Abstract
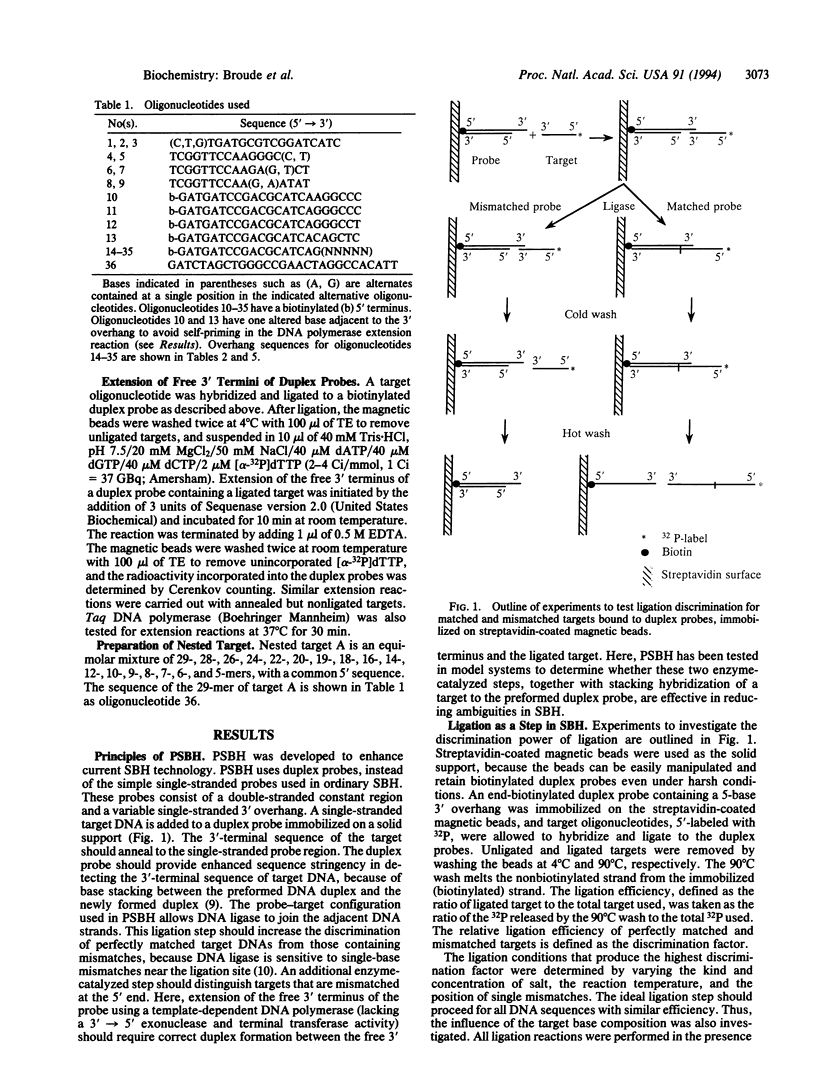
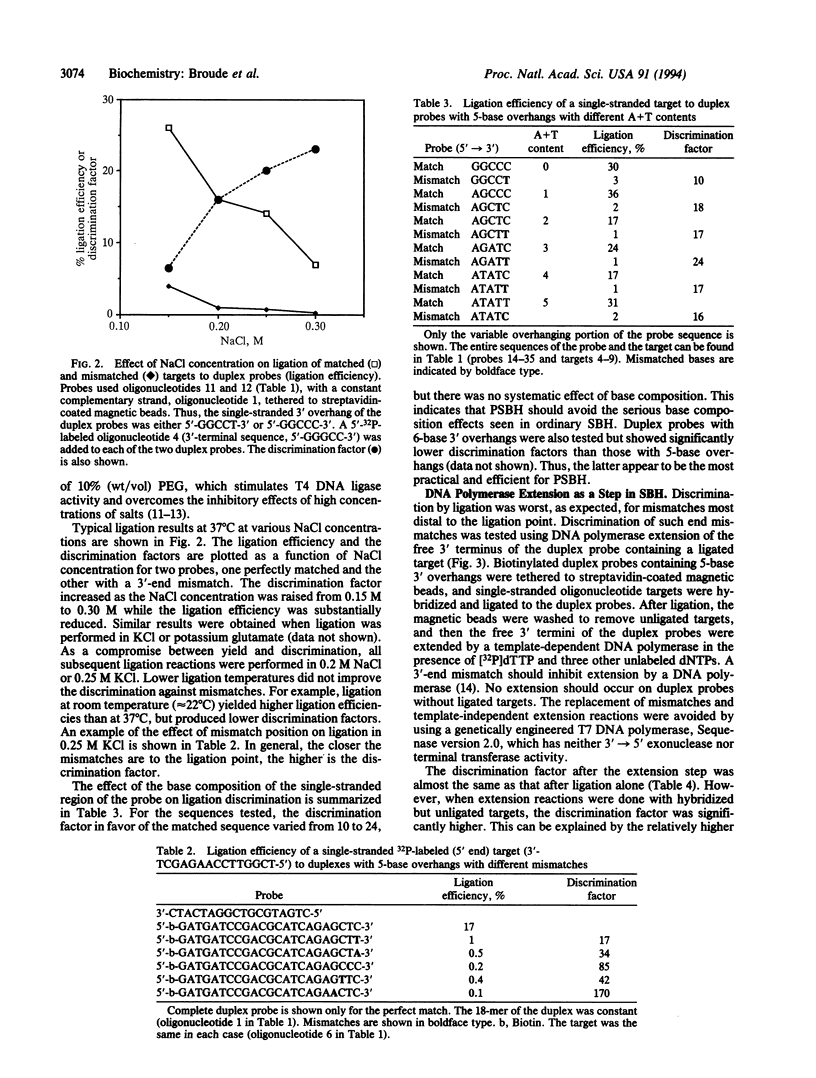
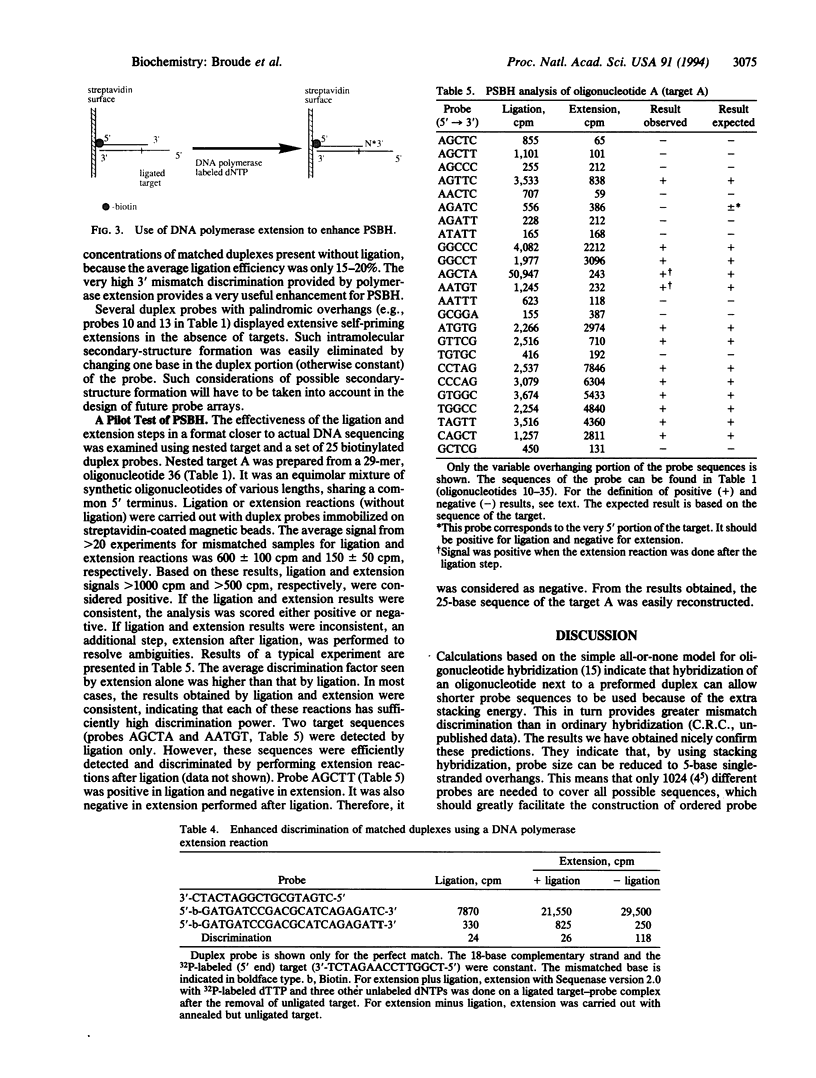
An enhanced version of DNA sequencing by hybridization (SBH), termed positional SBH (PSBH), has been developed. PSBH uses duplex probes containing single-stranded 3' overhangs, instead of simple single-stranded probes. Stacking interactions between the duplex probe and a single-stranded target should provide enhanced stringency in distinguishing perfectly matched 3' sequences. A second enhancement is the use of enzyme-catalyzed steps, instead of pure physical hybridization. The feasibility of this scheme has been investigated using biotinylated duplex probes containing single-stranded 5-base 3' overhangs, immobilized on streptavidin-coated magnetic beads. Ligation of a single-stranded target, hybridized to the single-stranded region of the duplex probes, provided enhanced discrimination of perfectly matched targets from those containing mismatches. In distinction to the serious complications caused by base composition effects in ordinary SBH, there was little effect of base composition in PSBH. The hardest mismatch to discriminate was the one furthest from the phosphodiester bond formed by ligation. However, mismatches in this position were efficiently discriminated by 3' extension of the duplex probe using a template-dependent DNA polymerase. These results demonstrate that PSBH offers considerable promise to facilitate actual implementations of SBH.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bains W. Hybridization methods for DNA sequencing. Genomics. 1991 Oct;11(2):294–301. doi: 10.1016/0888-7543(91)90135-2. [DOI] [PubMed] [Google Scholar]
- Bains W., Smith G. C. A novel method for nucleic acid sequence determination. J Theor Biol. 1988 Dec 7;135(3):303–307. doi: 10.1016/s0022-5193(88)80246-7. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Drmanac S., Strezoska Z., Paunesku T., Labat I., Zeremski M., Snoddy J., Funkhouser W. K., Koop B., Hood L. DNA sequence determination by hybridization: a strategy for efficient large-scale sequencing. Science. 1993 Jun 11;260(5114):1649–1652. doi: 10.1126/science.8503011. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Labat I., Brukner I., Crkvenjakov R. Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics. 1989 Feb;4(2):114–128. doi: 10.1016/0888-7543(89)90290-5. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Nakazawa M., Ishizaki Y., Obayashi A. Influence of monovalent cations on the activity of T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1985 May 10;13(9):3261–3271. doi: 10.1093/nar/13.9.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K. R., Lysov YuP, Khorlin A. A., Ivanov I. B., Yershov G. M., Vasilenko S. K., Florentiev V. L., Mirzabekov A. D. A method for DNA sequencing by hybridization with oligonucleotide matrix. DNA Seq. 1991;1(6):375–388. doi: 10.3109/10425179109020793. [DOI] [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988 Aug 26;241(4869):1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Lysov Iu P., Florent'ev V. L., Khorlin A. A., Khrapko K. R., Shik V. V. Opredelenie nukleotidnoi posledovatel'nosti DNK gibridizatsiei s oligonukleotidami. Novyi metod. Dokl Akad Nauk SSSR. 1988;303(6):1508–1511. [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P. A., Lysov YuP, Khrapko K. R., Belyavsky A. V., Florentiev V. L., Mirzabekov A. D. Improved chips for sequencing by hybridization. J Biomol Struct Dyn. 1991 Oct;9(2):399–410. doi: 10.1080/07391102.1991.10507920. [DOI] [PubMed] [Google Scholar]
- Raae A. J., Kleppe R. K., Kleppe K. Kinetics and effect of salts and polyamines on T4 polynucleotide ligase. Eur J Biochem. 1975 Dec 15;60(2):437–443. doi: 10.1111/j.1432-1033.1975.tb21021.x. [DOI] [PubMed] [Google Scholar]
- Strezoska Z., Paunesku T., Radosavljević D., Labat I., Drmanac R., Crkvenjakov R. DNA sequencing by hybridization: 100 bases read by a non-gel-based method. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10089–10093. doi: 10.1073/pnas.88.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G. DNA probes: applications of the principles of nucleic acid hybridization. Crit Rev Biochem Mol Biol. 1991;26(3-4):227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- Wu D. Y., Wallace R. B. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene. 1989;76(2):245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]