Abstract
Spontaneous damage to DNA is frequent and may lead to cell death, cell senescence, or mutations. DNA double-strand breaks (DSBs) are of special interest because they are highly toxic and have been implicated in neurodegeneration, cancer, and aging. Until now, there has not been a reliable system allowing tunable induction of random DSBs without affecting other macromolecules or cell functions. Here, we describe an adenoviral-based, doxycycline-mediated, and tamoxifen-dependent system for quantitative introduction of DSBs in mammalian cells. We generated a single adenoviral vector containing a tet-inducible, composite SacI restriction endonuclease/estrogen receptor (ERT2) gene, and a constitutively expressed reverse transactivator (rtTA) gene. Transduced mouse embryonic fibroblasts— as well as mouse liver cells in vivo—demonstrated a high level of DSBs in response to treatment with doxycycline and tamoxifen. We show that the amount of induced DSBs can be titrated by doxycycline dose and duration of treatment. This system should be useful for studying the processing of randomly induced DSBs and their effects on cell fate, without the side effects normally associated with radiation or chemical treatment.
Keywords: DNA damage, restriction endonuclease, double-strand breaks, aging
Introduction
Physical and chemical methods commonly used for generating DNA double-strand breaks (DSBs) in mammalian cells induce a spectrum of lesions and can affect other biological macromolecules as well as normal cell physiology. An alternative way of inducing DSBs in living cells is through ectopic expression of proteins possessing an endonuclease activity. The endonucleases recognize specific sequences, directly cleave DNA and, unlike physical and chemical methods, generate no other lesions and have no adverse physiological effects. Rare-cutting homing endonucleases I-SceI (1), I-CreI, and I-PpoI (2,3) have been successfully used for this purpose. The size of recognition sites for homing endonucleases (18, 24, and 15 bp, respectively) determines their low frequency in the mammalian genome and makes them ideal tools for studying DSB repair. However, the same feature makes these enzymes less useful for studying stochastically distributed DSBs (as in the natural situation) and the associated processing events and various cellular end points. This limitation can be overcome by the use of bacterial restriction endonucleases (REs). REs generally have shorter recognition sites (typically 4–8 bp) and are present in the genome at a much higher frequency. REs were shown to cause DSBs and induce chromosomal aberrations when directly electroporated or genetically expressed in eukaryotic cells (4–7). However, the utility of these approaches is seriously compromised by the necessity of special cell treatment to permit access to protein or plasmid DNA, and by the lack of RE activity regulation.
Here we describe an adenoviral-based, doxycycline-mediated, and tamoxifen-dependent system for the quantitative introduction of DSBs into genomic DNA. The generated construct contains both parts of the tet-inducible system: a tet-inducible promoter driving expression of SacI RE fused to mutated estrogen receptor gene ERT2 (8), and reverse transactivator (rtTA) driven by the CMV promoter in the backbone of an adenoviral vector. The SacI RE has a 6-bp recognition site (GAGCTC) and creates cohesive ends with a four- nucleotide overhang after cleaving DNA. Cultured mouse embryonic fibroblasts transduced with this virus, as well as mouse liver cells in vivo after tail vein injection of the virus suspension, demonstrate inducible expression of SacI in response to doxycycline treatment, which results in the increased expression of a DSB marker. We show that the level of DNA damage can be controlled by the dose of doxycycline and duration of drug application.
Materials and methods
Vector construction and virus production
SacIR coding sequence lacking the stop codon was amplified from genomic DNA of Streptomyces achromogenes (provided by New England BioLabs, Ipswich, MA, USA) with Pfx polymerase (Invitrogen, Carlsbad, CA, USA) and cloned in frame with a V5 epitope into an expression vector containing the CMV promoter to get pCMV-SacIV5. Primers used were 5′-CACCATGGGAATAACAATTAAAAAGAGCACGGCG- 3′ (forward) and 5′-CGTTTCAGGGAAGATCTCAGCCCA- 3′ (reverse). The SacI coding sequence was fused with the mutated estrogen receptor gene ERT2, excised from pCre-ERT2 plasmid (provided by P. Chambon, Illkirch, France) resulting in a tamoxifen-inducible variant of SacI. The SacI-ERT2 coding sequence was cloned together with V5 epitope into a pTRE-Tight vector (Clontech, Mountain View, CA, USA) to get pTight-SEV5.
Tight-SEV5-pA and CMV-rtTA-pA expression cassettes were PCR-amplified with Pfx polymerase (Invitrogen) from pTight-SEV5 and pTet-ON (Clontech), respectively, and cloned into pBluescript KS+ vector in head-to-tail orientation. Tight-SEV5-pA-CMV-rtTA-pA fragment was PCR-amplified with High Fidelity Platinum Taq Polymerase (Invitrogen) and sub-cloned into an entry vector using a pCR8/GW/TOPO TA Cloning Kit (Invitrogen). The resulting pGW/Tight-SEV5-CMV-rtTA entry vector was used to clone the Tight-SEV5-pA-CMV-rtTA-pA cassette into a pAd/PL-DEST vector (ViraPower Adenoviral Expression System Kit, Invitrogen) to get pAd/TSCR. All cloning work with plasmids containing the SacIR gene was performed in Stbl3 Escherichia coli strain (Invitrogen) transformed with a plasmid that expresses SacI methylase (provided by New England BioLabs).
Adenoviral stocks (A/TSCR) were produced in 293A/tTS cells according to the manufacturer’s instructions and titrated with Adenoviral Rapid Titer Kit (Clontech).
Western blotting
Protein extraction from cultured cells was performed with a Whole Cell Extraction Kit (Chemicon, Billerica, MA, USA) after separation on NuPAGE 4–12% Bis-Tris Gel (Invitrogen) in MOPS SDS running buffer (Invitrogen). Proteins were transferred on PVDF membrane using an iBlot Gel Transfer System (Invitrogen). The primary antibodies used were monoclonal anti-V5 (1:3000; Invitrogen), monoclonal anti-GAPDH (1:100000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and monoclonal anti-phospho-histone H2AX (1:3000; Upstate, Billerica, MA, USA). The secondary antibodies used were HRP-conjugated goat anti-mouse (1:5000; Santa Cruz Biotechnology). HRP was detected with Immun-Star HRP kit (Bio-Rad Laboratories, Hercules, CA, USA).
Cell culture
293FT cells (Invitrogen) were maintained in a humidified 10% CO2–containing atmosphere at 37°C in the medium recommended by the manufacturer. To get the 293A/tTS cell line for virus production, the 293A cells (Invitrogen) were transfected with ptTS-Neo (Clontech) and selected in Geneticin (500 μg/mL; Invitrogen). The stably transfected cell line was maintained in a humidified, 10% CO2–containing atmosphere at 37°C in the medium recommended by manufacturer for 293A cells and supplemented with Geneticin. Cell transfection was performed with Lipofectamine transfection reagent (Invitrogen) according to the manufacturer’s recommendations.
Primary mouse embryonic fibroblasts (MEFs) were obtained by trypsinization of D13.5 embryos of C57Bl/6 mouse and maintained in 10% CO2 and 3% O2 at 37°C in DMEM (GIBCO, Carlsbad, CA, USA), supplemented with 10% FBS (GIBCO). Tet-system–approved FBS (Clontech) and medium supplemented with doxycycline (Clontech) and 10 nM 4-hydroxy-tamoxifen (OHT; Sigma, St. Louis, MO, USA) were used when necessary.
Cell survival
MEFs were seeded onto 96-well plates (Cat. no. 3596, Corning, Corning, NY, USA) at a density of 5 × 103 cells/well and cultured as described in the “Results” section. The relative amount of cells was determined with the Vybrant MTT Cell Proliferation Assay Kit (Invitrogen) and the microplate reader Synergy 4 (BioTek Instruments, Winooski, VT, USA). Results were expressed as a percentage of control samples.
DSB detection and quantification
DSBs were detected by assessment of γ-H2AX expression (i.e., phosphorylated histone H2AX). Cells were cultured in 96-well plates (Cat. no. 165305; Nalge Nunc International, Rochester, NY, USA), fixed with ice-cold 100% methanol for 10 min, and stained for γ-H2AX and DNA. The primary antibody used was rabbit anti–phospho-histone H2AX (1:1000; Active Motif, Carlsbad, CA, USA) and the secondary antibody was Alexa Fluor 488–conjugated donkey anti-rabbit (1:1000; Molecular Probes, Eugene, OR, USA). The nuclear DNA was stained with propidium iodide/RNase staining solution (Chemicon). The intensity of fluorescence excited at 488 nm and emitted at 528 nm (γ-H2AX) and 620 nm (DNA) was measured with the microplate reader Synergy 4 (BioTek Instruments). The relative amount of DSBs was expressed as a ratio γ-H2AX/DNA.
Neutral comet assay
Direct visualization of DNA damage in MEFs was performed with the CometAssay Kit (R & D Systems, Minneapolis, MN, USA) according to the manufacturer’s protocol. Briefly, cells were embedded in low–melting point agarose and lysed in neutral conditions. After electrophoresis in TBE buffer, comets were visualized by staining with SYBR Green.
In vivo transduction and tissue collection
All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC). The 8-week-old C57BL/6 animals received a single tail vein injection of the A/TSCR virus resuspended in 0.1 mL PBS. Twenty-four hours after injection, animals were treated with doxycycline solution (2 mg/mL in 5% sucrose), administered in the drinking water, for 48 h. After 24 h of doxycycline administration, mice received one intraperitoneal injection of tamoxifen (Sigma-Aldrich) solution (1 mg/0.1 mL) in corn oil (Sigma-Aldrich). Animals were euthanized and perfused with formalin. Livers were isolated and postfixed in the same solution overnight.
Immunostaining
Cells for immunostaining were cultured on Lab-Tek II Chamber Slides (Nalge Nunc International) under conditions described in the text and fixed with ice-cold 100% methanol for 10 min. The primary antibodies used were mouse monoclonal anti-V5 (1:200; Invitrogen) and rabbit anti-phospho-histone H2AX (1:1000; Upstate). The secondary antibodies used were Alexa Fluor 488–conjugated donkey anti-mouse and Alexa Fluor 594–conjugated donkey anti-rabbit (both at 1:1000; Molecular Probes).
Sections of formalin-fixed, paraffin-embedded liver tissue were deparaffinized, rehydrated, incubated in 0.01 M citrate buffer (pH 6.0) at 95°C for 20 min, and immersed in PBS. The primary antibodies used were rabbit anti-phospho-histone H2AX (1:1000; Active Motif); the secondary antibodies used were HRP-conjugated goat anti-rabbit (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). The HRP was detected with DAB (Black) Substrate Kit (Invitrogen). Sections were counterstained with hematoxylin (Invitrogen) to visualize nuclei.
Statistical analysis
The p-values were calculated using a two-sample t-test (Microsoft Excel 2003; Microsoft Corp., Redmond, WA, USA). Each experiment was repeated a minimum of three times; representative results are shown.
Results
Expression of SacI restriction endonuclease in mammalian cells
The coding sequence of SacI restriction endonuclease lacking the stop codon was amplified from genomic DNA of S. achromogenes and cloned into an expression vector under control of the CMV promoter in frame with a V5 epitope to obtain a readout of SacI expression. The resulting vector was tested by transfection of 293FT cells. Twenty-four hours post-transfection, Western blot analysis of the whole-cell lysates revealed the presence of the SacI-V5 fusion and phosphorylated form of H2AX histone (γ-H2AX; not shown), a generally accepted indicator of DSBs (9).
Having demonstrated the functionality of the SacI construct, we set out to make this system regulatable in order to fine-tune its effects. For this purpose, we used primary mouse fibroblasts rather than cell lines, since in our opinion, DSBs are best studied in as close to the normal physiological situation as possible. The use of cell lines often confounds normal cellular responses to DNA damage. Hence, apart from being able to regulate the system, we considered it critical to make it applicable to primary mammalian cells.
Development of an inducible SacI expression system
To control SacI action, we combined two different strategies. First, we utilized the Tet-ON system in order to control levels of SacI transcription. Second, we generated a SacI–estrogen receptor (ERT2) fusion (8) to control the spatial separation of the enzyme and its substrate, genomic DNA. With this approach, we generated and tested an adenoviral vector carrying both SacI-ERT2-V5 (SEV5) controlled by the Tet-inducible promoter and the reverse transactivator (rtTA) driven by the CMV promoter. The resulting vector (pAd/Tight-SEV5-CMV-rtTA; Figure 1A) was used to produce adenoviral stock (A/TSCR).
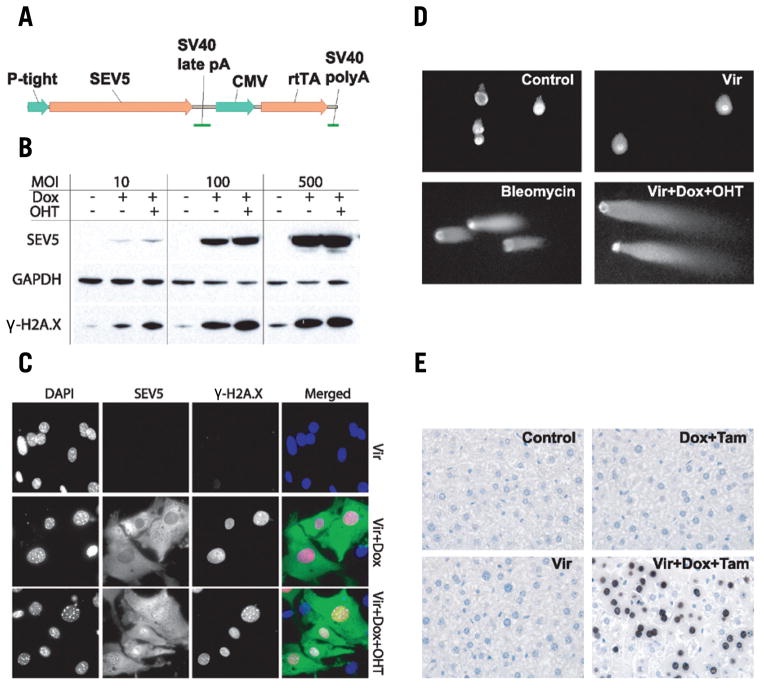
Figure 1. Inducible expression of SacI restriction endonuclease.
(A) Construct providing doxycycline/OHT-inducible expression of SacI restriction endonuclease. (B) Western blot analysis of the SacI-ERT2-V5 fusion (SEV5) and γ-H2AX expression patterns in MEFs transduced with A/TSCR and treated with doxycycline/OHT. (C) Immunostaining of A/TSCR-transduced MEFs without and with doxycycline/OHT treatment for SEV5 (green) and γ-H2AX (red). (D) Visualization of DNA damage caused by SacI by comet assay; (E) γ-H2AX immunostaining of liver tissue from control and A/TSCR-transduced mice.
Mouse embryonic fibroblasts (MEFs) obtained from E13.5 embryos were transduced with A/TSCR at different multiplicities of infection (MOIs) to check the functionality and regulatability of the generated construct in primary cells. Forty-eight hours post-transduction, culture medium was replaced by medium supplemented with doxycycline (1000 ng/mL) and cells were cultured for an additional 24 h. For the last 6 h of culture, medium containing doxycycline was supplemented with 4-hydroxy-tamoxifen (OHT). Transduced cells cultured in Tet/OHT-free conditions, as well as cells subjected only to doxycycline, served as control.
Western blot analysis revealed that expression of the SacI-ERT2-V5 fusion (SEV5) in cells transduced with A/TSCR is completely suppressed in the absence of doxycycline and greatly induced upon doxycycline treatment (Figure 1B). The level of expression depended on MOI. Induction of SEV5 expression by application of doxycycline led to elevated levels of DSBs in the genomic DNA of transduced cells, as indicated by the appearance of a γ-H2AX–reactive band (Figure 1B). The levels of γ-H2AX expression were greater when treatments with doxycycline and OHT were combined (Figure 1B). The effect of OHT application on γ-H2AX expression level was more prominent when lower MOIs were used (Figure 1B).
The results from Western blot analysis were confirmed by immunostaining MEFs transduced with A/TSCR (MOI 10) and treated with doxycycline and OHT. Cells were analyzed for the presence of SEV5 and appearance of γ-H2AX–positive foci in the nuclei. We found no traces of SEV5 expression in transduced cells cultured in the absence of doxycycline. Twenty-four hours after treatment with doxycycline, transduced cells demonstrated significant levels of SEV5 expression (Figure 1C). Although in the absence of OHT, SEV5 fusion localized preferentially in the cytoplasm, this did not completely protect genomic DNA from SacI action, as was evident from γ-H2AX staining.
The neutral comet assay is a useful method allowing direct visualization of DNA damage, mainly DSBs. The level of DNA damage was found to be the same in control cells and cells transduced with A/TSCR (Figure 1D). However, transduced cells demonstrated much higher levels of DSBs than control cells after treatment with doxycycline and OHT, as evident from the appearance of a significant “comet tail.” MEFs treated with bleomycin, a drug known to induce DSBs, served as a positive control.
To evaluate the utility of the system for in vivo applications, we analyzed the liver of mice injected in the tail vein with a single dose of A/TSCR. At 24 h after transduction, the regular drinking water was replaced by water supplemented with doxycycline. After 24 h of continuous doxycycline administration mice received an intraperitoneal injection of tamoxifen solution and were kept on doxycycline treatment for another 24 h, after which the experiment was ended. Liver tissue was stained for γ-H2AX, revealing a large number of hepatocytes with evident DSBs in mice transduced with A/TSCR and treated with doxycycline and tamoxifen (Figure 1E). Transduced animals that did not receive doxycycline, as well as non-transduced animals treated and not treated with doxycycline, served as controls.
Fine control of DSBs in cultured MEFs
Next, we tested the ability to control levels of DSBs in transduced cells by varying both the dose of applied doxycycline and the duration of treatment. The intensity of γ-H2AX expression was used to indicate the amount of DNA DSBs. The fraction of surviving cells 72 h after the end of the treatment served as an indicator of physiological effects of DSBs.
MEFs grown in 96-well plates were transduced with A/TSCR at MOI 50 and 500 the day after seeding. Forty-eight hours after transduction, culture medium was replaced with medium containing doxycycline at different concentrations (10, 100, and 1000 ng/mL) and the cells were cultured for an additional 24 h. For the final 6 h, medium was supplemented with OHT (10 nM). At this point one set of MEFs (MOI 50) was assessed for expression of γ-H2AX. The cells of two other sets (MOI 50 and 500) were trypsinized, split (1:2) and left in regular medium for 72 h to estimate cell survival. Non-transduced and transduced cells cultured in doxycycline-free conditions as well as cells treated with bleomycin (10 μg/mL for 2 h) and non-transduced cells treated with doxycycline/OHT (1000 ng/mL for 24 h and 10 nM for 6 h, respectively) served as a control.
Analysis of control samples demonstrated that doxycycline and OHT do not induce DSBs in cultured MEFs [as evidenced by the low level of γ-H2AX expression, indistinguishable from the non-treated control (Figure 2A)] and do not affect cell survival (Figure 2B). Cells transduced with A/TSCR and cultured in tet-free conditions did not show any significant increase in the level of γ-H2AX expression (Figure 2A), but proliferated more slowly (Figure 2B), most likely due to a nonspecific effect of viral transduction. As expected, treatment with bleomycin led to a significant elevation of γ-H2AX expression (Figure 2A) and had a substantial effect on cell survival (Figure 2B).
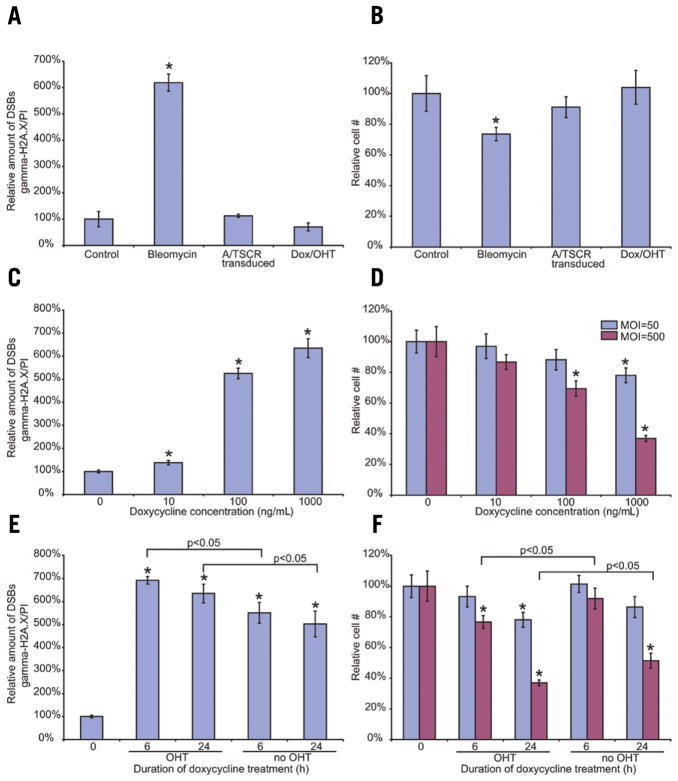
Figure 2. Controllable introduction of DSBs.
(A) γ-H2AX expression in control MEFs, MEFs treated with bleomycin, MEFs transduced with A/TSCR, or MEFs treated with doxycycline and OHT. (B) Cell survival after treatment with bleomycin, transduction with A/TSCR, or treatment with doxycycline and OHT. (C) γ-H2AX expression in MEFs transduced with A/TSCR (MOI 50) and treated with different doses of doxycycline. (D) Survival of cells transduced at different MOIs (50 and 500) and treated with different doses of doxycycline. (E) γ-H2AX expression in MEFs transduced with A/TSCR (MOI 50), treated with doxycycline for different periods of time in the presence or absence of OHT. (F) Survival of cells transduced at different MOIs (50 and 500) and treated with doxycycline for different periods of time in the presence or absence of OHT. The data was normalized and presented as a percentage of control; error bars, sd; *, statistically significant difference (P < 0.05) with corresponding control (panels A, C, and E: n = 6; panels B, D, and F: n = 10).
Activation of SacI in transduced MEFs by treatment with different concentrations of doxycycline followed by OHT treatment led to a significant increase in γ-H2AX expression, which was dependent on the concentration of doxycycline (Figure 2C). The survival of cells after the induction of DSBs was significantly decreased and this effect also correlated with doxycycline dose (Figure 2D). The dose-dependent cytotoxic effect of DSBs was more prominent when MEFs were transduced at a higher MOI (Figure 2D).
Next, we tested whether the level of DSB depended on the duration of the doxycycline treatment. In this set of experiments, transduced cells were subjected to doxycycline/OHT treatment for 6 h and compared with transduced cells, either maintained in doxycycline-free conditions or treated with doxycycline and OHT for 24 and 6 h, respectively. We found that 6 h of doxycycline treatment resulted in the induction of DSBs, as evidenced by the significant elevation of γ-H2AX expression in comparison with transduced non-treated control cells (Figure 2E). The level of γ-H2AX expression in transduced cells exposed to doxycycline for 6 h was similar to the level of expression in cells treated for 24 h. Nevertheless, the number of cells 3 days after 6-h doxycycline application was significantly higher than after 24-h treatment (Figure 2F). This observation suggests that the level of DSB induced by shorter doxycycline application was less severe.
Finally, we tested the functionality of the estrogen receptor protein fused to SacI. Transduced MEFs were exposed to doxycycline for 6 and 24 h without OHT. In both cases, absence of OHT treatment led to a statistically significant lesser amount of induced DSBs (Figure 2E) and a slight, but also statistically significant increase in cell survival 72 h after the last treatment when compared with the combined doxycycline/OHT treatment (Figure 2F).
Discussion
In this article we describe an adenoviral-based system allowing controllable introduction of DSBs into genomic DNA by inducing SacI restriction endonuclease expression. A dual activation system allowed us to achieve tight control over SacI action. Western blot analysis and immunostaining of MEFs transduced with an adenoviral vector expressing a reverse transactivator and containing a SacI-ERT2-V5 fusion protein under control of the tet-inducible promoter revealed that expression of SacI is completely suppressed in the absence of doxycycline and greatly induced upon doxycycline treatment. Induction of SacI expression by application of doxycycline led to elevated levels of DSBs in the genomic DNA in transduced cells. We demonstrated that the number of introduced DSBs can be controlled by the dose of doxycycline and duration of the treatment.
As an indicator of DSBs, we utilized the expression of γ-H2AX, a phosphorylated form of H2AX histone (9). Although it was recently revealed that phosphorylation of H2AX can be induced by DNA damages other than DSBs (10,11), γ-H2AX is widely recognized as a quantitative marker of DSBs (11,12). The phosphorylation of H2AX occurs as soon as 3 min after induction of DSBs by ionizing radiation, and reaches the maximum level within 10–30 min (9). The opposite process of H2AX dephosphorylation in the course of DSB repair is more prolonged: >50% of γ-H2AX becomes dephosphorylated in 3 h (13). Thus, the steady state level of γ-H2AX expression is a reflection of the dynamic equilibrium between DNA DSB generation and repair.
We found that the amount of DSBs induced in MEFs transduced with the adenoviral vector containing the regulatable SacI cassette can be titrated by applying different doses of doxycycline. The survival of transduced cells depended on the amount of DSBs and also correlated with the applied doxycycline dose. The observation that the higher doses of the drug decreased cell survival suggests that the cells can tolerate certain levels of DSBs, which makes the system suitable for induction of non-lethal numbers of DSBs.
The 6-h–long application of doxycycline on transduced cells resulted in the same level of induction of γ-H2AX expression as a 24 h long treatment, but survival of cells after shorter exposure to the drug was significantly better. This suggests that longer induction time leads to higher levels of DNA damage. Since the level of the γ-H2AX expression reflects a competition between SacI-mediated cleavage and DSB repair, it is conceivable that at some point this level will reach its maximum and will not grow any further despite the continuous generation of new DSBs. Additionally, prolonged SacI expression increases the risk of digestion of the TRE-SacI expression cassette, which contains a unique SacI recognition site in the tet-inducible promoter. This negative back-loop may serve as an additional limiting factor.
The choice of vector composition is critical and determines the functionality of the construct. In our study we utilized a single virus strategy where both rtTA and an inducible gene are incorporated in one vector. Two expression cassettes—one containing the tet-inducible promoter controlling the SacI gene and another containing the CMV promoter controlling the rtTA gene—were cloned in an adenoviral vector in head-to-tail orientation. This design was reported as optimal for tight regulation of transgene expression (14). We chose to use the constitutive CMV promoter to control rtTA over autoregulatory expression (where both the gene of interest and rtTA are TRE-regulated), because it allowed a more gradual activation of transgene expression in response to doxycycline treatment (15). Hence, this provides a better tool for quantitative induction of DNA DSBs.
The basal level of tet-inducible promoter activity in the off state (16) is undesirable, especially when a potentially cytotoxic protein is expressed. Two different strategies were utilized in our study to resolve this issue. First, we used the last generation of TRE-based promoter PTight, which has a modified TRE and demonstrates extremely low basal activity. Second, the SacI protein was fused to a mutated estrogen receptor ERT2 that prevents the transition of fused protein into the nuclei in absence of tamoxifen (8). Neither of these approaches allowed complete suppression of SacI action when used separately. We were also unable to produce a virus when a TRE-regulated construct lacking ERT2 or a construct containing SacI-ERT2 under control of a constitutive promoter was used.
The utility of mutated estrogen receptor for spatial separation of expressed chimeric protein and its target in the nuclei was previously validated and is widely used to control the activity of Cre recombinase (8). Later, the same approach was utilized for the generation of the inducible form of piggyBac transposase (17), I-SceI (18), and I-PpoI homing endonucleases (19). Although the ERT2 does not completely protect the genomic DNA from the SacI action in our system, the difference in the levels of DSBs induced with and without OHT treatment was statistically significant and we found this element of the system to be essential. The observed limited efficiency of the ERT2 in the described system can be due to characteristics of the in vitro model we utilized in our study. Since the MEFs are actively dividing cells, the chromosomes are exposed to the cytoplasm during M-phase of the cell cycle. It is unlikely that restriction enzymes will cleave when the DNA is in the form of highly condensed heterochromatin, but trace amounts of SacI can be incorporated into the nuclei during telophase and can attack specific sites when they become available.
Assuming an equal representation of each nucleotide in the genome, the probability of the 6-bp SacI recognition site is 0.256. In other words, one can expect a SacI site every 2.4 kb on average. Since the size of the mouse genome is ~3.2 Gb there are ~1.3 million SacI sites. By analogy with I-PpoI (3), about 10% of SacI recognition sites are expected to be available for cleavage. This leaves ~130,000 potential targets for restriction endonuclease action, ensuring stochastic distribution of induced DSBs along the DNA of the genome. Unlike other DNA-damaging factors, restriction endonuclease creates “clean” DSBs, which can be religated without pre-processing. Although it was reported previously that DNA bulk-damaged by α-irradiation is prone to large-scale motion (20), a more recent study on DSB repair inflicted by a I-SceI homing endonuclease suggests the positional immobility of broken DNA ends (21,22), which favors their rejoining. Furthermore, DSBs with cohesive DNA ends were previously found to be significantly less clastogenic than blunt-ended DSBs (5,23–25), most likely because the majority of cohesive ends are ligated immediately after cleavage. These features, together with the possibility to regulate the level of SacI expression, make the described system a valuable tool for introducing randomly distributed DSBs into genomic DNA without immediately fatal consequences. We envisage the use of this system in studying DSBs and their processing through genome maintenance pathways in various primary cell types as a function of cell fate. Such applications can easily be extended to the in vivo situation by adenoviral infection of specific organ systems, most notably the liver, or by the generation of tet/tamoxifen-regulatable transgenic animal models.
Acknowledgments
We thank Pierre Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) for the pCre-ERT2 plasmid, and Shuang-yong Xu (New England BioLabs) for S. achromogenes genomic DNA and pLG339-SacIM plasmid. This work was supported by the Ellison Medical Foundation (grant no. AG-SS-1496-05, to J.V.) and the National Institutes of Health (NIH; grant no. 7PO1-AG017242-11, to J.V.).
Footnotes
The authors declare no competing interests. This paper is subject to the NIH Public Access Policy.
References
- 1.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 3.Monnat RJ, Jr, Hackmann AF, Cantrell MA. Generation of highly site-specific DNA double-strand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem Biophys Res Commun. 1999;255:88–93. doi: 10.1006/bbrc.1999.0152. [DOI] [PubMed] [Google Scholar]
- 4.Barnes G, Rine J. Regulated expression of endonuclease EcoRI in Saccharomyces cerevisiae: nuclear entry and biological consequences. Proc Natl Acad Sci USA. 1985;82:1354–1358. doi: 10.1073/pnas.82.5.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa ND, Bryant PE. Differences in accumulation of blunt- and cohesive-ended double-strand breaks generated by restriction endonucleases in electroporated CHO cells. Mutat Res. 1991;254:239–246. doi: 10.1016/0921-8777(91)90062-t. [DOI] [PubMed] [Google Scholar]
- 6.Gustavino B, Johannes C, Obe G. Restriction endonuclease BamHI induces chromosomal aberrations in Chinese hamster ovary (CHO) cells. Mutat Res. 1986;175:91–95. doi: 10.1016/0165-7992(86)90130-2. [DOI] [PubMed] [Google Scholar]
- 7.Morgan WF, Fero ML, Land MC, Winegar RA. Inducible expression and cytogenetic effects of the EcoRI restriction endonuclease in Chinese hamster ovary cells. Mol Cell Biol. 1988;8:4204–4211. doi: 10.1128/mcb.8.10.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 10.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smart DJ, Halicka HD, Schmuck G, Traganos F, Darzynkiewicz Z, Williams GM. Assessment of DNA double-strand breaks and gammaH2AX induced by the topoisomerase II poisons etoposide and mitoxantrone. Mutat Res. 2008;641:43–47. doi: 10.1016/j.mrfmmm.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nazarov IB, Smirnova AN, Krutilina RI, Svetlova MP, Solovjeva LV, Nikiforov AA, Oei SL, Zalenskaya IA, et al. Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat Res. 2003;160:309–317. doi: 10.1667/rr3043. [DOI] [PubMed] [Google Scholar]
- 14.Chenuaud P, Larcher T, Rabinowitz JE, Provost N, Joussemet B, Bujard H, Samulski RJ, Favre D, Moullier P. Optimal design of a single recombinant adeno-associated virus derived from serotypes 1 and 2 to achieve more tightly regulated transgene expression from nonhuman primate muscle. Mol Ther. 2004;9:410–418. doi: 10.1016/j.ymthe.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Markusic D, Oude-Elferink R, Das AT, Berkhout B, Seppen J. Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter. Nucleic Acids Res. 2005;33:e63. doi: 10.1093/nar/gni062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fender P, Jeanson L, Ivanov MA, Colin P, Mallet J, Dedieu JF, Latta-Mahieu M. Controlled transgene expression by E1-E4-defective adenovirus vectors harbouring a “tet-on” switch system. J Gene Med. 2002;4:668–675. doi: 10.1002/jgm.315. [DOI] [PubMed] [Google Scholar]
- 17.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkovich E, Monnat RJ, Jr, Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protocols. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- 20.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 21.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soutoglou E, Misteli T. Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev. 2007;17:435–442. doi: 10.1016/j.gde.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu N, Bryant PE. Enhancement of frequencies of restriction endonuclease-induced chromatid breaks by arabinoside adenine in normal human and ataxia telangiectasia cells. Int J Radiat Biol. 1997;72:285–292. doi: 10.1080/095530097143275. [DOI] [PubMed] [Google Scholar]
- 24.Bryant PE, Johnston PJ. Restriction-endonuclease-induced DNA double-strand breaks and chromosomal aberrations in mammalian cells. Mutat Res. 1993;299:289–296. doi: 10.1016/0165-1218(93)90105-m. [DOI] [PubMed] [Google Scholar]
- 25.Lutze LH, Cleaver JE, Morgan WF, Winegar RA. Mechanisms involved in rejoining DNA double-strand breaks induced by ionizing radiation and restriction enzymes. Mutat Res. 1993;299:225–232. doi: 10.1016/0165-1218(93)90099-y. [DOI] [PubMed] [Google Scholar]