Abstract
Recent advancements and applications of nanofabrication have enabled the characterization and control of biological membranes at submicron scales. This review focuses on the application of nanofabrication towards the nanoscale observing, patterning, sorting, and concentrating membrane components. Membranes on living cells are a necessary component of many fundamental cellular processes that naturally incorporate nanoscale rearrangement of the membrane lipids and proteins. Nanofabrication has advanced these understandings, for example, by providing 30 nm resolution of membrane proteins with metal-enhanced fluorescence at the tip of a scanning probe on fixed cells. Naturally diffusing single molecules at high concentrations on live cells have been observed at 60 nm resolution by confining the fluorescence excitation light through nanoscale metallic apertures. The lateral reorganization on the plasma membrane during membrane-mediated signaling processes has been examined in response to nanoscale variations in the patterning and mobility of the signal-triggering molecules. Further, membrane components have been separated, concentrated, and extracted through on-chip electrophoretic and microfluidic methods. Nanofabrication provides numerous methods for examining and manipulating membranes for both greater understandings of membrane processes as well as for the application of membranes to other biophysical methods.
KEY TERMS: Supported Lipid Bilayer (SLB), Membrane Dynamics, Diffusion, Near-Field, Plasmonics, Electrophoresis, Patterning, Sorting, Concentrating, Nanobiotechnology
INTRODUCTION
Recently developed capabilities of nanofabrication have provided a means for unprecedented control and analysis of lipid membranes. Biological membranes are primarily composed of amphiphilic lipids and proteins which self-assemble into a planar sheet with a hydrophobic interior and hydrophilic exterior. The flow of molecules into and out of the cell is regulated by the membrane and its associated proteins. On the outer leaflet of the plasma membrane, proteins sense the extra-cellular environment and initiate cellular responses. On the inner leaflet of the plasma membrane, membrane-bound proteins commonly mediate cellular processes, such as initiating actin (de)polymerization, regulating second messenger molecules, or controlling plasma membrane endo/exocytosis.
The importance of membrane-mediated processes is demonstrated by the improvements in human health that have resulted from understanding membranes and their associated proteins; approximately 50% of all drugs on the market directly target membrane proteins, including drugs for hypertension, allergies, heart burn and mental illnesses.56 Despite this, many technical challenges in the manipulation and examination of membranes yield slow or entirely infeasible tests of particular membrane components. The plasma membrane contains thousands of different lipid species in addition to the complex membrane proteome. Since the functionality of membrane proteins often depends critically on their involvement in fragile protein complexes and their interaction with particular lipid species, isolating functional membrane proteins is particularly challenging. Significant progress has been made to purify membrane proteins, although often by the addition of detergents, genetic manipulations, and affecting the protein structure. Difficulty extracting, concentrating, sorting, and systematically observing membrane proteins in a functional state have slowed progress in understanding the means by which membranes influence human health.
Due to the high complexity and numerous technical complications associated with native cell membranes, model systems have been developed to probe particular membrane properties. Model membranes of known composition may be fabricated as spherical liposomes in solution, as a suspended bilayer separating two externally-accessible solutions, or as supported lipid bilayers (SLBs) coating a solid surface. SLBs have been integrated into numerous nanotechnology applications and mimic some key parameters of complex cellular membranes (e.g. lateral diffusion, elasticity, leaflet-leaflet interactions, and protein-lipid interactions). Further, diverse biophysical studies have utilized model membranes for minimizing non-specific protein adsorption, regulating cellular adhesion, or confining associated molecules to lateral diffusion on the membrane.
This review focuses on the current and future applications of nanofabrication towards controlling and analyzing nanoscale membrane composition, patterning, and/or dynamics. Nanofabrication is providing a means to isolate, concentrate, and study membranes with unprecedented control. We will focus on five key applications of nanotechnology to membranes and how these applications have advanced our understandings and uses of biological systems: 1) metallic apertures for confining fluorescence excitation light to sub-diffraction-limited areas, 2) surface plasmons on metallic nanostructures for optical field enhancement, 3) nanofabricated obstacles for restricting membrane diffusion, 4) patterning membrane composition, and 5) sorting membrane components. Micron-scale methods of patterning and probing membranes will not be addressed here in detail due to previous quality review articles on these techniques, as indicated throughout this manuscript. For example, review articles that focus on diffraction-limited optical techniques33,84 or field-effect transistors for electrical membrane sensing49 are available.
METALLIC APERTURES FOR OPTICAL CONFINEMENT
Cellular membranes perform a variety of complex tasks through dynamic regulation of their contents and lateral organization, but the detailed mechanisms by which they achieve their diverse functionality are not well understood. The governing principles of micron scale membrane dynamics include conceptually simple phenomena, such as Brownian motion, electrostatic bonding, and phase separation. However, the interplay of these phenomena at the nanoscale is difficult to resolve. Techniques such as freeze-fracture electron microscopy, nuclear magnetic resonance, calorimetry, and diffraction-limited microscopy are some of the key techniques used to date.81 But these techniques are unable to provide simultaneous nanometer and microsecond resolution of membrane dynamics.
Recent progress towards simultaneous nanoscale and microsecond resolution of membranes has been through expanding on the capabilities of optical microscopy and spectroscopy. Membrane dynamics are optically understood by monitoring the motion of membrane-bound particles, including observing individual particle trajectories, the equilibration of concentration gradient, or time correlation analysis of residency times.9 Among the most significant differences is the ability of each technique to observe non-Brownian diffusion. Many membrane-bound particles display anomalous diffusion at the nanoscale; the dominating mechanisms of anomalous membrane diffusion are frequently debated and rarely observed.16,81 The mechanisms of non-Brownian membrane diffusion are of considerable interest due to their involvement in various key cellular processes, such as receptor-mediated signaling, lipid-rafts, and actin-membrane interactions.
Numerous approaches to nanoscale patterning of metallic structures have provided nanoscale resolution of membrane dynamics via near-field light confinement/enhancement for decreased fluorophore illumination volumes. These approaches build upon the abundant tool-kit of fluorescence microscopy (e.g. membrane-bound fluorophores, chromatic filters, and sensitive detectors) while incorporating nanoscale metallic structures. Isolated metallic apertures have been used to prevent the majority of the illumination light from interacting with the membrane and limiting the excitation to the small area that is determined by the aperture dimensions rather than the diffraction-limit. In recent decades, a practical near-field imaging method has been developed as a near-field scanning optical microscope (NSOM).3,55 NSOMs have been used to provide 12 nm resolution for a variety of microscopy and spectroscopy applications.3 Recently, NSOMs have been used to examine the distribution of proteins and lipids on biological membranes.38,100 Since NSOM data are very sensitive to the probe-to-sample distance, the use of NSOMs for the examination of soft samples has been largely limited to biological samples that were chemically fixed, limiting the use of NSOM for measuring dynamics. However, recent advances in the control of the NSOM probe-to-sample distance have enabled near-field fluorescence correlation spectroscopy (FCS) on fluid SLBs93 and live cell membranes.58 FCS has proven to be a powerful optical technique for probing nanoscale membrane phenomena and will be even more useful when fully integrated with nanoscale optical methods.33 On live cells, 120 nm diameter apertures on NSOM tips have provided 20× faster residence time in the illumination spot than confocal optics and observed varying diffusion of fluorescently tagged phosphoethanolamine lipids and sphingolipids within the membrane.58 Continued developments in controlling the probe-to-sample distance will ensure minimal topographic artifacts or disruption of soft samples.
Nanoscale apertures in a planar aluminum film have been fabricated to provide optical confinement without the complications of incorporating a scanning probe (Fig. 1). Illumination volumes as small as 10−21 L have been made within arrays of hollow apertures in a planar metallic film, called zero-mode waveguides (ZMWs).54 This approach uses an opaque metallic film on the glass substrate to block most of the excitation light from penetrating into the bulk of the sample and only excites fluorophores at the bottom of the ZMWs (i.e. at the metal-glass interface). ZMWs provide 1,000× smaller illumination volumes than diffraction-limited total internal reflection microscopy and single molecule resolution at concentrations of up to 1 µM.60 As fluorophores diffuse on the membrane in and out of the ZMW, bursts of fluorescence emission are observed. The duration of the fluorescence emission for each fluorophore is dependent on the fluorophore diffusion rate and the ZMW dimensions, as seen with both single fluorescence burst analysis and autocorrelations in time via FCS. ZMWs have been used to examine model and living membranes, requiring that the membrane itself penetrates into the hollow aperture and membrane-bound fluorophores on the membrane are observed only at the bottom of the ZMW.15,94 The applicability of ZMWs to membranes can be complicated by the required membrane curvature for penetration into the ZMW and the lateral diffusion of large membrane-bound structures (e.g. lipid-rafts) may be altered.62 However, the binding of molecules from solution to the membrane is well suited for study in ZMWs; the binding constant of tetanus toxin C to the ganglioside GT1B has been determined to be 390 ± 40 nM−1 by monitoring single-molecule binding events at high concentrations (500 nM) of tetanus toxin C.77 This experiment utilizes the small illumination volume in ZMWs to obtain single-molecule resolution, whereas diffraction-limited optics would have too many fluorophores simultaneously illuminated to be useful.
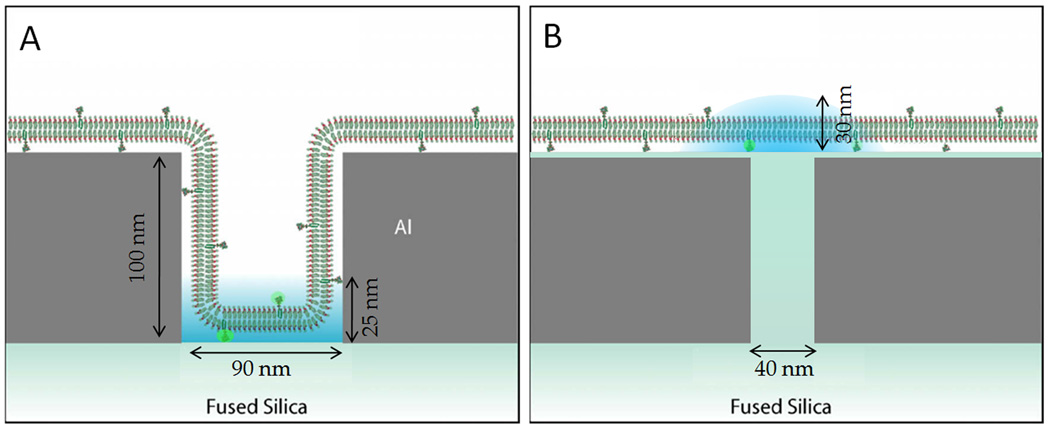
Figure 1.
(A) Zero-mode waveguides (ZMWs) and (B) planarized apertures utilize nanofabricated metallic structures to limit the illumination for fluorescence spectroscopy to sub-diffraction-limited areas. ZMWs illuminate the membrane at the bottom of the aperture while planarized apertures provide 60 nm resolution of planar membranes. (Adapted with permission from (A) Refs. 60 and 77 and (B) Ref. 50)
Recognition of the limitations of the scanning probe in NSOMs and the membrane curvature effects in ZMWs has encouraged the development of planarized apertures for near-field optical microscopy (PANOMs).50 PANOMs are conceptually equivalent to glass-filled ZMWs. However, the excitation light of PANOMs transmits through the aperture and the emission fluorescence is collected from above, without requiring the emission to transmit back through the aperture. PANOMs provide a confined, near-field illumination profile, like an NSOM, but can be fabricated on an array for numerous simultaneously illuminated apertures with reliably controlled aperture-to-membrane distances, like ZMWs (Fig. 1). The key aspects of PANOMs are their 50 nm diameter apertures, which provide 60 nm full width at half maximum illumination profiles, and their planarized surface, which provides unperturbed membrane diffusion, for both SLBs and live cells.50 Additionally, PANOMs provide the ability to examine apertures of varying sizes and numerous chromatically distinct fluorophores simultaneously with conventional excitation lasers and fluorophores, without inducing membrane curvature or incorporating a scanning probe.
SURFACE PLASMONS FOR OPTICAL ENHANCEMENT
Rather than using a metallic aperture to limit the regions of the sample exposed to the excitation light, it is possible to excite the electron plasmon within a metal to yield increased intensities of illumination in close proximity to the metal. Plasmon excitations have been used for wide-field spectroscopic sensors, near-field fluorescence enhancement, or surface-enhanced Raman spectroscopy (SERS). All of these techniques depend on the fabrication of nanoscale metallic structures. Because electric fields are enhanced at sharp metal corners and at the gaps between metallic structures, control of the metal size and shape are necessary for plasmonic devices.
With spectroscopic techniques, a change in surrounding the index of refraction (i.e. the binding and unbinding of biomolecules) affects the resonant frequency of light transmission through the metallic nanostructure. Nanoscale apertures in metallic films are the most commonly used structures to spectroscopically study membranes, however, metal nanoparticles,22 spherical micro-resonators,2 and conventional surface plasmon spectroscopy (SPR) have also been employed for label-free detection of biomolecules (un)binding to the metal or associated membranes. High-sensitivity plasmonic devices can be created with inexpensive colloidal lithography10 or precisely nanostructured surfaces for greater capabilities,23 and through-aperture microfluidics.43
Current technology now permits real-time, label-free analysis of membrane formation or absorption of proteins, such as α-hemolysin39 and head-group-specific binding proteins.22 Recent optimizations of biomolecules on plasmonic devices have enabled binding only to the most sensitive regions of the detector.11,20 The physical capabilities of spectroscopic plasmonic devices have been well reviewed26,59,76 and some review articles have highlighted plasmonic applications for membranes.34,75
Rather than monitoring the wavelength-dependent resonances of nanoscale metallic structures, designing a plasmonic device to couple into the excitation or emission of a fluorophore enables metal-enhanced fluorescence (MEF) or fluorescence quenching in the local proximity of the metal.52 MEF has been applied to membranes both with metallic structures on a surface as well as metallic structures on a scanning probe.67 On a scanning probe similar to NSOM, aperture-free near-field point probes enhance the excitation light to the 30 nm region surrounding the nanofabricated metallic antenna while illuminated and observed in the far-field (Fig. 2).101 Near-field point probes have been used to image individual membrane-bound calcium pumps35 and nanodomains of lymphocyte function-assisted antigen-1 (LFA-1).101 Scanning point probes may be easier to fabricate than apertures, but their use can be complicated by the higher background fluorescence due to far-field illumination. Obviously, the experimental setups for NSOMs and scanning point probes are related to those for atomic force microscopy (AFM). AFM has recently made significant improvements in high-resolution and high-speed imaging of membranes, including revealing nanoscale protein structures within membranes.5 Substantial technological advances are imminent as these technical improvements recently made for AFMs become better integrated with scanning probes utilizing near-field optics.
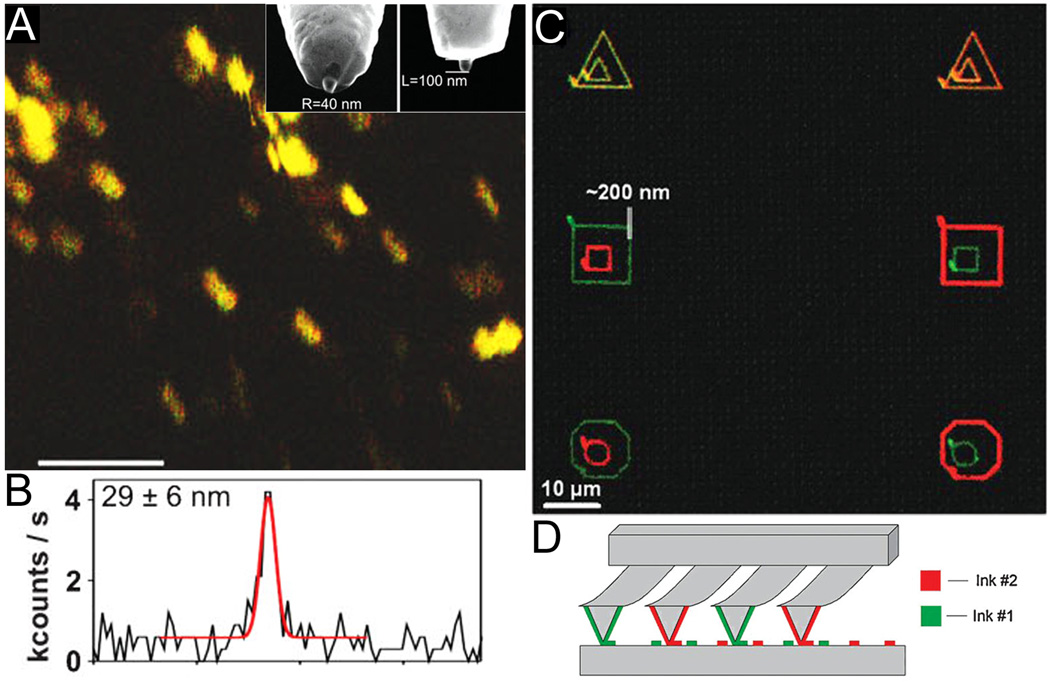
Figure 2.
Scanning probes have been used both for nanoscale imaging and patterning of membranes. (A) Fluorescence enhancement at the tip of nano-antenna on a scanning probe provides 30 nm resolution of membrane-bound integrin LFA-1; scale bar = 1 µm. A SEM image of the nano-antenna is shown in the inset of (A). (B) A line scan of fluorophore emission intensity vs. distance demonstrates 29 nm resolution. (C) SLBs of varying composition have been simultaneously patterned with dip-pen lithography at 200 nm resolution. (D) Multiplexed “pens” with varying composition of lipid-containing inks allow for simultaneous writing of many patterns and combinatorial mixing of the membrane components. (Reprinted with permission from (A,B) Ref. 101 and (C,D) Ref. 78.)
Just as strong electric fields from plasmon resonances can enhance or quench fluorescence, they can also be used for label-free SERS. By observing an energy change in scattered light due to molecular vibrations, Raman spectroscopy can provide molecular fingerprints of the sample.83 Coupling molecular vibrations with surface plasmon resonances increases the intensity of the Raman signal to better decipher it from the background fluorescence and scattering. Nanofabrication has advanced the use of SERS, but applications of SERS to membranes are rare.12 SERS of live cells has provided improved resolution in recent years and may develop into a powerful analytical tool as the capabilities of nanofabrication are better utilized.1
RESTRICTING MEMBRANE DIFFUSION
By imposing restrictions on membrane diffusion, nanofabrication has provided a tool for both understanding membranes as well as using membranes to understand other biophysical phenomena. For example, studying lipid diffusion around nanofabricated channels90,95 and nanogates66 has provided insights of membrane diffusion on complex surfaces. Carefully designed surface patterns may be applied for selective sorting of membrane components based on molecular mobility, size, or charge.71
Metallic barriers approximately 100 nm wide and 30 nm tall have been used to limit the extent of lipid diffusion by isolating supported lipid bilayers within rectangular corrals. These membrane corrals have been used to explore the how T cells interact with antigen presenting cells and rearrange their initially uniformly distributed membrane components for the formation of a structured immunological synapse (Fig. 3).57,65 Corralled SLB-bound signaling molecules have been used to mimic antigen presenting cells. Varying the concentration of antigens in the SLB and the dimensions of the membrane corrals provides a means for exploring key properties of antigen presenting cells for proper formation of the immunological synapse. This experimental method enables the investigation of the critical number, density, and mobility of signaling molecules necessary for these fundamental membrane phenomena.64,32
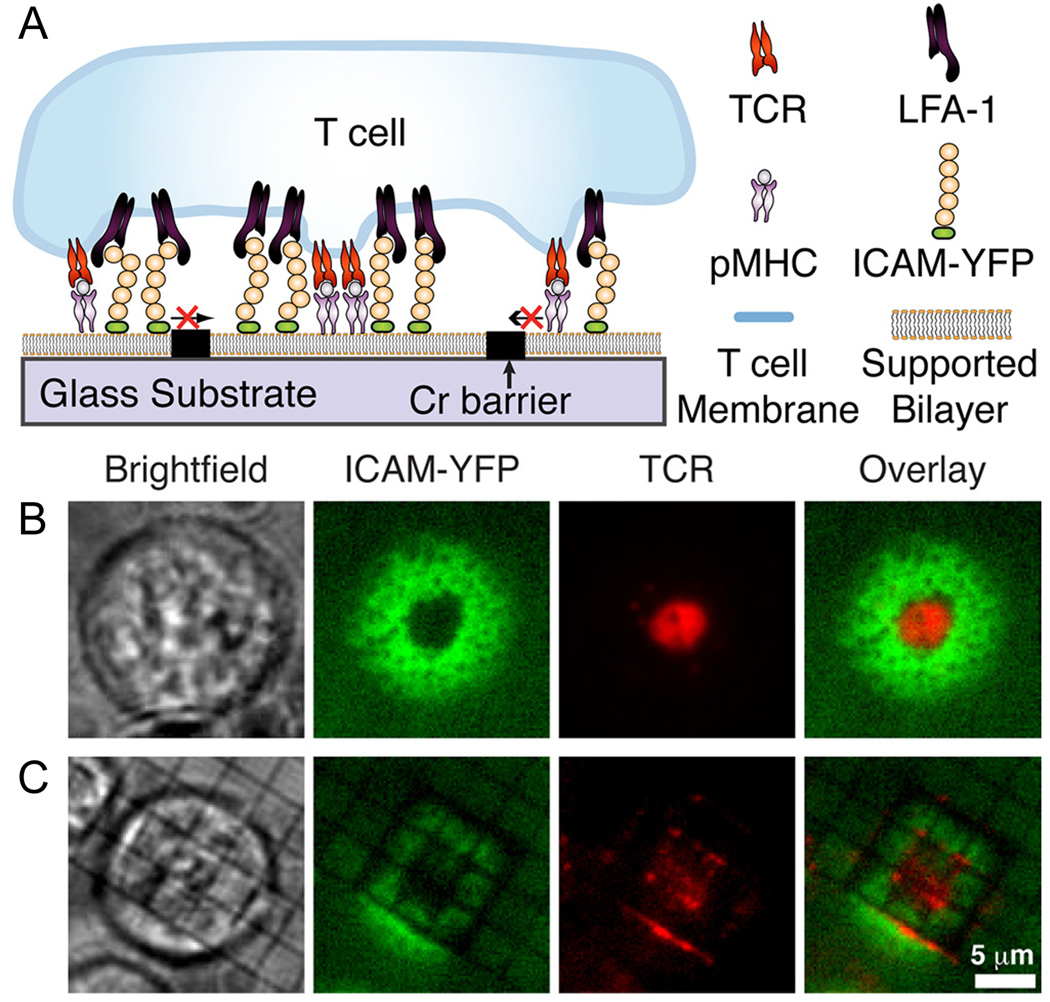
Figure 3.
Membrane reorganization is induced upon the binding of T cells to antigen presenting cells, forming an immunological synapse. (A) Key elements of antigen presenting cells (i.e. the peptide-major histocompatiability complex (pMHC) and the inter-cellular adhesion molecule (ICAM)) have been incorporated into a SLB and reorganized upon binding to T cell receptors (TCR) and to lymphocyte function-associated antigen 1 (LFA-1), respectively. The (B) presence or (C) absence of the metallic barriers to diffusion results in distinctly different morphology of the resulting immunological synapse as the T cell attempts to concentrate the TCRs within a ring of ICAMs. (Reprinted with permission from Ref. 32.)
With similar techniques, SLB-bound DNA has been utilized in a microfluidic channel to explore the biophysical properties of DNA and its associated proteins (Fig. 4).27 DNA was confined to the membrane with multivalent neutravidin after conjugation of DNA and lipids separately to biotin. Through careful control of the nanoscale metallic structures and hydrodynamic forces within the microfluidic channel, single molecules of DNA have been precisely localized for individual inspection.18,19,92 This technique has revealed the effects of hydrodynamic force on DNA-protein interactions,25 the energetics of nucleosome deposition,91 and protein disruption by DNA translocase.21 Similar techniques of immobilizing biomolecules to the plane of a SLB may prove to be a value tool for high-throughput, single-molecule experiments.
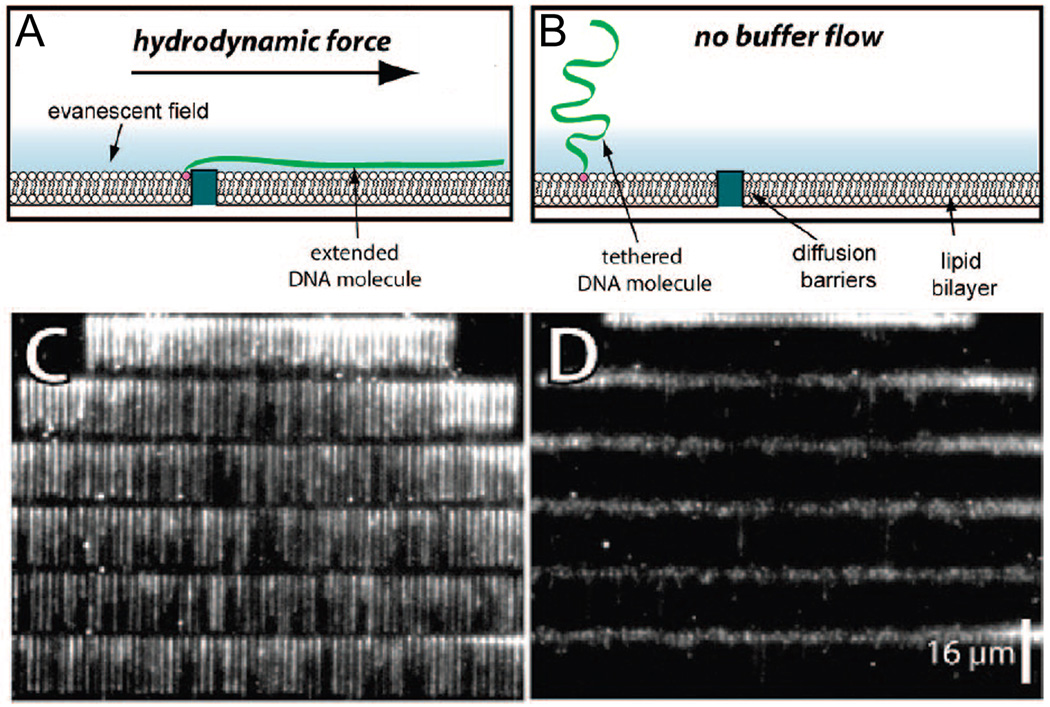
Figure 4.
Supported lipid bilayers and nanofabricated metallic barriers have been used for high-throughput single-molecule analysis of DNA. DNA molecules are bound to lipid molecules and analyzed with total-internal reflection fluorescence microscopy, as shown schematically (A) with and (B) without a hydrodynamic force confining them to metallic barriers. Isolated molecules 48.5 kb λ DNA stained with YOYO1 can be seen in a single image (C) with and (D) without flow. (Reprinted with permission from Ref. 92. Copyright 2008 American Chemical Society.)
In place of metallic barriers, polymerizable lipids and surface immobilized lipids have been incorporated into SLBs to restrict lipid diffusion. These barriers are formed post membrane formation and have primarily led to basic science explorations such as quantifying the effects of polymerization on diffusion and induced phase separation of membrane components at the micron scale.63,68 Surface-immobilized monolayers of n-octadecylsiloxanes37 and chlorotrimethylsilanes31 have provided a means of patterning just the bottom bilayer leaflet. With electron beam lithography, nanoscale membrane patterns have been created with silanes conjugated to amphiphilic lipid molecules45 or cholesterol derivatives30 through immobilization to nanoscale patterned noble metals. Sub-diffraction-limited, near-field illumination may provide a means of selectively polymerizing membrane components with nanoscale resolution on living samples. Methods of efficiently patterning and immobilizing nanoscale regions of SLBs would be applicable to a variety of biophysical experiments on SLBs, live cells, and other biomolecules.
PATTERNING MEMBRANE COMPOSITION
Selectively patterning membranes has permitted numerous studies in membrane biology and has been applied towards controlling cellular adhesion or exposure to biomolecules.69,75 Three general methods of patterning membranes have been broadly developed: (1) chemically altering regions of the homogeneous membrane to provide selective removal and exchange of the (un)altered regions, (2) forming the membrane on a pre-patterned substrate for selective membrane adsorption or removal, or (3) mechanically forming the membrane only in specific areas. These techniques have provided great advances at the micrometer scale, but membrane patterning at the submicron scale is still in its infancy. The techniques described here that provide the most potential for membrane patterning include polymer lift-off, pre-patterned substrate wells, microcontact stamping, and scanning-probe lithography.
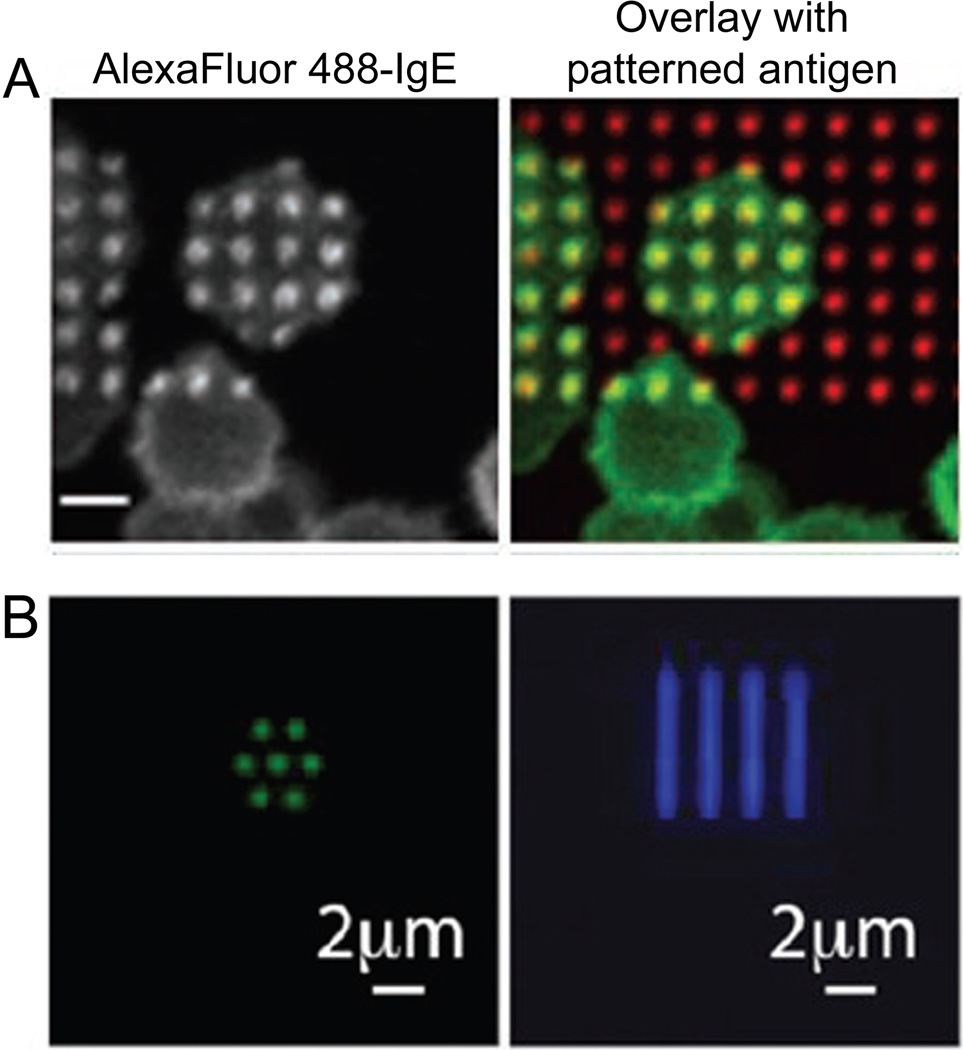
Among the most adept techniques for patterning membranes and biomolecules is polymer lift-off.85,89 The polymer lift-off method begins with conformally coating a substrate with a polymer, such as Parylene, and selectively etching through the polymer in specific areas. The entire substrate and polymer are then coated with lipids or other biomolecules and the polymer is peeled off to leave biomolecules only in the areas in which the polymer was previously etched. Membrane patterning via polymer lift-off at the micron scale has been used to create multiplexed method of monitoring membrane binding61 and local regions of antigen presentation to explore the signaling cascade in mast cells (Fig. 5).70 By confining the ligand presentation area to a patterned membrane, the downstream membrane-mediated IgE signaling cascade in mast cells was monitored, including the recruitment of signaling kinases,98 ordered-phase lipids,98 and cytoskeleton-adapting proteins,88 as well as localization of the resulting secretory granuals.97 Recent advances in polymer lift-off have demonstrated the patterning of biomolecules with parylene with 90 nm size patterns, but have yet to be applied to membranes.86 Advancements in polymer peel-off of membranes with nanoscale resolution would allow greater understandings of the membrane-mediated signaling cascade in addition to higher-throughput model membrane binding assays for drug discovery.
Figure 5.
Membranes and biomolecules have been patterned with polymer lift-off. (A) Micron scale patterns of antigen-incorporating SLBs recruit IgE on live cell membranes for spatial control of membrane-mediated cell signaling; scale bar = 10 µm. (B) Biomolecules, such as fluorescently-labeled antibodies, have been patterned with a resolution of 90 nm via polymer lift-off and electron beam lithography in nanoscale dots and lines. Polymer lift-off holds promise for nanoscale patterning of membranes with high-throughput and high-resolution. ((A) Reprinted with permission from Ref. 98. Copyright 2004 National Academy of Sciences, U.S.A. (B) Reprinted with permission from Ref. 86. Copyright 2010 American Chemical Society.)
Rather than lifting-off unwanted membrane material, recent developments have enabled a “squeegee”-based technique for scraping away membrane material that did not settle into predefined wells in the substrate.96 By pre-patterning 80 nm diameter wells into a silicon wafer and allowing small vesicles of native cell membrane extract to settle into the wells, Wittenberg et al. were able to remove membrane material not within the well via a squeegee technique. This yielded an array of membrane material within the array of wells that could be examined via fluorescence emission or label-free plasmonic spectra effects for high-throughput analysis of binding of analytes to the membrane.96
Rather than patterning a membrane after coating the full surface with a SLB, some techniques form the membrane only in defined regions, such as microcontact printing, robotic spotting, ink-jet printing, or scanning-probe-based lithography. Commercially available robotic arrayers and ink-jet printers have been used to pattern SLBs, but they traditionally have resolution significantly greater than 1 µm and moderate reproducibility.17 Microcontact printing on membranes has been well used for both adding and subtracting lipids in SLBs.36,46 Microcontact printing works much like a conventional rubber stamp; a textured stamp is pressed against a flat substrate and the substrate surface is modified with the addition or subtraction of material where the stamp and the surface made contact, depending on the initial materials on the stamp and the substrate. Microcontact printing provides the ability to efficiently pattern large substrates quickly with a reusable stamp. Additionally, microcontact printing has been used to define regions of adsorbed proteins as a barrier to membrane diffusion and the selective adhesion of live cells.47,51 However, achieving reproducible nanoscale features can be challenging due to wear on the sample and residue left on the substrate by some stamp materials, such as polydimethylsiloxane (PDMS).24
Scanning probe lithography has been demonstrated to 100 nm resolution with both the additive technique of dip-pen nanolithography72 and the subtractive technique of nanoshaving lithography.40,79 Lithography on membranes with a scanning probe utilizes a piezo-controlled pointed nanoscale tip to make contact with the surface only at specific locations. If lipids or biomolecules are initially coating the surfaces, the tip can remove material to pattern the surface. If the surface is initially clean and the tip is coated with a biomolecule-containing ink, the tip can be used to write the biomolecules on the surface at select locations. Massively parallel, multi-component arrays of lipids have been created at a resolution of 200 nm, and have been used to study nanoscale cellular adhesion (Fig. 2).53,78 Dip-pen nanolithography is among the best methods for patterning membranes at the nanoscale and future developments may allow scanning-probe lithography to be a standard, high-throughput nanofabrication technique.
SORTING MEMBRANE COMPONENTS
Many unknowns of membrane biology revolve around the stoichiometry and interactions of membrane components. Numerous bulk techniques have been used to separate membrane proteins based on their local membrane phase, such as detergent resistance and gel electrophoresis.80 However, these techniques provide little information as to the lipid-mediated effects on the protein function, often cause protein denaturation, and hinder analysis of the protein functionality.13 In recent years, now standard microscopy assays have been developed to determine co-localization with multi-colored fluorescence microscopy at diffraction-limited resolution. By fluorescently labeling both a molecule with an unknown membrane phase preference and a known liquid-order preferring membrane component with a spectrally distinctly fluorophores, the colocalization and similarity in phase preferences can be assessed. Most commonly, fluorescently labeled cholera toxin subunit B (CTxB) is used to label the liquid-ordered phase regions of the membrane since CTxB binds specifically to GM1, a liquid-order preferring ganglioside. However, this technique suffers from two key limitations. First, traditional fluorescence techniques are limited to diffraction-limited resolution and are unable to yield information on the size of membrane microdomains smaller than 200 nm. Secondly, the pentameric CTxB is known to induce phase separation of membrane components in some model systems29 and its binding to the membrane may perturb the natural balance of liquid-ordered and liquid-disordered components in living membranes.
Nanofabrication is overcoming these limitations by providing novel means of sorting membrane components based on their lipid tail order, electrophoretic mobility, and hydrodynamic mobility. The use of laminar flow to create a distribution of lipid phases within a microfluidic channel have provided a mechanism of separating lipids and membrane proteins based on their partition coefficient between liquid-ordered and liquid-disordered lipid phases in a microfluidic channel.7 Electrophoretically, membrane components have been separated based on size and charge on both model membranes42,74 and live cells.73 Numerous researchers have exploited nanofabricated metallic structures to apply strong electric fields across small membranes areas and have demonstrated sorting of fluorescent lipids,14,28 SLB-tethered vesicles,99 streptavidin,31 and polyhistidine-tagged proteins87 (Figs. 6 and 7). The removal of select separated membrane components is occasionally performed and these techniques have significant potential once complex cellular membranes are better integrated into these controlled SLB-based separations.14,48
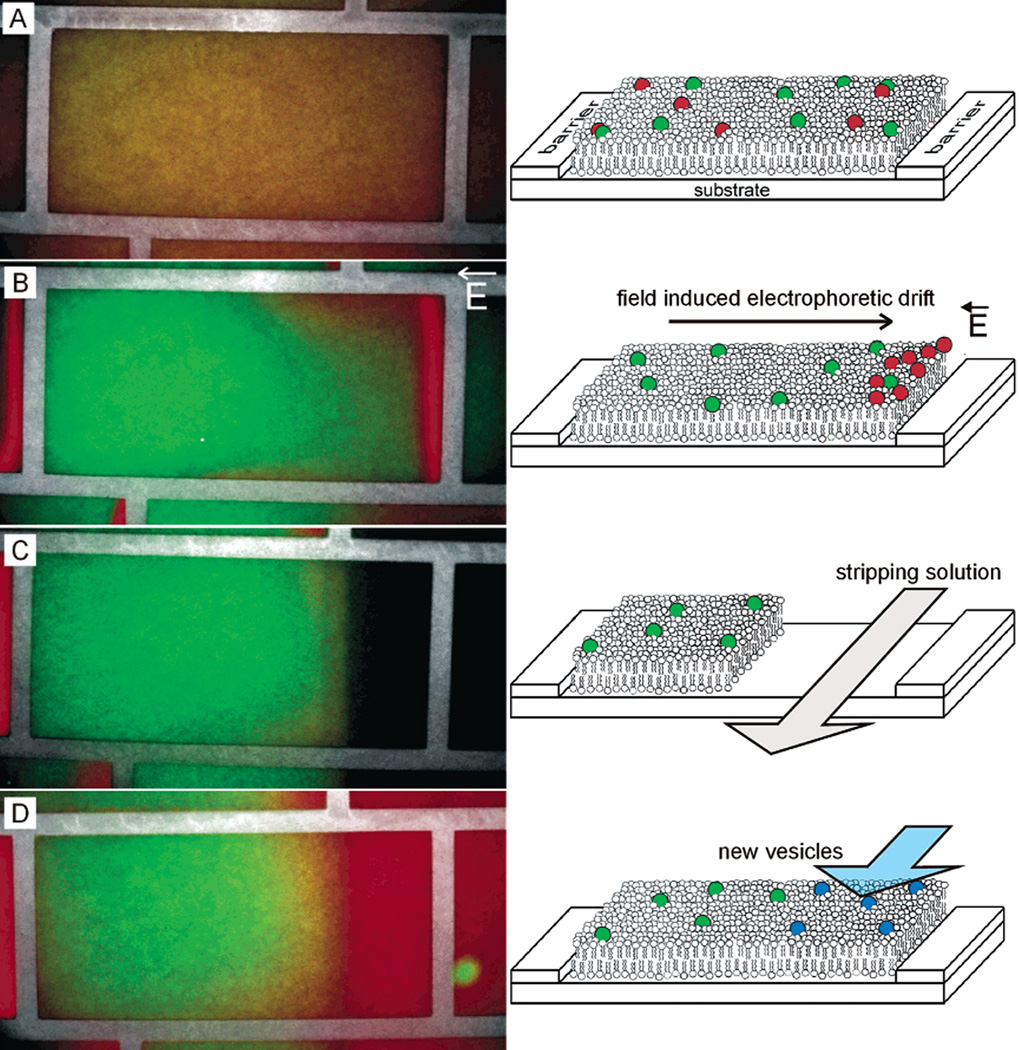
Figure 6.
The incorporation of electrophoresis and metallic barriers provide a mechanism of concentrating, separating, and purifying membrane components. (A) Initially mixed SLB components can be (B) separated by electrophoresis within the corral. (C) Controlled laminar flow can expose only a portion of the SLB to a stripping solution to remove the exposed SLB from the surface. (D) The partially stripped SLB can then be refilled via vesicle fusion to reform a complete SLB with potentially new components. SLBs are composed 97.5% zwitterionic egg phosphocholine lipids, 0.5% negatively charged Texas Red-phosphoethanolamine lipids (red), and 2% neutral NBD-phosphocholine lipids (green). (Reprinted with permission from Ref. 48. Copyright 2003 American Chemical Society.)
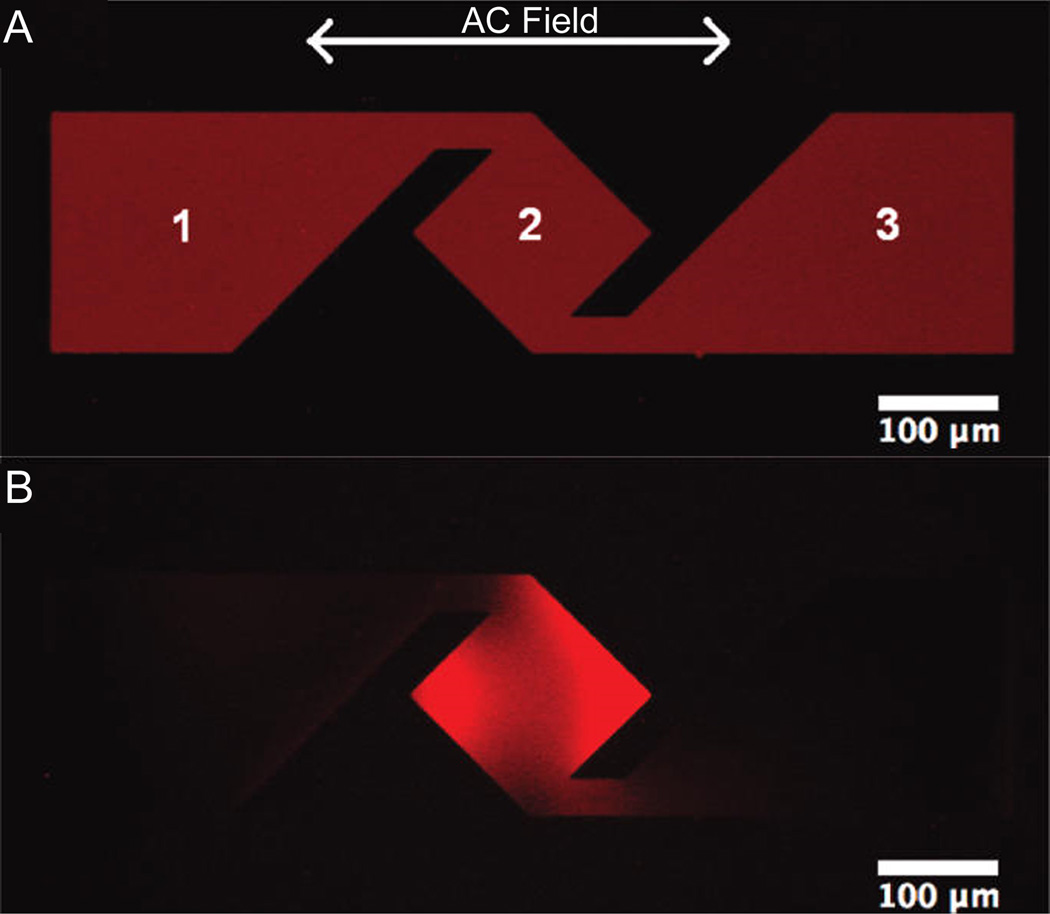
Figure 7.
Incorporating electric fields with complex metallic structures provides a means of concentrating membrane components. (A) Initially homogeneous SLBs diffuse uniformly around the metallic structures, but application of alternating electric fields may concentrate select components (B). SLBs are 99.8% zwitterionic phosphocholine lipids and 0.2% negatively charged Texas Red-phosphoethanolamine lipids. (Reprinted with permission from Ref. 8. Copyright 2011 American Chemical Society.)
Current trends in membrane separations include performing electrophoresis on substrates with complex fabricated barriers for more efficiently separating and concentrating membrane components,8,71 integrating electrodes into the substrate,41 or incorporating differences in hydrodynamic drag to separate membrane components.44 In particular, the recently developed methodology of incorporating native cell membranes into continuous SLBs will allow for analysis and sorting of components within complex membranes with these nanofabricated structures.82 These techniques have great potential to advance membrane biology through the on-chip manipulation of membrane components for diverse scientific and medical applications.
CONCLUSIONS AND OUTLOOK
When these nanofabrication methodologies prove to be effective and reliable, they may quickly become incorporated into numerous methods for drug and biomarker discovery. For example, surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) is currently being employed for biomarker discovery from biological fluids, but is limited in its resolution of scarce markers with detection limits of µg/mL.6 Similarly, spectrally sensitive plasmonic structures have the potential to detect the binding of regents to membranes while maintaining label-free targets and binding candidates. But controlling the application of native membranes onto plasmonic structures has yet to be reliably demonstrated. Techniques such as these are on the threshold of being widely used as nanofabrication and have the potential to incorporate native membrane and concentrate proteins of interest, opening the door to integration with other analysis and characterization tools.
The use of nanopatterning has permitted observation of membranes with high spatial resolution and high sensitivity to slight changes in the membrane. Single-molecule fluorescence experiments can now be performed with 30 nm resolution using conventional fluorophores and without high-powered lasers. As these techniques for sub-diffraction-limited resolution advance, greater detail of the organization and dynamics of model and live cell membranes will be revealed. Similarly, nanopatterned substrates for controlled diffusion of membrane-bound lipids and proteins will continue to be utilized as a tool for understanding a variety of biological processes. Current techniques for patterning membrane components are of frequent use in biosensors and controlling cellular adhesion, but are primarily limited to micron dimensions. Nanoscale membrane patterning will permit higher resolution, greater throughput, more efficient material use, and better membrane localization for biosensors. As nanofabrication techniques are further developed, biologists, engineers, physicians, and patients will all benefit by improved abilities to observe and control of membranes.
ACKNOWLEDGMENTS
C.V.K. is supported by a NIH Kirschstein National Research Service Award postdoctoral fellowship (F32-GM092106). Barbara Baird and David Holowka are gratefully acknowledged for their frequent support and advice.
REFERENCES
- 1.Ba H, Rodríguez-Fernández J, Stefani FD, Feldmann J. Immobilization of Gold Nanoparticles on Living Cell Membranes upon Controlled Lipid Binding. Nano Lett. 2010;10:3006–3012. doi: 10.1021/nl101454a. [DOI] [PubMed] [Google Scholar]
- 2.Beche B, Potel A, Barbe J, Vie V, Zyss J, Godet C, Huby N, Pluchon D, Gaviot E. Resonant coupling into hybrid 3D micro-resonator devices on organic/biomolecular film/glass photonic structures. Opt Commun. 2010;283:164–168. [Google Scholar]
- 3.Betzig E, Trautman JK, Harris TD, Weiner JS, Kostelak RL. Breaking the Diffraction Barrier: Optical Microscopy on a Nanometric Scale. Science. 1991;251:1468–1470. doi: 10.1126/science.251.5000.1468. [DOI] [PubMed] [Google Scholar]
- 4.Betzig E, Trautman JK. Near-Field Optics: Microscopy, Spectroscopy, and Surface Modification Beyond the Diffraction Limit. Science. 1992;257:189–195. doi: 10.1126/science.257.5067.189. [DOI] [PubMed] [Google Scholar]
- 5.Bippes CA, Muller DJ. High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Reports on Progress in Physics. 2011;74:086601. [Google Scholar]
- 6.De Bock M, de Seny D, Meuwis M-A, Chapelle J-P, Louis E, Malaise M, Merville M-P, Fillet M. Challenges for biomarker discovery in body fluids using SELDI-TOF-MS. J. Biomed. Biotechnol. 2010;2010:906082. doi: 10.1155/2010/906082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao L, Daniel S. Measuring the Partitioning Kinetics of Membrane Biomolecules using Patterned Two-phase Coexistant Lipid Bilayers. J Am Chem Soc. 2011;133:15635–15643. doi: 10.1021/ja205274g. [DOI] [PubMed] [Google Scholar]
- 8.Cheetham MR, Bramble JP, McMillan DGG, Krzeminski L, Han X, Johnson BRG, Bushby RJ, Olmsted PD, Jeuken LJC, Marritt SJ, Butt JN, Evans SD. Concentrating Membrane Proteins Using Asymmetric Traps and AC Electric Fields. J Am Chem Soc. 2011;133:6521–6524. doi: 10.1021/ja2007615. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Lagerholm BC, Yang B, Jacobson K. Methods to measure the lateral diffusion of membrane lipids and proteins. Methods. 2006;39:147–153. doi: 10.1016/j.ymeth.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Dahlin AB, Chen S, Jonsson MP, Gunnarsson L, Kall M, Hook F. High-Resolution Microspectroscopy of Plasmonic Nanostructures for Miniaturized Biosensing. Anal Chem. 2009;81:6572–6580. doi: 10.1021/ac901175k. [DOI] [PubMed] [Google Scholar]
- 11.Dahlin AB, Jonsson MP, Hook F. Specific self-assembly of single lipid vesicles in nanoplasmonic apertures in gold. Adv Mater. 2008;20:1436-+. [Google Scholar]
- 12.Dahlin A, Zach M, Rindzevicius T, Kall M, Sutherland DS, Hook F. Localized surface plasmon resonance sensing of lipid-membrane-mediated biorecognition events. J Am Chem Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- 13.Daniel S, Chao L. Interfaces and Interphases in Analytical Chemistry. American Chemical Society; 2011. Supported Lipid Bilayer Electrophoresis for Separation and Analytical Studies of Cell Membrane Biomolecules; pp. 99–121. [Google Scholar]
- 14.Daniel S, Diaz AJ, Martinez KM, Bench BJ, Albertorio F, Cremer PS. Separation of membrane-bound compounds by solid-supported bilayer electrophoresis. J Am Chem Soc. 2007;129:8072-+. doi: 10.1021/ja0720816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edel JB, Wu M, Baird B, Craighead HG. High spatial resolution observation of single-molecule dynamics in living cell membranes. Biophys J. 2005;88:L43–L45. doi: 10.1529/biophysj.105.061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457 doi: 10.1038/nature07596. 1159-U121. [DOI] [PubMed] [Google Scholar]
- 17.Fang Y, Frutos AG, Lahiri J. Membrane protein microarrays. J Am Chem Soc. 2002;124:2394–2395. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 18.Fazio TA, Visnapuu M-L, Greene EC, Wind SJ. Fabrication of nanascale “curtain rods” for DNA curtains using nanoimprint lithography. J Vac Sci Technol B. 2009;27:3095–3098. doi: 10.1116/1.3259951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazio T, Visnapuu M-L, Wind S, Greene EC. DNA Curtains and Nanoscale Curtain Rods: High-Throughput Tools for Single Molecule Imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuz L, Jönsson P, Jonsson MP, Höök F. Improving the Limit of Detection of Nanoscale Sensors by Directed Binding to High-Sensitivity Areas. ACS Nano. 2010;4:2167–2177. doi: 10.1021/nn901457f. [DOI] [PubMed] [Google Scholar]
- 21.Finkelstein IJ, Visnapuu M-L, Greene EC. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468:983–987. doi: 10.1038/nature09561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galush WJ, Shelby SA, Mulvihill MJ, Tao A, Yang P, Groves JT. A Nanocube Plasmonic Sensor for Molecular Binding on Membrane Surfaces. Nano Lett. 2009;9:2077–2082. doi: 10.1021/nl900513k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Yang J-C, Lin JY, Stuparu AD, Lee MH, Mrksich M, Odom TW. Using the Angle-Dependent Resonances of Molded Plasmonic Crystals To Improve the Sensitivities of Biosensors. Nano Lett. 2010;10:2549–2554. doi: 10.1021/nl101165r. [DOI] [PubMed] [Google Scholar]
- 24.Glasmastar K, Gold J, Andersson AS, Sutherland D, Kasemo B. Silicone transfer during microcontact printing. Langmuir. 2003;19:5475–5483. [Google Scholar]
- 25.Gorman J, Fazio T, Wang F, Wind S, Greene EC. Nanofabricated Racks of Aligned and Anchored DNA Substrates for Single-Molecule Imaging. Langmuir. 2010;26:1372–1379. doi: 10.1021/la902443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gramotnev DK, Bozhevolnyi SI. Plasmonics beyond the diffraction limit. Nat Photon. 2010;4:83–91. [Google Scholar]
- 27.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu M-L. Dna Curtains for High-Throughput Single-Molecule Optical Imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groves JT, Wülfing C, Boxer SG. Electrical manipulation of glycan-phosphatidyl inositol-tethered proteins in planar supported bilayers. Biophys J. 1996;71:2716–2723. doi: 10.1016/S0006-3495(96)79462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc Natl Acad Sci U S A. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han XJ, Achalkumar AS, Bushby RJ, Evans SD. A Cholesterol-Based Tether for Creating Photopatterned Lipid Membrane Arrays on both a Silica and Gold Surface. Chemistry. 2009;15:6363–6370. doi: 10.1002/chem.200900404. [DOI] [PubMed] [Google Scholar]
- 31.Han X, Cheetham MR, Sheikh K, Olmsted PD, Bushby RJ, Evans SD. Manipulation and charge determination of proteins in photopatterned solid supported bilayers. Integr Biol. 2009;1:205. doi: 10.1039/b815601h. [DOI] [PubMed] [Google Scholar]
- 32.Hartman NC, Nye JA, Groves JT. Cluster size regulates protein sorting in the immunological synapse. Proc Natl Acad Sci U S A. 2009;106:12729–12734. doi: 10.1073/pnas.0902621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He H-T, Marguet D. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu Rev Phys Chem. 2011;62:417–436. doi: 10.1146/annurev-physchem-032210-103402. [DOI] [PubMed] [Google Scholar]
- 34.Höök F, Stengel G, Dahlin AB, Gunnarsson A, Jonsson MP, Jönsson P, Reimhult E, Simonsson L, Svedhem S. Supported lipid bilayers, tethered lipid vesicles, and vesicle fusion investigated using gravimetric, plasmonic, and microscopy techniques. Biointerphases. 2008;3:FA108. doi: 10.1116/1.2948313. [DOI] [PubMed] [Google Scholar]
- 35.Höppener C, Novotny L. Antenna-based optical imaging of single Ca2+ transmembrane proteins in liquids. Nano Lett. 2008;8:642–646. doi: 10.1021/nl073057t. [DOI] [PubMed] [Google Scholar]
- 36.Hovis JS, Boxer SG. Patterning barriers to lateral diffusion in supported lipid bilayer membranes by blotting and stamping. Langmuir. 2000;16:894–897. [Google Scholar]
- 37.Howland M, Sapuri-Butti A, Dixit S, Dattelbaum A, Shreve A, Parikh A. Phospholipid morphologies on photochemically patterned silane monolayers. J Am Chem Soc. 2005;127:6752–6765. doi: 10.1021/ja043439q. [DOI] [PubMed] [Google Scholar]
- 38.Hwang J, Gheber LA, Margolis L, Edidin M. Domains in cell plasma membranes investigated by near-field scanning optical microscopy. Biophys J. 1998;74:2184–2190. doi: 10.1016/S0006-3495(98)77927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh SH. Membrane protein biosensing with plasmonic nanopore arrays and pore-spanning lipid membranes. Chem Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson BL, Groves JT. Scanning probe lithography on fluid lipid membranes. J Am Chem Soc. 2004;126:13878–13879. doi: 10.1021/ja046040a. [DOI] [PubMed] [Google Scholar]
- 41.Jackson BL, Nye JA, Groves JT. Electrical Manipulation of Supported Lipid Membranes by Embedded Electrodes. Langmuir. 2008;24:6189–6193. doi: 10.1021/la800040w. [DOI] [PubMed] [Google Scholar]
- 42.Jaffe LF. Electrophoresis along cell membranes. Nature. 1977;265:600–602. doi: 10.1038/265600a0. [DOI] [PubMed] [Google Scholar]
- 43.Jonsson MP, Dahlin AB, Feuz L, Petronis S, Höök F. Locally Functionalized Short-Range Ordered Nanoplasmonic Pores for Bioanalytical Sensing. Anal Chem. 2010;82:2087–2094. doi: 10.1021/ac902925e. [DOI] [PubMed] [Google Scholar]
- 44.Jönsson P, Gunnarsson A, Höök F. Accumulation and Separation of Membrane-Bound Proteins Using Hydrodynamic Forces. Anal Chem. 2011;83:604–611. doi: 10.1021/ac102979b. [DOI] [PubMed] [Google Scholar]
- 45.Jung M, Vogel N, Koper I. Nanoscale Patterning of Solid-Supported Membranes by Integrated Diffusion Barriers. Langmuir. 2011;27:7008–7015. doi: 10.1021/la200027e. [DOI] [PubMed] [Google Scholar]
- 46.Jung SY, Holden MA, Cremer PS, Collier CP. Two-component membrane lithography via lipid backfilling. Chem Phys Chem. 2005;6:423–426. doi: 10.1002/cphc.200400540. [DOI] [PubMed] [Google Scholar]
- 47.Kam L, Boxer SG. Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. J Biomed Mater Res. 2001;55:487–495. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 48.Kam L, Boxer SG. Spatially Selective Manipulation of Supported Lipid Bilayers by Laminar Flow: Steps Toward Biomembrane Microfluidics. Langmuir. 2003;19:1624–1631. [Google Scholar]
- 49.Kataoka-Hamai C, Miyahara Y. Field-effect detection using phospholipid membranes. Sci. Technol. Adv. Mater. 2010;11:033001. doi: 10.1088/1468-6996/11/3/033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelly CV, Baird BA, Craighead HG. An Array of Planar Apertures for Near-Field Fluorescence Correlation Spectroscopy. Biophys J. 2011;100:L34–L36. doi: 10.1016/j.bpj.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kung LA, Kam L, Hovis JS, Boxer SG. Patterning hybrid surfaces of proteins and supported lipid bilayers. Langmuir. 2000;16:6773–6776. [Google Scholar]
- 52.Lakowicz JR, Ray K, Chowdhury M, Szmacinski H, Fu Y, Zhang J, Nowaczyk K. Plasmon-controlled fluorescence: a new paradigm in fluorescence spectroscopy. Analyst. 2008;133:1308–1346. doi: 10.1039/b802918k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenhert S, Sun P, Wang YH, Fuchs H, Mirkin CA. Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 54.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 55.Lewis A, Isaacson M, Harootunian A, Muray A. Development of a 500 Å spatial resolution light microscope: I. light is efficiently transmitted through [lambda]/16 diameter apertures. Ultramicroscopy. 1984;13:227–231. [Google Scholar]
- 56.Ma P, Zemmel R. Value of novelty? Nat Rev Drug Discov. 2002;1:571–572. doi: 10.1038/nrd884. [DOI] [PubMed] [Google Scholar]
- 57.Manz BN, Groves JT. Spatial organization and signal transduction at intercellular junctions. Nat Rev Mol Cell Biol. 2010;11:342–352. doi: 10.1038/nrm2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manzo C, van Zanten TS, Garcia-Parajo MF. Nanoscale Fluorescence Correlation Spectroscopy on Intact Living Cell Membranes with NSOM Probes. Biophys J. 2011;100:L8–L10. doi: 10.1016/j.bpj.2010.12.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer K, Hafner J. Localized Surface Plasmon Resonance Sensors. Chem Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 60.Moran-Mirabal JM, Craighead HG. Zero-mode waveguides: Sub-wavelength nanostructures for single molecule studies at high concentrations. Methods. 2008;46:11–17. doi: 10.1016/j.ymeth.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Moran-Mirabal JM, Edel JB, Meyer GD, Throckmorton D, Singh AK, Craighead HG. Micrometer-Sized Supported Lipid Bilayer Arrays for Bacterial Toxin Binding Studies through Total Internal Reflection Fluorescence Microscopy. Biophys J. 2005;89:296–305. doi: 10.1529/biophysj.104.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moran-Mirabal JM, Torres AJ, Samiee KT, Baird BA, Craighead HG. Cell investigation of nanostructures: zero-mode waveguides for plasma membrane studies with single molecule resolution. Nanotechnology. 2007;18:195101. [Google Scholar]
- 63.Morigaki K, Baumgart T, Jonas U, Offenhäusser A, Knoll W. Photopolymerization of Diacetylene Lipid Bilayers and Its Application to the Construction of Micropatterned Biomimetic Membranes. Langmuir. 2002;18:4082–4089. [Google Scholar]
- 64.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 65.Mossman K, Groves J. Micropatterned supported membranes as tools for quantitative studies of the immunological synapse. Chem Soc Rev. 2007;36:46–54. doi: 10.1039/b605319j. [DOI] [PubMed] [Google Scholar]
- 66.Nabika H, Iijima N, Takimoto B, Ueno K, Misawa H, Murakoshi K. Segregation of Molecules in Lipid Bilayer Spreading through Metal Nanogates. Anal Chem. 2009;81:699–704. doi: 10.1021/ac802130e. [DOI] [PubMed] [Google Scholar]
- 67.Novotny L, van Hulst N. Antennas for light. Nat Photon. 2011;5:83–90. [Google Scholar]
- 68.Okazaki T, Tatsu Y, Morigaki K. Phase Separation of Lipid Microdomains Controlled by Polymerized Lipid Bilayer Matrices. Langmuir. 2010;26:4126–4129. doi: 10.1021/la9032892. [DOI] [PubMed] [Google Scholar]
- 69.Oliver AE, Parikh AN. Templating membrane assembly, structure, and dynamics using engineered interfaces. Biochim Biophys Acta. 2010;1798:839–850. doi: 10.1016/j.bbamem.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 70.Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA. Mast cell activation on patterned lipid bilayers of subcellular dimensions. Langmuir. 2003;19:1599–1605. [Google Scholar]
- 71.van Oudenaarden A, Boxer SG. Brownian ratchets: molecular separations in lipid bilayers supported on patterned arrays. Science. 1999;285:1046–1048. doi: 10.1126/science.285.5430.1046. [DOI] [PubMed] [Google Scholar]
- 72.Piner RD, Zhu J, Xu F, Hong SH, Mirkin CA. “Dip-pen” lithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 73.Poo M. In situ electrophoresis of membrane components. Annu Rev Biophys Bioeng. 1981;10:245–276. doi: 10.1146/annurev.bb.10.060181.001333. [DOI] [PubMed] [Google Scholar]
- 74.Poo M, Robinson KR. Electrophoresis of concanavalin A receptors along embryonic muscle cell membrane. Nature. 1977;265:602–605. doi: 10.1038/265602a0. [DOI] [PubMed] [Google Scholar]
- 75.Reimhult E, Baumann MK, Kaufmann S, Kumar K, Spycher PR. Advances in nanopatterned and nanostructured supported lipid membranes and their applications. Biotechnol Genet Eng Rev. 2010;27:185–216. doi: 10.1080/02648725.2010.10648150. [DOI] [PubMed] [Google Scholar]
- 76.Roh S, Chung T, Lee B. Overview of the Characteristics of Micro- and Nano-Structured Surface Plasmon Resonance Sensors. Sensors. 2011;11:1565–1588. doi: 10.3390/s110201565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samiee KT, Moran-Mirabal JM, Cheung YK, Craighead HG. Zero mode waveguides for single-molecule spectroscopy on lipid membranes. Biophys J. 2006;90:3288–3299. doi: 10.1529/biophysj.105.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sekula S, Fuchs J, Weg-Remers S, Nagel P, Schuppler S, Fragala J, Theilacker N, Franzreb M, Wingren C, Ellmark P, Borrebaeck CAK, Mirkin CA, Fuchs H, Lenhert S. Multiplexed Lipid Dip-Pen Nanolithography on Subcellular Sacles for the Templating of Functional Proteins and Cell Culture. Small. 2008;4:1785–1793. doi: 10.1002/smll.200800949. [DOI] [PubMed] [Google Scholar]
- 79.Shi JJ, Chen JX, Cremer PS. Sub-100 nm Patterning of supported bilayers by nanoshaving lithography. J Am Chem Soc. 2008;130:2718-+. doi: 10.1021/ja077730s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 81.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 82.Simonsson L, Gunnarsson A, Wallin P, Jönsson P, Höök F. Continuous Lipid Bilayers Derived from Cell Membranes for Spatial Molecular Manipulation. J Am Chem Soc. 2011;133:14027–14032. doi: 10.1021/ja204589a. [DOI] [PubMed] [Google Scholar]
- 83.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 84.Stöckl MT, Herrmann A. Detection of lipid domains in model and cell membranes by fluorescence lifetime imaging microscopy. Biochim. Biophys. Acta. 2010;1798:1444–1456. doi: 10.1016/j.bbamem.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 85.Tan CP, Craighead HG. Surface Engineering and Patterning Using Parylene for Biological Applications. Materials. 2010;3:1803–1832. [Google Scholar]
- 86.Tan C, Cipriany B, Lin D, Craighead HG. Nanoscale Resolution, Multicomponent Biomolecular Arrays Generated By Aligned Printing With Parylene Peel-Off. Nano Lett. 2010;10:719–725. doi: 10.1021/nl903968s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanaka M, Hermann J, Haase I, Fischer M, Boxer SG. Frictional drag and electrical manipulation of recombinant proteins in polymer-supported membranes. Langmuir. 2007;23:5638–5644. doi: 10.1021/la0628219. [DOI] [PubMed] [Google Scholar]
- 88.Torres AJ, Vasudevan L, Holowka D, Baird BA. Nanomaterials in Medicine Special Feature Sackler Colloquium: Focal adhesion proteins connect IgE receptors to the cytoskeleton as revealed by micropatterned ligand arrays. Proc Natl Acad Sci U S A. 2008;105:17238–17244. doi: 10.1073/pnas.0802138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Torres AJ, Wu M, Holowka D, Baird B. Nanobiotechnology and Cell Biology: Micro- and Nanofabricated Surfaces to Investigate Receptor-Mediated Signaling. Annu Rev Biophys. 2008;37:265–288. doi: 10.1146/annurev.biophys.36.040306.132651. [DOI] [PubMed] [Google Scholar]
- 90.Tsai J, Sun E, Gao Y, Hone JC, Kam LC. Non-Brownian diffusion of membrane molecules in nanopatterned supported lipid bilayers. Nano Lett. 2008;8:425–430. doi: 10.1021/nl072304q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Visnapuu M-L, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16:U1056–U1075. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Visnapuu M-L, Fazio T, Wind S, Greene EC. Parallel Arrays of Geometric Nanowells for Assembling Curtains of DNA with Controlled Lateral Dispersion. Langmuir. 2008;24:11293–11299. doi: 10.1021/la8017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vobornik D, Banks DS, Lu ZF, Fradin C, Taylor R, Johnston LJ. Fluorescence correlation spectroscopy with sub-diffraction-limited resolution using near-field optical probes. Appl Phys Lett. 2008;93:163904. [Google Scholar]
- 94.Wenger J, Rigneault H, Dintinger J, Marguet D, Lenne PF. Single-fluorophore diffusion in a lipid membrane over a subwavelength aperture. J Biol Phys. 2006;32:SN1–SN4. doi: 10.1007/s10867-006-2909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Werner JH, Montaño GA, Garcia AL, Zurek NA, Akhadov EA, Lopez GP, Shreve AP. Formation and Dynamics of Supported Phospholipid Membranes on a Periodic Nanotextured Substrate. Langmuir. 2009;25:2986–2993. doi: 10.1021/la802249f. [DOI] [PubMed] [Google Scholar]
- 96.Wittenberg NJ, Im H, Johnson TW, Xu X, Warrington AE, Rodriguez M, Oh S-H. Facile Assembly of Micro- and Nanoarrays for Sensing with Natural Cell Membranes. ACS Nano. 2011;5:7555–7564. doi: 10.1021/nn202554t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu M, Baumgart T, Hammond S, Holowka D, Baird B. Differential targeting of secretory lysosomes and recycling endosomes in mast cells revealed by patterned antigen arrays. J Cell Sci. 2007;120:3147–3154. doi: 10.1242/jcs.007260. [DOI] [PubMed] [Google Scholar]
- 98.Wu M, Holowka DA, Craighead HG, Baird BA. Visualization of plasma membrane compartmentalization with patterned lipid bilayers. Proc Natl Acad Sci U S A. 2004;101:13798–13803. doi: 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoshina-Ishii C, Boxer SG. Controlling two-dimensional tethered vesicle motion using an electric field: interplay of electrophoresis and electro-osmosis. Langmuir. 2006;22:2384–2391. doi: 10.1021/la0526277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Zanten TS, Gomez J, Manzo C, Cambi A, Buceta J, Reigada R, Garcia-Parajo MF. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc Natl Acad Sci U S A. 2010;107:15437–15442. doi: 10.1073/pnas.1003876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Zanten TS, Lopez-Bosque MJ, Garcia-Parajo MF. Imaging Individual Proteins and Nanodomains on Intact Cell Membranes with a Probe-Based Optical Antenna. Small. 2010;6:270–275. doi: 10.1002/smll.200901204. [DOI] [PubMed] [Google Scholar]