 7f, 7i, and miR-98, were roughly similar to each other (Figure 1a). Meanwhile, the different expression levels of the eight let-7 members in the intact nerve (referred to as 0 hour, control) were also detected (Figure 1b).
Figure 1.
Time-dependent expression of let-7 miRNAs in injured nerve after sciatic nerve injury. (a) Heatmap and clustering from Solexa sequencing showing the expression changes of eight members of let-7 family in proximal sciatic nerve segment following sciatic nerve transection. (b) The expression level (relative to that of U6) of eight family members of let-7 miRNAs in intact sciatic nerve (0 hour after nerve injury). (c,d) The expression change of let-7 members (c) in proximal sciatic nerve segment following sciatic nerve transection or (d) in injured sciatic nerve following sciatic nerve crush, as detected by qPCR. *P < 0.05, **P < 0.01 versus control (0 hour after nerve injury).
Quantitative real-time RT-PCR (qPCR) confirmed the data from Solexa sequencing. After sciatic nerve transection, the expression of six members of let-7 family (except let-7a and 7b) in the proximal nerve increased at 1 day after nerve injury, and decreased at 4 and 7 days after nerve injury, followed by rebounding at 14 days after nerve injury, as compared to control (0 hour) (Figure 1c). After sciatic nerve crush, the expression of all the eight members of let-7 family increased immediately with a peak value at 1 or 2 days after nerve injury, and then decreased with a valley value at 4 and 7 days after nerve injury, as compared to control (0 hour) (Figure 1d).
The temporal expression profiles of most members of let-7 family had a nearly common feature of first increasing, then decreasing, and again increasing (just like an inverted S-shaped time-dependence). Considering that the time-dependent expression changes of both let-7d and miR-98 were quite typical, we selected these two let-7 miRNAs as representatives in the following tests.
Effects of let-7d/miR-98 on SCs proliferation and migration
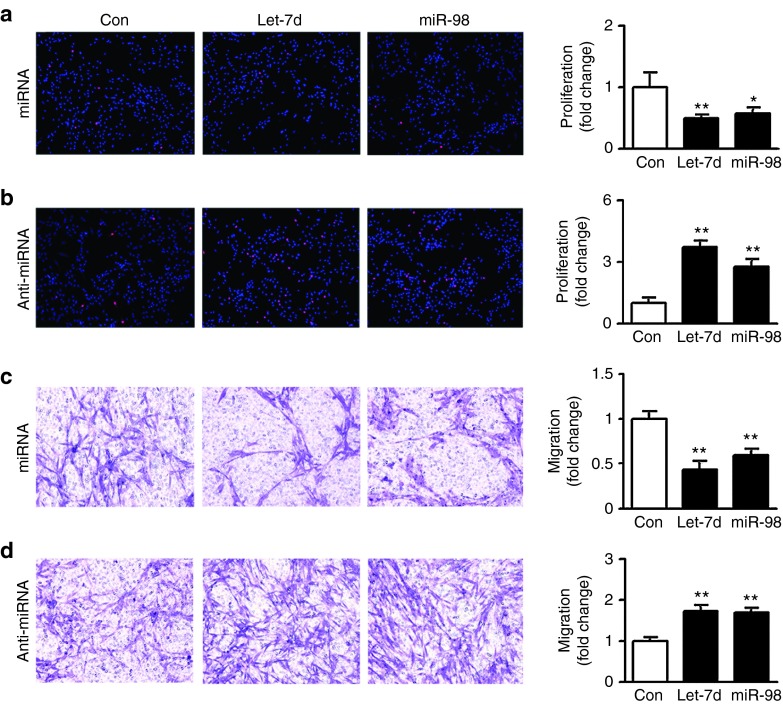
Primary SCs were transfected with let-7d/miR-98 mimics, let-7d/miR-98 inhibitors, and nontargeting negative control, respectively. EdU incorporation indicated that the proliferation rate of SCs transfected with let-7d/miR-98 mimics was significantly decreased to ~50% of control (Figure 2a), while the proliferation rate of SCs transfected with let-7d/miR-98 inhibitors was significantly increased to approximately four- or threefold of control, respectively (Figure 2b).
Figure 2.
Effects of let-7d/miR-98 on SCs proliferation and migration. (a,b) Merges of EdU staining (red) and Hoechst 33342 staining (blue) for primary SCs transfected (a) with let-7d mimic (let-7d+miRNA), miR-98 mimic (miR-98+miRNA) and mimic control (Con+miRNA), respectively or (b) with let-7d inhibitor (let-7d+anti-miRNA), miR-98 inhibitor (miR-98+anti-miRNA), and inhibitor control (Con+miRNA), respectively. Also shown are histograms showing the cell proliferation rate (expressed as the percentage of EdU-positive cells to all SCs, and normalized to control) of primary SCs that had been differently treated. (c,d) Images showing that SCs migrated to the bottom of the transwell chamber after being transfected with (c) let-7d mimic (let-7d+miRNA), miR-98 mimic (miR-98+miRNA) and mimic control (Con+miRNA), respectively or (d) with let-7d inhibitor (let-7d+anti-miRNA), miR-98 inhibitor (miR-98+anti-miRNA), or inhibitor control (Con+miRNA), respectively. Also shown are histograms showing the cell migration ability (normalized to control) of primary SCs that had been differently treated. In all histograms, *P < 0.05 and **P < 0.01 versus control (Con).
Transwell migration assay showed that transfection with let-7d/miR-98 mimics significantly suppressed the migratory ability of SCs as compared to control (Figure 2c), while transfection with let-7d/miR-98 inhibitors significantly promote cell migration of SCs as compared to control (Figure 2d), suggesting that let-7d/miR-98 was able to reduce proliferation and migration of SCs.
Suppression of NGF translation by let-7d/miR-98 targeting its 3′-untranslated region
A dual-luciferase reporter assay was conducted to determine whether NGF was regulated by let-7 miRNAs through direct binding to 3′-untranslated region (3′-UTR) of NGF. The let-7 binding sequences at 3′-UTR of NGF are highly conserved across species. The wild-type or mutant 3′-UTR of NGF was constructed and inserted into the downstream region of the luciferase reporter gene, respectively (Figure 3a). When the wild-type NGF 3′-UTR-containing plasmid was cotransfected with let-7d/miR-98 mimics, the expression of luciferase significantly decreased, while this decrease was abrogated in mutant 3′-UTR-containing plasmid that was cotransfected with let-7d/miR-98 mimics (Figure 3b). The results suggested that let-7 miRNAs directly inhibited NGF expression by binding to a defined target sequence. Moreover, let-7d/miR-98 mimics or inhibitors were respectively transfected into primary SCs, and the mRNA and protein expressions of NGF were respectively determined. The mRNA expression of NGF in primary SCs was not changed after the cells were transfected with let-7d/miR-98 mimics or inhibitors (Figure 3c). In contrast, the protein expression of NGF was significantly suppressed by overexpression of let-7d/miR-98, but was significantly enhanced by silencing of let-7d/miR-98 (Figure 3d). These results implied that let-7 miRNAs led to translational suppression of NGF rather than mRNA degradation of NGF.
Figure 3.
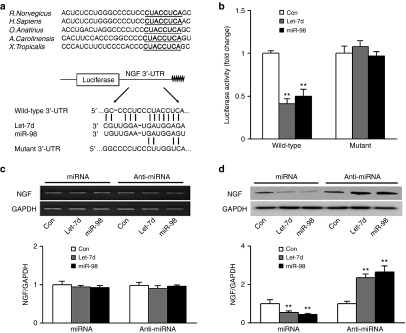
NGF being a target gene of let-7 miRNAs. (a) Sequence alignment of the putative let-7d/miR-98 binding sites across species, and sketch of the construction of wild-type or mutant p-Luc-UTR vectors. The conserved binding site is shown in bold and underlined. (b) The relative luciferase activity was analyzed after the p-Luc-UTR vectors were cotransfected into 293T cells with let-7d/miR-98 mimics (let-7d/miR-98) or mimic control (Con). The (c) mRNA and (d) protein expression of NGF in SCs transfected with let-7d/miR-98 mimics (miRNA) or with let-7d/miR-98 inhibitors (anti-miRNA). Also shown in c and d are representative semiquantitative RT-PCR gels and Western blot bands, respectively. GAPDH served as internal reference. **P < 0.01 versus control (Con).
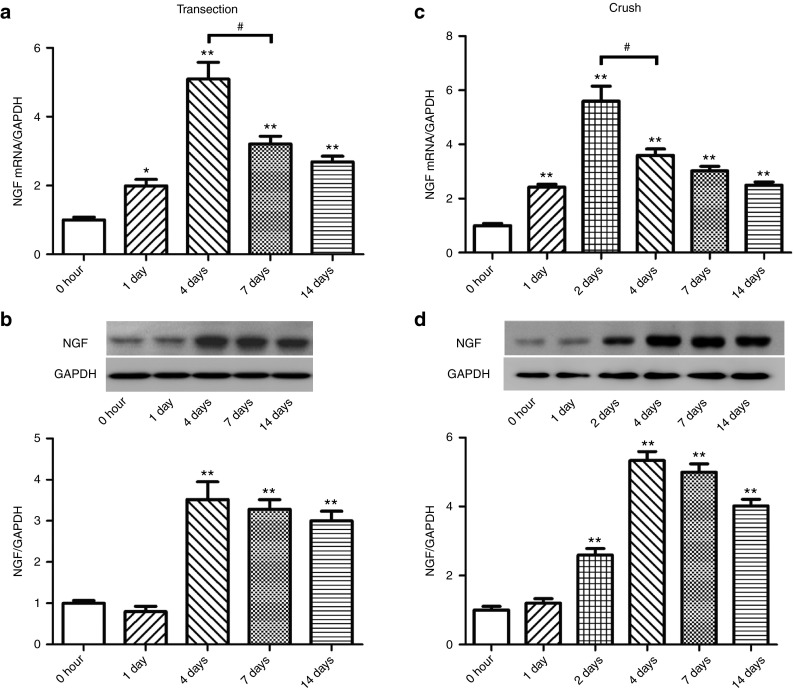
Temporal changes in NGF expression after sciatic nerve injury
After rat sciatic nerve transection, the mRNA expression of NGF in the proximal nerve segment significantly increased at 1, 4, 7, and 14 days after nerve injury as compared to control (0 hour) with a peak value occurring at 4 days after nerve injury (Figure 4a). The protein expression of NGF in the proximal nerve segment slightly decreased at 1 day after nerve injury as compared to control (0 hour) without significant difference from control (0 hour), and then significantly increased at 4, 7, and 14 days after nerve injury as compared to control (0 hour) with a peak value occurring at 4 days after nerve transection (Figure 4b).
Figure 4.
Time-dependent mRNA and protein expression of NGF in injured nerve after sciatic nerve injury. The expression change of NGF at (a,c) mRNA and (b,d) protein levels (a,b) in proximal sciatic nerve segment following nerve transection or (c,d) in injured sciatic nerve following nerve crush, respectively. *P < 0.05 and **P < 0.01 versus control (0 hour after nerve injury), and #P < 0.05. Also shown in b and d are representative Western blot bands. GAPDH served as internal reference.
After sciatic nerve crush, the mRNA expression of NGF in the injured nerve significantly increased at 1, 2, 4, 7, and 14 days after nerve crush as compared to control (0 hour) with a peak value occurring at 2 days after nerve crush (Figure 4c). The protein expression of NGF in the injured nerve remained nearly unchanged at 1 day after nerve crush as compared to control (0 hour), but significantly increased at 2, 4, 7, and 14 days after nerve crush as compared to control (0 hour) with a peak value occurring at 4 days after nerve crush (Figure 4d). It was easily known that following transection or crush injury to rat sciatic nerve, the protein expression profile of NGF was not exactly parallel to the mRNA expression profile of NGF.
Interestingly, the comparison between Figures 1 and 4 suggested that the temporal expression profile of let-7 miRNAs was negatively correlated with that of NGF, providing further evidence that let-7 miRNAs negatively regulated the expression of NGF, the target of let-7 miRNAs.
Attenuation of let-7 inhibition-induced increases in SCs proliferation and migration by NGF knockdown
After stable knockdown of NGF in primary SCs (Figure 5a,b), cell proliferation and migration of SCs were significantly inhibited (Figure 5c,d), indicating the effects of NGF knockdown on SCs were similar to those of let-7d/miR-98.
Figure 5.
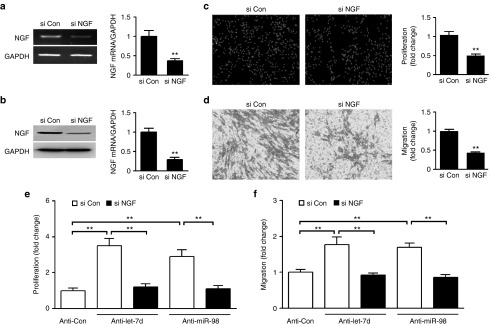
Attenuation of let-7d/miR-98 inhibitors-induced increase in SCs proliferation and migration by NGF knockdown. (a,b) The mRNA or protein expression of NGF in SCs transfected with NGF siRNA (si NGF) was significantly decreased as compared to that in SCs transfected with siRNA control (si Con). In addition to representative RT-PCR gels Western blot bands, histograms were used for the expression comparisons, in which **P < 0.01 versus control siRNA (si Con). (c,d) NGF siRNA (si NGF) significantly inhibited SCs (c) proliferation and (d) migration as compared to control siRNA (si con). **P < 0.01 versus control siRNA (si Con). (e,f) Increase in cell (e) proliferation or (f) migration of SCs transfected with let-7d/miR-98 inhibitors (anti-let-7d/miR-98) was rescued by cotransfection with NGF siRNA (si NGF), in which **P < 0.01.
Because NGF was regulated posttranscriptionally by let-7 miRNAs and NGF knockdown inhibited SCs proliferation and migration, we reasonably assumed that let-7d/miR-98 regulation of proliferation and migration of SCs was likely mediated by NGF. To test this assumption, primary SCs were transfected with let-7d/miR-98 inhibitors in the presence or absence of NGF siRNA. After SCs were transfected with let-7d/miR-98 inhibitors, cell proliferation and migration of SCs significantly increased as compared to control, while the increases in SCs proliferation and migration could be significantly attenuated by cotransfection with both let-7d/miR-98 inhibitors and NGF siRNA (Figure 5e,f). It was clear that NGF knockdown blocked the enhancing effect of let-7d/miR-98 inhibitors on SCs proliferation and migration, thus providing additional evidence that NGF was a functional mediator for let-7 miRNAs in SCs.
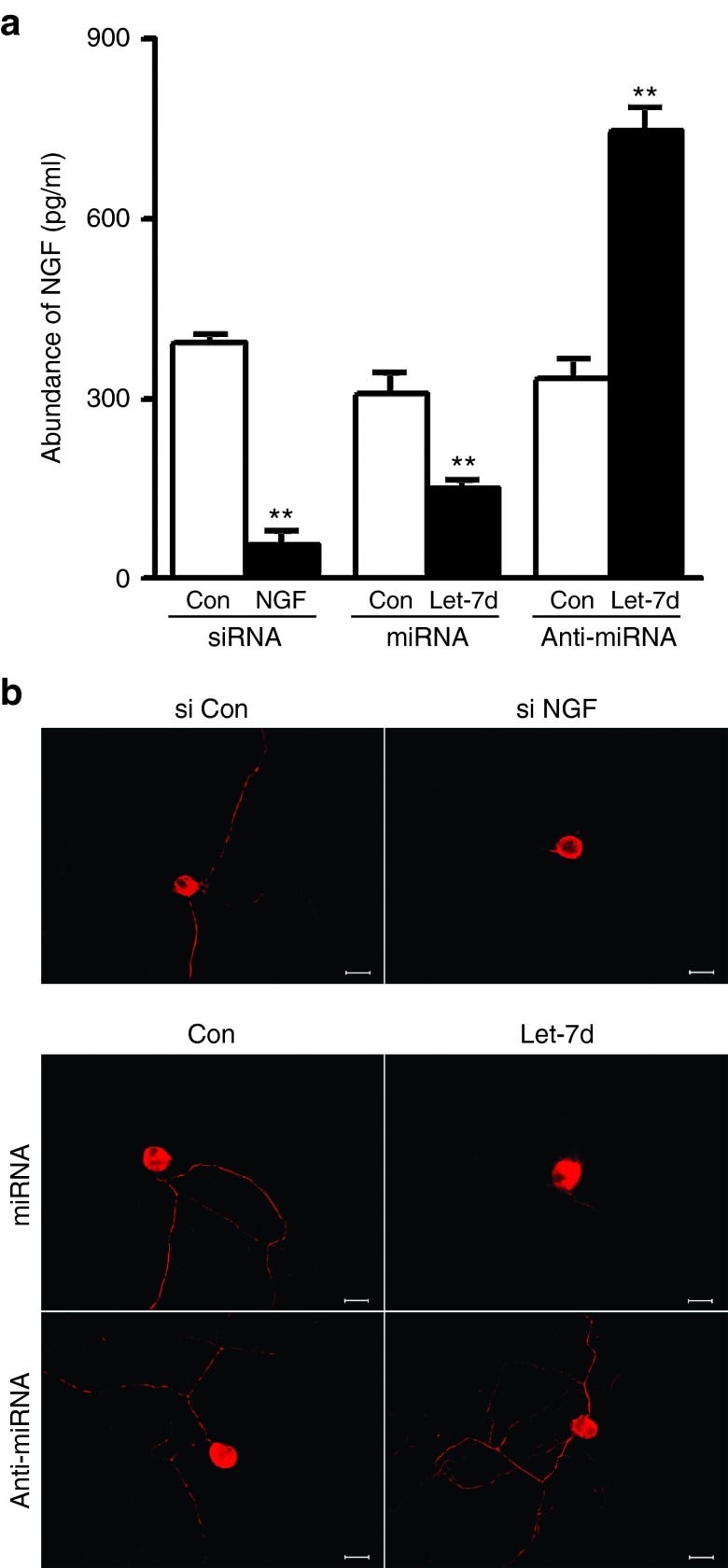
Effects of let-7 miRNAs on NGF secretion from SCs and on axon outgrowth
NGF is mainly secreted by SCs, and its secreted form is an active form.26 Enzyme-linked immunosorbent assay was used to determine the effects of let-7d on NGF secretion from SCs. NGF siRNA significantly reduced NGF secretion from SCs to ~20% of control. Moreover, let-7d mimic also significantly decreased NGF secretion from SCs to about 50% of control, while let-7d inhibitor increased NGF secretion from SCs to ~2.5-fold of control (Figure 6a).
Figure 6.
Effects of Let-7d on NGF secretion from SCs and axon outgrowth. (a) NGF secretion was reduced from SCs transfected with NGF siRNA or with let-7d mimic (let-7d+miRNA), but was increased from SCs transfected with let-7d inhibitor (let-7d+anti-miRNA). **P < 0.01 versus respective controls (Con). (b) Immunostaining with anti-NF200 showing that axon outgrowth was decreased in coculture consisting of SCs, which had been transfected with NGF siRNA or with let-7d mimic (let-7d+miRNA), and DRG neurons, but increased in coculture consisting of SCs, which had been transfected with let-7d inhibitor (let-7d+anti-miRNA), and DRG neurons. Scale bar, 50 μm.
Axon outgrowth was remarkably reduced in coculture consisting of SCs, which had been transfected with let-7d mimic or with NGF siRNA, and dorsal root ganglion (DRG) neurons. On the contrary, axon outgrowth was obviously enhanced in coculture consisting of SCs, which had been transfected with let-7d inhibitor, and DRG neurons (Figure 6b).
In vivo effects of let-7 miRNAs on SCs migration and axon outgrowth
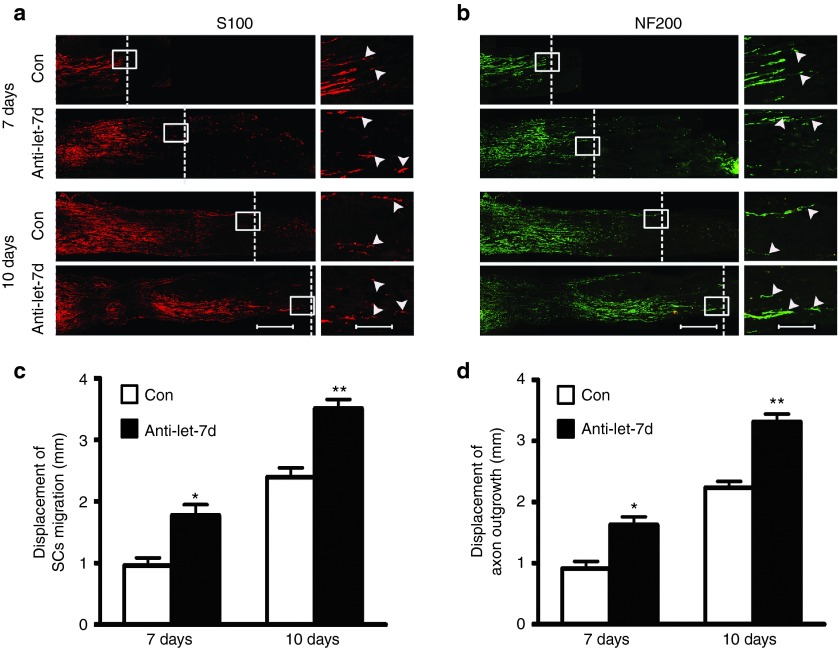
Adult rats with sciatic nerve transection were used as an animal model to determine in vivo effects of let-7d on cell behaviors of neural cells during sciatic nerve regeneration. Following nerve injury, a silicone tube was implanted into the sciatic nerve gap, and let-7d inhibitor (antagomir) was injected into the silicone tube. Immunostaining with anti-S100 or with anti-neurofilament-200 (anti-NF200) showed that let-7d inhibitor significantly increased SCs migration and axonal growth in rats at 7 or 10 days after sciatic nerve injury, respectively (Figure 7), suggesting in vivo promoting effects of let-7d inhibitor on peripheral nerve regeneration. Meanwhile, we also observed that injection with let-7d mimic (agomir) into the silicone tube decreased SCs migration and axon outgrowth in rats at 7 or 10 days after sciatic nerve injury, respectively (data not shown).
Figure 7.
In vivo effects of let-7d on SCs migration and axon outgrowth. (a,b) Immunostaining with anti-S100β or with anti-NF200 showing that let-7d inhibitor encouraged SCs migration and axon outgrowth at 7 and 10 days after nerve injury, respectively. SCs migration and axon outgrowth was assessed by cell displacement from the proximal toward distal nerve stump within a silicone tube that had been implanted into sciatic nerve gap of rats, which were divided into two groups to receive injection of a mixture containing let-7d inhibitor (anti-let-7d) or control (Con), respectively. The white dotted line in micrographs is used to mark the frontier of SCs migration and axon outgrowth. Scale bar: 500 μm. Also shown are the higher magnifications of boxed areas, in which Scale bar: 125 μm. (c,d) Histograms showing quantitative comparisons of cell displacement of SCs and axons at 7 and 10 days after implantation as the above mentioned. *P < 0.05, **P < 0.01 versus respective controls (Con).
Effects of let-7 miRNAs on cell apoptosis of SCs
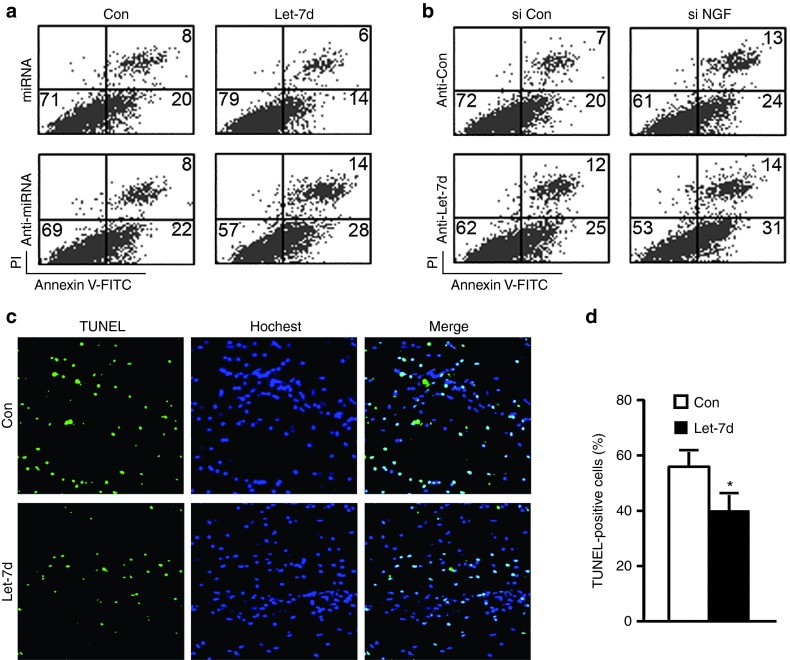
Cell proliferation and apoptosis are two fundamental processes in multicellular organisms, and the balance between these two processes is important for cell development and tissue homeostasis. After sciatic nerve injury, exposure to an oxidative stress leads to early apoptosis of SCs at the lesion site.27 In this study, H2O2 stimulation for 16 hours was adopted to mimic the oxidative stress following peripheral nerve injury. Upon exposure to H2O2 stimulation, primary SCs demonstrated about 20% apoptosis (data not shown), which was used as control in subsequent experiments. Flow cytometry indicated that as compared to control, let-7d mimic significantly decreased cell apoptosis of SCs, while let-7d inhibitor observably promoted cell apoptosis of SCs (Figure 8a). To examine the possible involvement of NGF in the inhibitory effect of let-7 miRNAs on SCs apoptosis, we found that NGF knockdown promoted cell apoptosis of primary SCs transfected with control, but failed to attenuate let-7d inhibitor-induced increase in SCs apoptosis (Figure 8b). The results implied that the inhibition of SCs apoptosis by let-7d might not be through a NGF-dependent pathway. TUNEL analysis provided in vivo evidence that let-7d significantly reduced cell apoptosis in the crushed sciatic nerve of rats as compared to control (Figure 8c,d).
Figure 8.
In vitro and in vivo effects of let-7d on cell apoptosis of SCs. (a) Flow cytometry showing the apoptosis rate of SCs after transfection with let-7d mimic (let-7d+miRNA), mimic control (Con+miRNA), let-7d inhibitor (let-7d+anti-miRNA) or inhibitor control (Con+miRNA). (b) Flow cytometry showing the apoptosis rate of SCs after transfection with let-7d inhibitor (anti-let-7d) with or without NGF siRNA (si NGF). (c) TUNEL analysis was used to test cell apoptosis in crushed sciatic nerve, into which a mixture containing let-7d agomir (let-7d) or control agomir (Con) had been injected respectively. Fluorescence micrographs showing TUNEL staining and Hoechst 33258 nuclear counterstaining of cells in crushed sciatic nerve that had been harvested at 1 day after nerve injury. The colocalization of TUNEL and Hoechst staining is also shown. (d) Histogram showing the number percentage of TUNEL-positive cells in total cell population in crushed sciatic nerve that had been harvested at 1 day after nerve injury. *P < 0.05 versus crushed sciatic nerve injected with control agomir (Con).
Discussion
Recently, many therapeutic strategies have been developed to improve the functional outcomes of peripheral nerve regeneration. This study focused on the possible regulatory role of let-7 miRNAs during peripheral nerve injury and regeneration. The expressions of eight members of let-7 miRNA family in the injured sciatic nerve changed with time following sciatic nerve transection or crush, and their temporal expression profiles were similar to each other. This result suggested that let-7 miRNAs might be involved in peripheral nerve regeneration.
SCs provide a permissive environment for peripheral nerve regeneration.28 Following sciatic nerve injury, SCs dedifferentiate and proliferate, migrate to form bands of Bünger, and promote axon regeneration.29,30 Since primary culture of SCs represents a well-defined model for studying cell behaviors of SCs in the injured peripheral nerve,31 this study firstly investigated the effects of let-7 miRNAs on phenotypic modulation of primary SCs. We observed that let-7d/miR-98 inhibited cell proliferation and migration of primary SCs. On the other hand, in vivo investigation further confirmed that at 7 or 10 days after sciatic nerve injury in adult rats, let-7 inhibitor could promote SCs migration and axon outgrowth within a nerve regenerative microenvironment.
To determine the action mechanisms of let-7d/miR-98, target prediction algorithm suggested that NGF might be a potential target gene of let-7 miRNAs, and our data from a dual-luciferase reporter assay confirmed that NGF was a real target gene of let-7d/miR-98 through directly binding to NGF 3′-UTR. Furthermore, we observed that let-7d/miR-98 repressed NGF expression possibly through inhibiting NGF translation rather than causing NGF mRNA degradation. To investigate in vivo regulation of NGF by let-7 miRNAs, we noted that NGF mRNA expression increased in the injured nerve after sciatic nerve injury. This finding was in agreement with previously reported results.32 After sciatic nerve injury, however, the temporal change of NGF protein expression in the injured nerve was not completely consistent with that of NGF mRNA expression. For example, at 1 day after nerve transection, NGF mRNA expression increased to about twofold of control (0 hour), but NGF protein expression slightly decreased as compared to control (0 hour). At the same time point, the expression of let-7d/miR-98 significantly increased as compared to control (0 hour). As another example, from 4 to 7 days after nerve transaction, NGF mRNA expression significantly decreased as compared to control (0 hour), but NGF protein expression remained nearly unchanged. Over the same time period, the expression of let-7d/miR-98 significantly decreased as compared to control (0 hour). Collectively, the above results suggested that NGF was likely to be negatively regulated by let-7 miRNAs at posttranscriptional level. Likewise, the negative regulation of NGF protein expression by let-7 miRNAs was also observed following sciatic nerve crush.
NGF knockdown in primary SCs was carried out to examine the effects of NGF downregulation on phenotype modulation of SCs. We found that NGF knockdown attenuated an increase in SCs proliferation and migration induced by let-7d/miR-98 inhibitors, suggesting that NGF knockdown could recapitulate the suppressing effects of let-7 miRNAs on SCs phenotypic modulation. This interesting finding provided further evidence that NGF was a functional mediator of let-7 miRNAs. Our previous study reported that miR-221/222 promoted cell proliferation and migration of SCs,17 and other researchers' result indicated that NGF increased the expression of miR-221/222.33 It seems that let-7 miRNAs target NGF expression, which, in turn, regulate the expression of miR-221/222, and thereby affect cell proliferation and migration of SCs. The cascade of let-7 miRNAs to NGF to miR-221/222 may represent a bypass of let-7 regulation of SCs phenotypic modulation.
In fact, the role of let-7 miRNAs targeting NGF seems to be in agreement with the neurotrophic action of NGF for peripheral nerve regeneration. NGF occupies a unique position in neurotrophin family and is often viewed as a prototype of all other neurotrophins,34 and it plays a critical role in the development and survival of neurons in the PNS and CNS.35 In addition, NGF, as a signaling molecule, is involved in immune and inflammatory responses.34 So far, recombinant human NGF (rhNGF) has been produced and tested for therapeutic goals. The clinical use of rhNGF for peripheral nerve regeneration, however, is limited by difficulties in overcoming side effects of rhNGF on human subjects, including myalgias and injection site hyperalgesia, short half-life, inability to cross the blood–nerve barrier, and possible tumorigenicity at high dosage.2,3 In consequence, endogenous regulation of NGF secretion from neural cells could perhaps provide a therapeutic alternative to administration of exogenous NGF. Based on this consideration, we investigated the effects of let-7d on NGF production by primary SCs, and found that downregulation of let-7d stimulated SCs to produce an increased amount of NGF, and vise visa. We also noted that downregulation of let-7d enhanced axon outgrowth at least partly through increasing NGF supply. Accordingly, the promoting role of let-7d inhibitors in peripheral nerve regeneration was similar to that of the application of NGF.
Previous evidence has indicated that let-7 miRNAs regulate cell apoptosis in several cell lines,36,37 and that NGF also impacts cell apoptosis in several cell lines, including SCs.38 To interpret the increased expression of most let-7 family members at 1 or 2 days after sciatic nerve injury as compared to control (0 hour), we examined the in vitro and in vivo effects of let-7d and NGF on SCs apoptosis. The results showed that both let-7d and NGF inhibited cell apoptosis of primary SCs, but NGF knockdown did not rescue the proapoptotic effect of let-7d inhibitor on primary SCs, suggesting that let-7d regulated SCs apoptosis possibly not through a NGF-dependent pathway. It has been proposed that some pathways, involving Fas and caspase 3,36,37 inflammatory cytokines (IL-6 and IL-10),39,40 or Bcl-xL, HMGA2, and Myc,41,42,43 may be responsible for let-7 miRNAs regulation of cell apoptosis. Likewise, in vivo examination also showed that let-7d could reduce cell apoptosis in injured sciatic nerve.
The inhibitory effect of let-7 miRNAs on SCs apoptosis may be considered as an early stress response to peripheral nerve injury. Soon after nerve injury, axons in the proximal stump degenerate for some distance back from the injury site. Meanwhile, inflammatory response is triggered and cell apoptosis is induced.29,30 Although the proper inflammatory response may facilitate nerve regeneration, it is still necessary to inhibit excessive inflammation and prevent apoptosis of neural cells. Our results indicated that the expression of most let-7 miRNAs was increased at 1 or 2 days following sciatic nerve injury, which might be related to anti-apoptosis actions of let-7 miRNAs. Also, the results might probably explain why cell apoptosis of SCs was not obviously observed at an early stage of peripheral nerve degeneration.
In summary, the time-dependent expression profiles of eight let-7 family members in the injured nerve were observed following peripheral nerve transection or crush injury. SCs proliferation and migration were specifically regulated by let-7d/mir-98 through targeting NGF in vitro and in vivo. The downregulation of let-7d stimulated SCs to increase NGF production, which further encouraged axon regrowth. In addition, let-7 miRNAs could modify the regenerative microenvironment by decreasing SCs apoptosis at an early stage following sciatic nerve injury. Accordingly, this study not only provides new insight into let-7 miRNAs regulation of peripheral nerve regeneration by robust phenotypic modulation of neural cells, but also opens a novel therapeutic window for peripheral nerve injury by mediating NGF production.
Materials and Methods
Animal surgery. Adult, male Sprague-Dawley (SD) rats (180–220 g) received surgery to induce sciatic nerve transection as described previously.44 Proximal nerve segments were harvested at 0, 1, 4, 7, and 14 days after nerve transaction. RNA samples were extracted from harvested nerves and then subjected to Solexa sequencing as described previously.44
Similarly, another cohort of SD rats was anaesthetized before the sciatic nerve was exposed through an incision on the mid-thigh of left hind limb, and a 3-mm long nerve was crushed two times (15 seconds/each time, 3 seconds interval) with a hemostatic forceps. A 3 mm long crushed nerve, together with both nerve ends (1 mm long), was harvested at 0, 1, 2, 4, 7, and 14 days after nerve crush.
Primary culture of SCs and cell transfection. SCs were isolated from the sciatic nerve of 1-day-old SD rats, and purified by removing fibroblasts with anti-Thy1.1 antibody and rabbit complement (Sigma, St Louis, MO) as previously described.45 The cell preparation consisted of 98% SCs, as assessed by immunocytochemistry with anti-S100 (DAKO, Carpinteria, CA). The resultant SCs were cultured in DMEM containing 10% fetal bovine serum (FBS) at 37 °C in humidified 5% CO2 in air. The cells were passaged no more than two times prior to use.
Primary cultured SCs were transfected with miRNA mimics, miRNA inhibitors or NGF siRNAs (Ribobio, Guangzhou, China), respectively, using Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA) according to manufacturer's instructions.
qPCR and semiquantitative RT-PCR. To determine let-7 expression, a total amount of 20 ng RNAs was reversely transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and stem-loop RT primers (Ribobio) according to manufacturer's instructions. To detect the mRNA level of NGF, RNA samples were reversely transcribed to cDNA by using a Prime-Script reagent Kit (TaKaRa, Dalian, China) according to manufacturer's instructions. qPCR was performed using SYBR Green Premix Ex Taq (TaKaRa) on an Applied Biosystems Stepone real-time PCR System. All reactions were run in triplicate. The relative expression was calculated using the comparative 2−ΔΔCt method. For semiquantitative RT-PCR, the cDNA generated was used as a template for PCR reaction. The thermocycler program was as follows: 5 minutes at 94 °C; 30 cycles of 30 seconds at 94 °C; 45 seconds at 58 °C; 30 seconds at 72 °C; and 5 minutes at 72 °C. The sequences of NGF primer are as follows (5′-3′): forward, CCAAGGACGCAGCTTTCTATC; reverse, CTGTGTCAAGGGAATGCTGAAG.
Western blot analysis. Protein extracts were prepared from nerve tissues or cell cultures. Equal amounts of protein were separated on 10% SDS–polyacrylamide gels and transferred to PVDF membrane (Millipore, Bedford, MA). The membrane was blocked with PBS containing 0.1% Tween-20 (PBST) and 5% nonfat dry milk, reacted with primary NGF antibodies (Abcam, Cambridge, MA), followed by incubation with horseradish peroxidase (HRP)-conjugated second antibodies with wash between each step. The membrane was developed with enhanced chemiluminescence reagent (Cell Signaling, Beverly, MA) and exposure to Kodak X-Omat Blue film (NEN life science, Boston, MA). Quantitative analysis was performed using Grab-it 2.5 and Gelwork software.
Cell proliferation assay. Primary SCs were resuspended in fresh prewarmed (37 °C) complete medium, counted, and plated at a density of 2 × 105 cells/ml on poly-L-lysine-coated 96-well plates. Following cell treatment, 50 μmol/l EdU was applied to cell culture, and SCs were grown for additional 12 hours. The cells were fixed with 4% paraformaldehyde, and cell proliferation of primary SCs was determined with Cell-Light EdU DNA Cell Proliferation Kit (Ribobio) according to the manufacturer's protocol. The ratio of EdU-positive cells to total cells was calculated by counting cells from randomly selected fields of images obtained using DMR fluorescence microscopy (Leica Microsystems, Wetzlar, Germany). Assays were done three times using triplicate wells.
Cell migration assay. Migration of SCs was examined using 6.5 mm Transwell chambers with 8 μm pores (Costar, Cambridge, MA) as described previously.46 The bottom surface of each membrane was coated with 10 μg/ml fibronectin. 100 μl SCs (106 cells/ml) resuspended in DMEM were transferred to the top chambers of each transwell and allowed to migrate at 37 °C in 5% CO2, and 600 μl of complete medium was injected into the lower chambers. The upper surface of each membrane was cleaned with a cotton swab at the indicated time point. Cells adhering to the bottom surface of each membrane were stained with 0.1% crystal violet, imaged, and counted using a DMR inverted microscope (Leica). Assays were done three times using triplicate wells.
Plasmid construction and luciferase assay. The 3′-UTR of NGF was amplified by PCR using rat genomic DNA as a template. The PCR products were subcloned into the region directly downstream of the stop codon in the luciferase gene in the luciferase reporter vector to generate p-Luc-UTR reporter plasmid. Overlap PCR was used to construct 3′-UTR mutant reporter plasmid. The sequences of wild-type and mutant 3′-UTR were confirmed by sequencing. HEK 293T cells were seeded in 24-well plates and transfected with a mixture of 120 ng p-Luc-UTR, 20 pmol miRNA mimics, and 20 ng Renilla luciferae vector pRL-CMV (Promega, Madison, WI) following the recommended protocol for the Lipofectamine 2000 transfection system (Invitrogen). After incubation for 36 hours, firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Promega) from the cell lysates.
ELISA. Primary SCs were transfected with NGF siRNA and negative control, let-7 mimic and control, let-7 inhibitor and control, respectively. Afterwards, the medium was replaced with FBS-free DMEM for additional incubation. The medium was then taken out and filtered through a 0.22 μm filter (Millipore) to furnish the supernatant. The protein level of NGF in the medium was measured with a NGF ELISA Kit (RayBiotech, Norcross, GA) as per manufacturer's instructions. The data were measured and averaged from three independent cultures, each comprising triplicate wells.
Coculture of DRG neurons and SCs. Primary SCs were transfected with NGF siRNA and control, let-7d mimic and control, let-7d inhibitor and control, respectively, by the above described protocols, and then cocultured with rat DRG neurons, which had been prepared as described previously47 and did not undergo any transfection. Briefly, DRGs were removed from 1-day-old postnatal SD rats and digested with 1% collagenase for 30 minutes at 37 °C, and further dissection with 0.25% trypsin for 10 minutes. After incubation, DRGs were rinsed with DMEM supplemented with 10% FBS, mechanically dissociated, and purified through a differential velocity adherent technique, and seeded on poly-L-lysine-coated glass covers lips at a cell density of 5 × 103/cm2.
After 48 hours coculture of SCs with DRGs, DRG neurons were isolated, and fixed with 4% paraformaldehyde to undergo immunocytochemistry with anti-NF200 antibody (Sigma) to observe axon outgrowth.
In vivo experiments. Adult male SD rats were anaesthetized before the sciatic nerve was exposed through an incision on the left hind limb and transected to create a gap. A silicone tube (i.d. 1.0 mm) was implanted to bridge the nerve gap with the proximal nerve stump anastomosed to the tube at the junction. The rats were randomly divided into two groups (n = 3 each) to receive injection of a mixture of Matrigel (BD Biosciences, Billerica, MA) with let-7d inhibitor (antagomir, Ribobio) or a mixture of Matrigel with corresponding control (Ribobio) both at a volume ratio of 1:1, respectively. Similarly, another cohort of rats were also divided into two groups to receive injection of a mixture of Matrigel with let-7d mimic (agomir, Ribobio) or a mixture of Matrigel with corresponding control (Ribobio) both at a volume ratio of 1:1, respectively. The injection was performed from the opposite opening of the silicone tube into the tube lumen using a precooled micropipette, followed by anastomosis of the tube to the distal nerve stump at the junction. The injection was done as slowly as possible to prevent the formation of air bubbles. Afterwards, the surgical incision was closed in a routine fashion, and animals were housed in large cages. At 7 and 10 days after surgery, rats were killed and the silicone tube, together with regenerated nerves, was harvested for cutting into sections, which were subjected to immunohistochemistry anti-S100β and anti-NF200 (both from Sigma), respectively. To assess migration of SCs and axons within the nerve gap, the edge of the proximal nerve stump was labeled, SCs or axons were identified as S100β- or NF200-positive cells, respectively, and then the cell displacement of SCs or axons from the proximal to distal nerve stump was measured.
Cell apoptosis assay. Cell apoptosis of primary SCs was assayed by using annexin-V/propidium iodide double staining. In brief, primary SCs were trypsinized and harvested, followed by resuspension and centrifugation. After the supernatant was discarded, the cell culture was stained with FITC-conjugated annexin-V antibody and propidium iodide as per kit instructions. The resulting mixture was blended by vortexing, and incubated for 30 minutes at 4 °C, followed by analysis by flow cytometry (BD Bioscience, San Jose, CA) using 488 nm excitation and a 515-nm bandpass filter for fluorescein detection. Three independent flow cytometric experiments were performed.
Adult male SD rats underwent sciatic nerve crush as above mentioned. A mixture of Matrigel and let-7d agomir and a mixture of Matrigel and control agomir (both at a volume ratio of 1:1) were respectively injected into the crushed sciatic nerve. At 1 day after surgery, the injured nerve were harvested from killed rats, fixed, dehydrated and then cut into sections. The sections were fixed in 4.0% paraformaldehyde, and then subjected to terminal deoxynucleotidyl transferase (TdT)-mediated deoxyuridine triphosphate (dUTP)-biotin nick end labeling of fragmented DNA (TUNEL) assay with In Situ Cell Death Detection Kit, Fluorescein (Roche, Basel, Switzerland) according to the manufacturer's specifications. In brief, DNA strand breaks were labeled with fluorescein-dUTP and TdT in dark at 37 °C for 1 hour. Subsequently, the sections were counter-stained with Hoechst 33258 dye. TUNEL-positive (apoptotic) cells were detected as localized bright green cells by using scanning laser confocal microscopy (Leica, Heidelberg, Germany). Data were expressed as the ratio of apoptotic cells to total cells.
Statistical analysis. All data are expressed as means ± SD. The Student's t-test was used for statistical analysis between groups by the aid of SPSS 15.0 software. P < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by National Key Basic Research Program of China (973 and 863 programs, nos. 2014CB542202, 2012AA020502), National Natural Science Foundation of China (nos. 81130080, 31300879), Jiangsu Provincial Natural Science Foundation (no. BK2012230), Collegiate Natural Science Foundation of Jiangsu Province (no. 11KJB180009), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The authors declare no conflict of interest.
References
- Taylor CA, Braza D, Rice JB, Dillingham T. The incidence of peripheral nerve injury in extremity trauma. Am J Phys Med Rehabil. 2008;87:381–385. doi: 10.1097/PHM.0b013e31815e6370. [DOI] [PubMed] [Google Scholar]
- Aloe L, Rocco ML, Bianchi P, Manni L. Nerve growth factor: from the early discoveries to the potential clinical use. J Transl Med. 2012;10:239. doi: 10.1186/1479-5876-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni L, Rocco ML, Bianchi P, Soligo M, Guaragna M, Barbaro SP.et al. (2013Nerve growth factor: basic studies and possible therapeutic applications Growth Factors 31115–122. [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Liu K, Liu Y, Mo W, Qiu R, Wang X, Wu JY.et al. (2011MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1 Nucleic Acids Res 392869–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Wood MJ. Therapeutic targeting of non-coding RNAs. Essays Biochem. 2013;54:127–145. doi: 10.1042/bse0540127. [DOI] [PubMed] [Google Scholar]
- Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, McDonald D, Cheng C, Magnowski B, Durand J, Zochodne DW. Axon and Schwann cell partnership during nerve regrowth. J Neuropathol Exp Neurol. 2005;64:613–622. doi: 10.1097/01.jnen.0000171650.94341.46. [DOI] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmén C, Somandin C, Miehe M, Jacob C.et al. (2010Dicer in Schwann cells is required for myelination and axonal integrity J Neurosci 306763–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. J Neurosci. 2010;30:7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yu B, Wang S, Gu Y, Yao D, Wang Y.et al. (2012Identification and functional analysis of novel micro-RNAs in rat dorsal root ganglia after sciatic nerve resection J Neurosci Res 90791–801. [DOI] [PubMed] [Google Scholar]
- Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding F.et al. (2012miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury J Cell Sci 125Pt 112675–2683. [DOI] [PubMed] [Google Scholar]
- Zhou S, Gao R, Hu W, Qian T, Wang N, Ding G.et al. (2014MiR-9 inhibits Schwann cell migration by targeting Cthrc1 following sciatic nerve injury J Cell Sci 127Pt 5967–976. [DOI] [PubMed] [Google Scholar]
- Gu X, Ding F, Williams DF. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials. 2014;35:6143–6156. doi: 10.1016/j.biomaterials.2014.04.064. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE.et al. (2000The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans Nature 403901–906. [DOI] [PubMed] [Google Scholar]
- Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J.et al. (2012An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration Nat Neurosci 15827–835. [DOI] [PubMed] [Google Scholar]
- Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012;31:4511–4523. doi: 10.1038/emboj.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 2012;23:202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang CF, Chang C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340:372–376. doi: 10.1126/science.1231321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit ME, Tenner AJ. Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J Neurosci. 2011;31:3459–3469. doi: 10.1523/JNEUROSCI.3932-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosaki T, Kamiya H, Yasuda Y, Naruse K, Kato K, Kozakae M.et al. (2008Reduced NGF secretion by Schwann cells under the high glucose condition decreases neurite outgrowth of DRG neurons Exp Neurol 213381–387. [DOI] [PubMed] [Google Scholar]
- Luo X, Chen B, Zheng R, Lin P, Li J, Chen H. Hydrogen peroxide induces apoptosis through the mitochondrial pathway in rat Schwann cells. Neurosci Lett. 2010;485:60–64. doi: 10.1016/j.neulet.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Frostick SP, Yin Q, Kemp GJ. Schwann cells, neurotrophic factors, and peripheral nerve regeneration. Microsurgery. 1998;18:397–405. doi: 10.1002/(sici)1098-2752(1998)18:7<397::aid-micr2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Lee SK, Wolfe SW. Peripheral nerve injury and repair. J Am Acad Orthop Surg. 2000;8:243–252. doi: 10.5435/00124635-200007000-00005. [DOI] [PubMed] [Google Scholar]
- Webber C, Zochodne D. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp Neurol. 2010;223:51–59. doi: 10.1016/j.expneurol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB.et al. (2009Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity Nat Neurosci 12839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann R, Korsching S, Bandtlow C, Thoenen H. Changes of nerve growth factor synthesis in nonneuronal cells in response to sciatic nerve transection. J Cell Biol. 1987;104:1623–1631. doi: 10.1083/jcb.104.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276:3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- Fiore M, Chaldakov GN, Aloe L. Nerve growth factor as a signaling molecule for nerve cells and also for the neuroendocrine-immune systems. Rev Neurosci. 2009;20:133–145. doi: 10.1515/revneuro.2009.20.2.133. [DOI] [PubMed] [Google Scholar]
- Ebendal T. Function and evolution in the NGF family and its receptors. J Neurosci Res. 1992;32:461–470. doi: 10.1002/jnr.490320402. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- Wang S, Tang Y, Cui H, Zhao X, Luo X, Pan W.et al. (2011Let-7/miR-98 regulate Fas and Fas-mediated apoptosis Genes Immun 12149–154. [DOI] [PubMed] [Google Scholar]
- Petratos S, Butzkueven H, Shipham K, Cooper H, Bucci T, Reid K.et al. (2003Schwann cell apoptosis in the postnatal axotomized sciatic nerve is mediated via NGF through the low-affinity neurotrophin receptor J Neuropathol Exp Neurol 62398–411. [DOI] [PubMed] [Google Scholar]
- Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL.et al. (2012Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression J Immunol 1886238–6246. [DOI] [PubMed] [Google Scholar]
- Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P.et al. (2008Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma Mol Cancer Res 6663–673. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A.et al. (2010The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma J Hepatol 52698–704. [DOI] [PubMed] [Google Scholar]
- Wong TS, Man OY, Tsang CM, Tsao SW, Tsang RK, Chan JY.et al. (2011MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression J Cancer Res Clin Oncol 137415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yu B, Wang Y, Yao D, Zhang Z, Gu X. Identification and functional annotation of novel microRNAs in the proximal sciatic nerve after sciatic nerve transection. Sci China Life Sci. 2011;54:806–812. doi: 10.1007/s11427-011-4213-7. [DOI] [PubMed] [Google Scholar]
- Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F.et al. (2012miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury Nucleic Acids Res 4010356–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantuano E, Inoue G, Li X, Takahashi K, Gaultier A, Gonias SL.et al. (2008The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein J Neurosci 2811571–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang J, Ding F, Hu N, Wang Y, Gu X. Neurotrophic actions of bone marrow stromal cells on primary culture of dorsal root ganglion tissues and neurons. J Mol Neurosci. 2010;40:332–341. doi: 10.1007/s12031-009-9304-6. [DOI] [PubMed] [Google Scholar]