Abstract
Rationale: Using microarray data, we previously identified gene expression–based subclasses of septic shock with important phenotypic differences. The subclass-defining genes correspond to adaptive immunity and glucocorticoid receptor signaling. Identifying the subclasses in real time has theranostic implications, given the potential for immune-enhancing therapies and controversies surrounding adjunctive corticosteroids for septic shock.
Objectives: To develop and validate a real-time subclassification method for septic shock.
Methods: Gene expression data for the 100 subclass-defining genes were generated using a multiplex messenger RNA quantification platform (NanoString nCounter) and visualized using gene expression mosaics. Study subjects (n = 168) were allocated to the subclasses using computer-assisted image analysis and microarray-based reference mosaics. A gene expression score was calculated to reduce the gene expression patterns to a single metric. The method was tested prospectively in a separate cohort (n = 132).
Measurements and Main Results: The NanoString-based data reproduced two septic shock subclasses. As previously, one subclass had decreased expression of the subclass-defining genes. The gene expression score identified this subclass with an area under the curve of 0.98 (95% confidence interval [CI95] = 0.96–0.99). Prospective testing of the subclassification method corroborated these findings. Allocation to this subclass was independently associated with mortality (odds ratio = 2.7; CI95 = 1.2–6.0; P = 0.016), and adjunctive corticosteroids prescribed at physician discretion were independently associated with mortality in this subclass (odds ratio = 4.1; CI95 = 1.4–12.0; P = 0.011).
Conclusions: We developed and tested a gene expression–based classification method for pediatric septic shock that meets the time constraints of the critical care environment, and can potentially inform therapeutic decisions.
Keywords: sepsis, gene expression, subclassification, adaptive immunity, glucocorticoids
At a Glance Commentary
Scientific Knowledge on the Subject
Using whole-genome expression profiling, we previously identified subclasses of pediatric septic shock differentiated by distinct gene expression patterns corresponding to relevant biological processes and clinical phenotypes. Although very powerful for discovery, whole-genome approaches do not meet the time-sensitive theranostic demands of critically ill patients with septic shock.
What This Study Adds to the Field
We have adapted complex, time-consuming genomic methods for use in the critical care environment. Using a multiplex, digital messenger RNA quantification platform that can generate gene expression data in about 8–12 hours, we have developed and validated a method to subclassify children with septic shock based on a 100-gene expression signature. The subclasses have clinically relevant phenotypes, and the class-defining genes correspond to adaptive immunity and glucocorticoid receptor signaling, thus raising the possibility of a theranostic approach to pediatric septic shock.
Septic shock is a heterogeneous syndrome with widely ranging physiological and biological manifestations (1–3). This heterogeneity represents a major challenge for the development of new therapies and for targeting therapies to patients most likely to benefit. An approach to meet this challenge is to define subclasses (i.e., endotypes) of septic shock differentiated by distinct gene expression patterns corresponding to relevant biological processes and clinical phenotypes.
Using genome-wide expression profiling and unsupervised hierarchical clustering, we previously reported three putative gene expression–based subclasses of pediatric septic shock, and found that one of the subclasses, subclass A, had greater organ failure burden and mortality (4). We subsequently validated the existence of these subclasses and condensed the subclass-defining gene expression signature to the top 100 class-predictor genes (5, 6). The class-predictor genes correspond to adaptive immunity and glucocorticoid receptor signaling, as determined by objective pathway analysis. This opens the possibility of a theranostic approach, because immune-enhancing therapies are being considered for septic shock (1, 7), and the role of adjunctive corticosteroids for septic shock remains unclear due to heterogeneous patient responses (8).
All of this previous work was based on microarray technology. Although this is a powerful tool for discovery, it does not meet the time-sensitive demands for theranostic information in critically ill patients with septic shock (9). In the current study, we adapt these complex, time-consuming, whole-genome methods for use in the critical care environment. We measured expression of the 100 subclass-defining genes using a multiplex messenger RNA (mRNA) quantification platform (NanoString nCounter; NanoString Technologies, Seattle, WA) capable of generating expression data in about 8–12 hours (10). Using these data, we first determined if NanoString-based gene expression mosaics could reproduce the previously reported gene expression–based subclasses of pediatric septic shock and their respective phenotypes. Second, we determined if the gene expression signature could be reduced to a simple score that reliably distinguishes the subclasses. Third, we prospectively tested the newly developed classification method in a separate validation cohort of patients, while simultaneously considering the effects of interassay variability. Finally, to demonstrate proof of theranostic principle, we determined if associations between adjunctive corticosteroids and septic shock outcomes are dependent on subclass allocation, as would be suggested when discriminating between patients based on genes associated with adaptive immune function and the glucocorticoid receptor signaling pathway.
Methods
Study Subjects and Data Collection
The study subjects for developing the subclassification method were previously reported (4–6), whereas the study subjects for prospectively testing the subclassification method have not. The study protocol and the procedures for generating microarray data have been described in detail (4–6, 11, 12), and the previous microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession nos. GSE26440 and GSE26378).
Briefly, children 10 years of age or less admitted to the pediatric intensive care unit (PICU) and meeting pediatric-specific consensus criteria for septic shock were enrolled after informed consent from parents or legal guardians. Blood samples were obtained within 24 hours of initial presentation to the PICU with septic shock. Total RNA was isolated from whole blood using the PaxGene Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA). Clinical and laboratory data were collected daily while in the PICU. Mortality and organ failure were tracked for 28 days after enrollment. Illness severity was measured using Pediatric Risk of Mortality (PRISM) scores (13).
Multiplex mRNA Quantification
A custom NanoString nCounter codeset was generated for the 100 subclass-defining genes. The technology is based on standard hybridization between the target gene, and target-specific capture and reporter probes (14). All NanoString-based measurements were conducted at the University of Minnesota Genomics Center Core Facility. Four housekeeping genes were used to normalize the NanoString-derived expression data: β-2-microglobulin, folylpolyglutamate synthase, 2,4-dienoyl coenzyme A reductase 1, and peptidylprolyl isomerase B. These were selected from our transcriptomic database, because they showed minimal expression variation across 180 subjects with septic shock. Expression values were normalized to the geometric mean of the housekeeping genes.
Gene Expression Mosaics and Computer-assisted Image Analysis
Gene expression mosaics representing the expression patterns of the 100 subclass-defining genes were generated using the Gene Expression Dynamics Inspector, as previously described (5, 6, 15, 16). The gene expression mosaics from individual patients were compared with subclass reference mosaics using a public analysis platform (ImageJ; National Institutes of Health, Bethesda, MD), as previously described (5). The absolute difference in RGB pixel-to-pixel intensity was calculated for each individual patient mosaic, relative to the reference mosaics. Final subclass allocation was based on the least difference between the individual patient mosaic and one of the reference mosaics representing the septic shock subclasses.
Gene Expression Score
Gene expression data for the 100 class-defining genes was reduced to a single metric, the gene expression score (GES). The GES was defined to quantify the range of variability in expression of the 100 subclass-defining genes by calculating the sum of the squared differences between the expression levels of each gene and the geometric mean of all genes using the equation:
where ei is the gene expression level for an individual gene and μg is the geometric mean expression value of the 100 subclass-defining genes for a given patient. The sum was scaled by a factor of 1 × 106.
Statistical Analysis
Statistical procedures used SigmaStat Software (Systat Software, Inc., San Jose, CA). General clinical and demographic data are described using medians, interquartile ranges, frequencies, and percentages. Comparisons between groups used the Mann-Whitney U test, chi-square, or Fisher’s exact tests, as appropriate. The performance of the GES for distinguishing subclasses was measured by constructing receiver operating characteristics (ROCs) curves and calculating diagnostic test characteristics. The association between subclass allocation and outcome was modeled using multivariable logistic regression. The primary outcome variable for the regression procedures was all-cause 28-day mortality. Because persistent, multiple organ failure is a major antecedent of death secondary to sepsis, we also modeled a complicated course, defined as the persistence of two or more organ failures at Day 7 of septic shock or 28-day mortality (17–20). This allows the exploration of association between gene expression patterns and a nuance of sepsis severity beyond the dichotomy of “alive” versus “dead.”
Results
Identification of Pediatric Septic Shock Subclasses via Multiplex mRNA Quantification
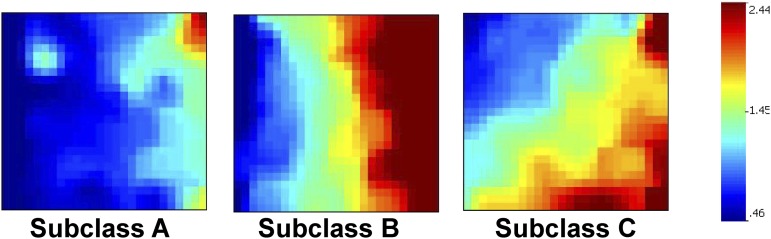
Using microarray data, we previously generated individual gene expression mosaics for the 100 septic shock subclass–defining genes in two separate cohorts (4–6). In the current study, we used the microarray data from all 180 subjects in both cohorts to generate composite mosaics for each of the three subclasses. The composite mosaics represent the mean expression values of the 100 subclass-defining genes within each subclass. Figure 1 shows the composite mosaics for the three subclasses. Table E1 in the online supplement shows the 100 septic shock subclass–defining genes.
Figure 1.
Composite gene expression mosaics for the 100 class-defining genes based on previous microarray data. The composite mosaics represent the mean expression values of the 100 subclass-defining genes within each subclass. Red intensity correlates with increased gene expression, and blue intensity correlates with decreased gene expression. The composite mosaics were used as a reference to classify subjects based on NanoString-generated data for the 100 subclass-defining genes. Classification was performed using computer-assisted image analysis.
Among the original 180 subjects used to generate the microarray-based composite mosaics, there were 168 (93%) with remaining RNA samples. These samples were used to generate new NanoString-based expression data for the 100 subclass-defining genes and individual patient gene expression mosaics. Using computer-assisted image analysis, the new NanoString-based expression mosaics were compared with the microarray-based composite mosaics as a reference, and the 168 subjects were re-allocated into one of the three septic shock subclasses.
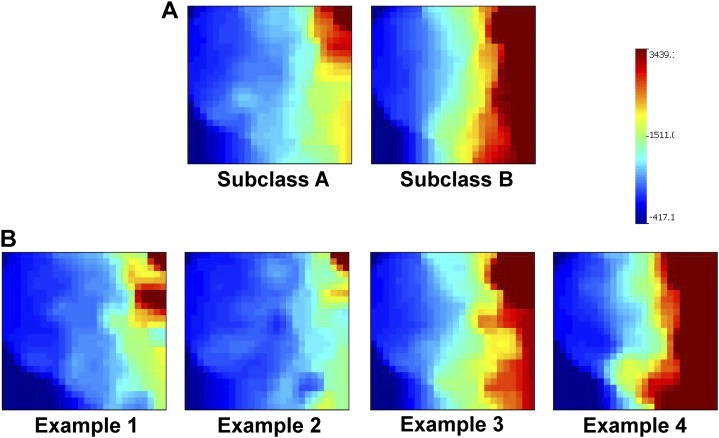
A total of 57 subjects (34%) were allocated to subclass A, and 111 subjects were allocated to subclass B. No subjects were allocated to subclass C. Figure 2A shows the composite gene expression mosaics for the subjects in subclasses A and B based on NanoString-generated data. Figure 2B shows examples of individual patient gene expression mosaics. Table 1 shows the clinical and demographic data for the subjects in subclasses A and B. At baseline, the subjects in subclass A had higher median PRISM scores, were younger, and a lower proportion had a comorbidity compared with those in subclass B. Subjects in subclass A had lower total white blood cell and absolute neutrophil counts, but higher absolute lymphocyte counts, compared with those in subclass B. No other differences were noted at baseline. With respect to outcomes, subjects in subclass A had a higher mortality rate and a higher rate of a complicated course compared with those in subclass B.
Figure 2.
(A) Composite gene expression mosaics for the 100 subclass-defining genes based on NanoString-derived expression data. The composite mosaics represent the mean expression values of the 100 subclass-defining genes within each subclass. Red intensity correlates with increased gene expression, and blue intensity correlates with decreased gene expression. (B) Examples of individual patient gene expression mosaics for subjects in the test cohort based on NanoString-derived expression data. The individual patient gene expression mosaics were compared with the reference composite mosaics (A) to prospectively allocate the test cohort subjects into subclass A or B. Image comparisons were performed using computer-assisted image analysis. Examples 1 and 2 were allocated to subclass A, whereas examples 3 and 4 were allocated to subclass B.
Table 1.
Clinical and Demographic Data for the Derivation Cohort
| Subclass A | Subclass B | |
|---|---|---|
| n | 57 | 111 |
| Median age (IQR), yr | 1.4 (0.2–2.9) | 3.0 (1.5–7.3)* |
| Males, n (%) | 36 (63) | 64 (58) |
| 28-d mortality, n (%) | 12 (21) | 11 (10)† |
| Complicated course, n (%) | 24 (42) | 26 (23)† |
| Median PRISM score (IQR) | 16 (12–23) | 13 (9–20)* |
| Median WBC count ×103/mm3 (IQR) | 10.0 (3.8–16.9) | 14.6 (7.7–19.9)* |
| Median neutrophil count ×103/mm3 (IQR) | 6.1 (2.4–11.4) | 10.9 (4.5–16.8)* |
| Median lymphocyte count ×103/mm3 (IQR) | 1.8 (0.9–3.5) | 1.5 (0.7–2.5)* |
| Median monocyte count ×103/mm3 (IQR) | 0.6 (0.1–1.4) | 0.6 (0.2–1.3) |
| No. with gram-negative bacteria (%) | 11 (19) | 26 (23) |
| No. with gram-positive bacteria (%) | 16 (28) | 28 (25) |
| No. with other pathogen isolated (%) | 6 (11) | 5 (5) |
| No. with no pathogen identified (%) | 24 (42) | 52 (47) |
| No. with comorbidity (%) | 11 (19) | 46 (41)† |
| No. with malignancy (%) | 1 (2) | 8 (7) |
| No. with immune suppression (%) | 3 (5) | 11 (10) |
| No. with bone marrow transplantation (%) | 1 (2) | 5 (5) |
Definition of abbreviations: IQR = interquartile range; PRISM = Pediatric Risk of Mortality; WBC = white blood cell.
P < 0.05 versus subclass A, rank sum test.
P < 0.05 versus subclass A, chi-square.
Collectively, these data demonstrate that a multiplex RNA quantification platform that is amenable to rapid turnaround in the acute care setting can subclassify patients with septic shock based on gene expression data, and that subjects in subclass A have worse outcomes.
Development of a GES to Subclassify Septic Shock
As an alternative to subclassification based on gene expression mosaics and computer-assisted image analysis, we constructed a GES to allocate subjects into subclass A or B. Based on visual inspection of the composite gene expression mosaics depicted in Figure 2A, we reasoned that a score that quantifies the variability of gene expression intensity relative to some central reference value within a given patient would provide utility in differentiating the subclasses. Because patients in subclass A tended to have less variability than those in subclass B, we expected that the GES value would be lower in subjects in subclass A compared with those in subclass B.
As shown in Figure 3A, the median GES for the subjects in subclass A was significantly lower compared with those in subclass B. We next generated an ROC curve to determine the ability of the GES to discriminate subclass A from subclass B, using the gene expression mosaic–based classification method as the gold standard (NanoString-based data). Figure 3B shows the ROC curve with an area under the curve of 0.98 (95% confidence interval [CI95] = 0.96–0.99). The 90th percentile GES for the subjects in subclass A was 401. Using a GES of 401 or lower, we calculated the diagnostic test characteristics of the GES for identifying subjects in subclass A, which were: sensitivity, 91% (CI95 = 80–97); specificity, 92% (CI95 = 85–96); positive predictive value, 85% (73 to 93); negative predictive value, 95% (CI95 = 89–98); positive likelihood ratio (+LR), 11.3 (CI95 = 6.0–21.2); and −LR, 0.1 (CI95 = 0.01–0.2).
Figure 3.
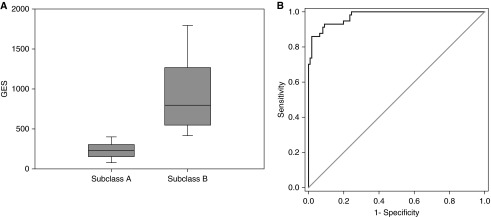
(A) Box-and-whisker plots depicting the gene expression score (GES) values for subjects in subclasses A and B. (B) Receiver operating characteristic curve demonstrating the performance of the GES for distinguishing subclass A from subclass B. Classifications based on the gene expression mosaics (NanoString-based data) were used as the gold standard classification.
These data demonstrate that the GES can distinguish subclass A from subclass B with a high level of accuracy in the derivation cohort.
Prospective Subclassification of Patients with Septic Shock
We next tested this subclassification method prospectively in a separate cohort of 132 subjects. With the goal of developing a clinically reproducible method, we took into account the potential for interassay variability by generating the prospective mRNA expression data a priori in eight separate batches over a 1-year period. We measured expression of the 100 subclass-defining genes for each subject in the test cohort using the NanoString platform, and generated individual patient gene expression mosaics. We then compared the individual patient gene expression mosaics to the composite mosaics shown in Figure 2A as a reference, and allocated each subject into subclass A or B using computer-assisted image analysis and the GES.
A total of 63 subjects (48%) were allocated to subclass A and 69 subjects were allocated to subclass B by computer-assisted image analysis in the test cohort. Table 2 shows the clinical and demographic data for the subjects in subclasses A and B in the test cohort. At baseline, the subjects in subclass A were younger than those in subclass B, and the distribution of white blood cells was similar to that seen in the derivation cohort. No other baseline differences were noted. Patients in subclass A had a higher mortality rate and a higher rate of a complicated course compared with those in subclass B.
Table 2.
Clinical and Demographic Data for the Test Cohort
| Subclass A | Subclass B | |
|---|---|---|
| n | 63 | 69 |
| Median age (IQR), yr | 1.4 (0.3–3.9) | 4.1 (1.3–6.6)* |
| Males, n (%) | 34 (54) | 39 (57) |
| 28-d mortality, n (%) | 11 (17) | 4 (5)† |
| Complicated course, n (%) | 27 (43) | 11 (16)† |
| Median PRISM score (IQR) | 11 (6–18) | 11 (8–19) |
| Median WBC count ×103/mm3 (IQR) | 8.6 (2.9–14.7) | 13.4 (6.2–20.8)* |
| Median neutrophil count ×103/mm3 (IQR) | 4.6 (0.8–8.6) | 11.9 (4.8–16.6)* |
| Median lymphocyte count ×103/mm3 (IQR) | 2.3 (1.3–4.3) | 1.2 (0.5–2.1)* |
| Median monocyte count ×103/mm3 (IQR) | 0.5 (0.1–0.9) | 0.5 (0.3–1.2) |
| No. with gram negative bacteria (%) | 17 (27) | 12 (17) |
| No. with gram positive bacteria (%) | 10 (16) | 13 (19) |
| No. with other pathogen isolated (%) | 5 (8) | 13 (19) |
| No. with no pathogen identified (%) | 31 (49) | 31 (45) |
| No. with comorbidity (%) | 16 (25) | 26 (38) |
| No. with malignancy (%) | 6 (10) | 5 (7) |
| No. with immune suppression (%) | 6 (10) | 9 (13) |
| No. with bone marrow transplantation (%) | 0 (0) | 3 (4) |
Definition of abbreviations: IQR = interquartile range; PRISM = Pediatric Risk of Mortality; WBC = white blood cell.
P < 0.05 versus subclass A, rank sum test.
P < 0.05 versus subclass A, chi-square.
For determining subclassification based on the GES, the area under the curve for the ROC curve was 0.98 (CI95 = 0.96–1.00) in the test cohort. The diagnostic test characteristics of the GES for identifying subjects in subclass A in the test cohort were: sensitivity, 94% (CI95 = 84–98); specificity, 93% (CI95 = 83–97); positive predictive value, 92% (CI95 = 82–97); negative predictive value, 94% (CI95 = 85–98); +LR, 12.9 (CI95 = 5.5–30.1); and −LR, 0.06 (CI95 = 0.02–0.2).
The observation that subjects in subclass A in both the derivation cohort and the test cohort had higher mortality rates and higher rates of a complicated course could reflect confounding by illness severity, presence of comorbidity, and age, rather than class allocation per se. To investigate this possibility, we used logistic regression to test the association between class allocation and outcome adjusted for age, presence of comorbidity, and illness severity (PRISM scores). For all subjects (n = 300), allocation to subclass A was independently associated with increased risk of mortality (odds ratio [OR] = 2.7; CI95 = 1.2–6.0; P = 0.016) and complicated course (OR = 2.7; CI95 = 1.5–4.8; P < 0.001). When we restricted the analysis to subjects with positive microbiological cultures (n = 162), allocation to subclass A was independently associated with increased risk of a complicated course (OR = 2.2; CI95 = 1.0–4.7; P = 0.043), and there was a tendency for subjects in subclass A to have higher mortality (OR = 2.7; CI95 = 1.0–7.3; P = 0.054).
These data demonstrate that prospective application of the gene expression–based classification method in a test cohort identifies two septic shock classes, and that allocation to subclass A is independently associated with worse outcomes. In addition, prospective application of the GES demonstrated excellent discrimination between the two subclasses.
Adjunctive Treatment with Corticosteroids and Outcomes
The 100-gene expression signature that defines the two septic shock classes is enriched for genes corresponding to glucocorticoid receptor signaling, and these genes are repressed in the patients in subclass A relative to those in subclass B (4–6). This raises the possibility that the two subclasses could have different responses to adjunctive treatment with corticosteroids.
We combined all subjects and tested the association between adjunctive corticosteroids and outcomes within the two septic shock subclasses. Prescription of adjunctive corticosteroids was at the discretion of the physicians caring for the study subjects. Among the 120 subjects in subclass A, 52 (43%) received adjunctive corticosteroids, and, among the 180 subjects in subclass B, 104 (58%) received adjunctive corticosteroids. We used logistic regression to test the association between adjunctive corticosteroids and outcomes within each subclass. Table E2 shows the results. After adjusting for illness severity (PRISM score), presence of comorbidity, and age, adjunctive corticosteroids were independently associated with an increased risk of mortality in the subjects in subclass A (OR = 4.1; CI95 = 1.4–12.0; P = 0.011), but not the subjects in subclass B. When testing the interaction between subclass and adjunctive corticosteroids, the effect of corticosteroids on the OR for mortality was nearly four times higher in the patients in subclass A than in those in subclass B (OR = 3.9; CI95 = 0.8–18.3; P = 0.089). Adjunctive corticosteroids were not independently associated with an increased risk of a complicated course in either subclass.
Discussion
Using a multiplex mRNA quantification platform and gene expression mosaics, we confirmed the presence of gene expression–based subclasses of pediatric septic shock (4–6). The original, microarray-based subclassification identified three subclasses (A, B, and C), whereas the current NanoString-based subclassification identified two subclasses (A and B). This discrepancy is not problematic from a clinical standpoint, because subjects in subclasses B and C did not differ with respect to clinical phenotype (4–6). In contrast, subjects in subclass A had worse clinical outcomes in our original, microarray-based studies, and this was corroborated using the NanoString-based subclassification method. As an alternative to gene expression mosaics and computer-assisted image analyses for subclass identification, we also derived a metric, the GES, which reliably distinguishes subclasses A and B.
We prospectively tested this subclassification method in a different cohort, and simultaneously accounted for interassay variability by analyzing the test cohort over a 1-year period consisting of eight different batches of samples. This prospective test of the subclassification method corroborated the key finding, that subjects in subclass A have worse clinical outcomes, and validated the diagnostic test characteristics of the GES. Taking into account the potential confounders of age, comorbidity, and illness severity, we show that allocation to subclass A is independently associated with poor outcomes.
Taken together, these data demonstrate the feasibility of using complex gene expression data within the time-sensitive constraints of the critical care environment and at a direct assay cost of about $100 per patient. This contention is supported by a recent study that used similar approaches to predict poor outcomes in critically ill adults suffering from major trauma (10, 21). The development of microfluidics-based technologies to rapidly isolate RNA from blood samples further supports the idea that the this subclassification method can be translated into the time-sensitive critical care environment (22, 23).
The subclass-defining genes correspond to the adaptive immune system and glucocorticoid receptor signaling (4–6), and these genes are repressed in the subjects in subclass A relative to the those in subclass B. Repression of adaptive immunity–related genes does not appear to be an artifact of lymphopenia, because the absolute lymphocyte counts were higher in the subjects in subclass A compared with those in subclass B. Accordingly, our subclassification method has the potential to promote personalized medicine in that allocation to subclass A or B could potentially direct therapy. For example, the use of adjunctive corticosteroids for septic shock continues to be controversial in the field of critical care medicine (8). Clinical trials based on cortisol levels and adrenocorticotropic hormone stimulation tests have yielded conflicting results (24, 25). This reflects, in part, the challenge of identifying which patients with septic shock will benefit from adjunctive corticosteroids and which will not (26). Our subclassification method identifies a group of patients (subclass A) who may not respond favorably to adjunctive corticosteroids. Although adjunctive corticosteroids are independently associated with increased mortality in the subjects in subclass A, it is not clear whether this association is because subjects in subclass A fail to respond to corticosteroids, or if corticosteroids directly worsen outcome in subjects in subclass A. Both scenarios are biologically plausible and require further exploration. In a similar vein, leading investigators in the field are proposing immune-enhancing therapies to improve sepsis outcomes (1, 7, 27). Our subclassification method indicates that subjects in subclass A might benefit the most from such an approach.
We note the limitations of our study. As an observational study, clinical care was not under protocol. Accordingly, outcomes can reflect variability in the care processes, independent of the gene expression–based classifications. We did not directly measure adaptive immune function or glucocorticoid receptor function. Dysfunction of these systems is inferred based on gene expression patterns, which may not necessarily correlate with changes in function. In addition, we do not have complete data regarding baseline cortisol levels or results of adrenocorticotropic hormone stimulation tests. Finally, the use of adjunctive corticosteroids was not under protocol. Therefore, the associations between adjunctive corticosteroids and poor outcome should be interpreted with caution.
In conclusion, we have developed and successfully tested a gene expression–based subclassification method for pediatric septic shock. The method has the potential to meet the time constraints inherent to clinical decision making in critically ill patients, and to inform therapy-related clinical decision making.
Acknowledgments
Acknowledgment
The authors thank the following clinical research coordinators for enrolling patients at the various study sites: Debra Spears, Jenny Bush, Mary Ann De Liberto, Adelle Barreiro, Trisha Williams, Amber Hughes, Michelle Goldsworthy, Christi Rider, Mary Ellen Riodan, Veronica DeLeon, Hyacinth Bryant, Tiffany Patterson, Ofelia Vargas, Monica Weber, Lauren Hoadley, Heather Anthony, Lisa Steele, Angela Doucette, and Katherine Woods.
Footnotes
Supported by National Institutes of Health grants RO1GM064619, RO1GM099773, and R01GM108025, and in part by Institutional Clinical and Translational Science Award NIH/NCRR 8 UL1 TR000077.
Author Contributions: H.R.W. conceived and developed the study, obtained funding for the study, conducted the analysis, and edited the manuscript; N.Z.C., N.A., G.L.A., N.J.T., M.T.B., S.L.W., J.F., P.A.C., K.M., T.P.S., M.Q., M.H., R.G., R.J.F., J.N., R.S.S., S.G., and E.D. enrolled subjects at the participating institutions, provided clinical data and biological samples, and edited the manuscript; K. Howard, E.B., and E.F. maintained the clinical database and coordinated all interinstitutional research activity; K. Harmon maintained the biological repository and processed all biological samples; C.J.L. developed the study, assisted with analysis, and edited the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201410-1864OC on December 9, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 3.Hanna W, Wong HR. Pediatric sepsis: challenges and adjunctive therapies. Crit Care Clin. 2013;29:203–222. doi: 10.1016/j.ccc.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, et al. Validation of a gene expression–based subclassification strategy for pediatric septic shock. Crit Care Med. 2011;39:2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong HR, Wheeler DS, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Doughty LA. Toward a clinically feasible gene expression–based subclassification strategy for septic shock: proof of concept. Crit Care Med. 2010;38:1955–1961. doi: 10.1097/CCM.0b013e3181eb924f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchins NA, Unsinger J, Hotchkiss RS, Ayala A. The new normal: immunomodulatory agents against sepsis immune suppression. Trends Mol Med. 2014;20:224–233. doi: 10.1016/j.molmed.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel GP, Balk RA. Systemic steroids in severe sepsis and septic shock. Am J Respir Crit Care Med. 2012;185:133–139. doi: 10.1164/rccm.201011-1897CI. [DOI] [PubMed] [Google Scholar]
- 9.Maslove DM, Wong HR. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuenca AG, Gentile LF, Lopez MC, Ungaro R, Liu H, Xiao W, Seok J, Mindrinos MN, Ang D, Baslanti TO, et al. Inflammation and Host Response to Injury Collaborative Research Program. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41:1175–1185. doi: 10.1097/CCM.0b013e318277131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, et al. Genomics of Pediatric SIRS/Septic Shock Investigators. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013;73:564–569. doi: 10.1038/pr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III–Acute Physiology Score (PRISM III–APS): a method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr. 1997;131:575–581. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- 14.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 15.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S. Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J Biomed Biotechnol. 2006;2006:69141. doi: 10.1155/JBB/2006/69141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Weiss SL, Shanley TP, et al. Corticosteroids are associated with repression of adaptive immunity gene programs in pediatric septic shock. Am J Respir Crit Care Med. 2014;189:940–946. doi: 10.1164/rccm.201401-0171OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abulebda K, Cvijanovich NZ, Thomas NJ, Allen GL, Anas N, Bigham MT, Hall M, Freishtat RJ, Sen A, Meyer K, et al. Post-ICU admission fluid balance and pediatric septic shock outcomes: a risk-stratified analysis. Crit Care Med. 2014;42:397–403. doi: 10.1097/CCM.0b013e3182a64607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mickiewicz B, Vogel HJ, Wong HR, Winston BW. Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. Am J Respir Crit Care Med. 2013;187:967–976. doi: 10.1164/rccm.201209-1726OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkinson SJ, Cvijanovich NZ, Thomas NJ, Allen GL, Anas N, Bigham MT, Hall M, Freishtat RJ, Sen A, Meyer K, et al. Corticosteroids and pediatric septic shock outcomes: a risk stratified analysis. PLoS ONE. 2014;9:e112702. doi: 10.1371/journal.pone.0112702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, Moldawer LL, Moore EE, Harbrecht BG, Pelak K, et al. Inflammation and Host Response to Injury Large Scale Collaborative Research Program. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15:220–227. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, et al. Inflammation and the Host Response to Injury Collaborative Research Program. Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010;16:1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warner EA, Kotz KT, Ungaro RF, Abouhamze AS, Lopez MC, Cuenca AG, Kelly-Scumpia KM, Moreno C, O’Malley KA, Lanz JD, et al. Microfluidics-based capture of human neutrophils for expression analysis in blood and bronchoalveolar lavage. Lab Invest. 2011;91:1787–1795. doi: 10.1038/labinvest.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Annane D, Sébille V, Charpentier C, Bollaert PE, François B, Korach JM, Capellier G, Cohen Y, Azoulay E, Troché G, et al. Effect of treatment with low doses of hydrocortisone and fludrocortisone on mortality in patients with septic shock JAMA 2002288862–871.[Published erratum appears in JAMA 300:1652. Chaumet-Riffaut, Philippe corrected to Chaumet-Riffaud, Philippe]. [DOI] [PubMed] [Google Scholar]
- 25.Sprung CL, Annane D, Keh D, Moreno R, Singer M, Freivogel K, Weiss YG, Benbenishty J, Kalenka A, Forst H, et al. CORTICUS Study Group. Hydrocortisone therapy for patients with septic shock. N Engl J Med. 2008;358:111–124. doi: 10.1056/NEJMoa071366. [DOI] [PubMed] [Google Scholar]
- 26.Atkinson SJ, Wong HR. Identifying critically ill patients who may benefit from adjunctive corticosteroids: not as easy as we thought. Pediatr Crit Care Med. 2014;15:769–771. doi: 10.1097/PCC.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hotchkiss RS, Opal S. Immunotherapy for sepsis—a new approach against an ancient foe. N Engl J Med. 2010;363:87–89. doi: 10.1056/NEJMcibr1004371. [DOI] [PMC free article] [PubMed] [Google Scholar]