SUMMARY
The fatty-acid ethanolamides (FAEs) are lipid mediators present in all organisms and involved in highly conserved biological functions such as innate immunity, energy balance and stress control. They are produced from membrane N-acylphosphatidylethanolamines (NAPEs) and include agonists for G protein-coupled receptors (e.g. cannabinoid receptors) and nuclear receptors (e.g. PPAR-α). Here we report the crystal structure of human NAPE-hydrolyzing phospholipase D (NAPE-PLD) at 2.65 Å resolution, a membrane enzyme that catalyzes FAE formation in mammals. NAPE-PLD forms homodimers partly separated by an internal ~9 Å-wide channel and uniquely adapted to associate with phospholipids. A hydrophobic cavity provides an entryway for NAPE into the active site, where a binuclear Zn2+ center orchestrates its hydrolysis. Bile acids bind with high affinity to selective pockets in this cavity, enhancing dimer assembly and enabling catalysis. These elements offer multiple targets for the design of small-molecule NAPE-PLD modulators with potential applications in inflammation and metabolic disorders.
INTRODUCTION
The FAEs are a family of bioactive lipids with conserved cellular signaling functions throughout phylogeny (Chapman, 2004; Kang et al., 2008; Lucanic et al., 2011; Wellner et al., 2013). In mammals, monounsaturated and saturated FAEs (e.g., oleoylethanolamide and palmitoylethanolamide) are agonists for peroxisome proliferator-activated nuclear receptors (PPAR-α) (Fu et al., 2003) and G protein-coupled receptors (GPR119 and GPR55) (Godlewski et al., 2009). Oleylethanolamide regulates satiety, body weight, and cancer cell proliferation (Piomelli, 2013; Masoodi et al., 2014); palmitoylethanolamide serves as an early stop signal that contrasts the progress of inflammation (Solorzano et al., 2009). On the other hand, polyunsaturated FAEs (e.g., anandamide) activate cannabinoid receptors (CBs) – the same G protein-coupled receptors targeted by the psychotropic component Δ9-tetrahydrocannabinol in marijuana – and contribute in important ways to the regulation of food intake, stress and pain response, and synaptic function (Kathuria et al., 2003; Hohmann et al., 2005; Di Marzo and Matias, 2005; Di Marzo, 2008; Piomelli and Sasso, 2014). Despite the relevant signaling roles of these lipid molecules, little is known about the physiological stimuli that lead to their production, and about the mechanism and regulation of their biogenesis.
The first step in mammalian FAE biosynthesis is the transfer of a fatty acyl group from the sn-1 position of phosphatidylcholine to the amine of phosphatidylethanolamine (PE) (Schmid et al., 1996). This process produces a family of NAPEs, which are unusual among glycerophospholipids in that they contain three fatty acyl chains embedded in the lipid bilayer (Lafrance et al., 1997). A membrane-associated phospholipase, NAPE-PLD, cleaves the distal phosphodiester bond of NAPEs to generate bioactive FAEs and phosphatidic acid (PA) (Okamoto et al., 2004). The sequence of the NAPEPLD gene predicts a member of the metallo-β-lactamases (MβLs) (Wang et al., 2006), a protein superfamily that includes hydrolases involved in antibiotic resistance, DNA repair and RNA maturation (Garau et al., 2005; Dominski Z, 2007). NAPE-PLD is unrelated to other mammalian phospholipases D (PLD1 and PLD2) (Stuckey and Dixon, 1999), and further differs from these enzymes in its ability to selectively hydrolyze NAPEs (Okamoto et al., 2004). Importantly, NAPE-PLD recognizes all NAPEs, irrespective of their N-acyl substituents (Wang et al., 2006), and is able therefore to produce FAEs with diverse biological activities. To understand the molecular bases of FAE biosynthesis, we have determined the structure of human membrane NAPE-PLD at a resolution of 2.65 Å (Table 1), crystallized in the presence of the detergent sodium deoxycholate (DC). Our findings reveal unexpected binding sites and a potential key role of this natural bile acid in enzyme regulation.
Table 1.
Data collection and refinement statistics.
| NAPE-PLD | ||
|---|---|---|
| Data Collection | Native | SeMet |
| Synchrotron, Beamline | ELETTRA, XRD1 | ESRF, ID23h2 |
| Wavelength (Å) | 0.9795 | 0.8729 |
| Space group | P6522 | P6522 |
| Cell dimensions | ||
| a, b, c (Å) | 95.10, 95.10, 444.17 | 95.19, 95.19, 444.27 |
| α, β, γ (deg) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 88.83 - 2.65 (2.72 - 2.65) | 55.53 - 3.20 (3.42 - 3.20) |
| Rsym | 0.05 (0.55) | 0.16 (0.56) |
| I / σI | 26.7 (3.8) | 22.6 (9.3) |
| Completeness (%) | 100.0 (99.9) | 100.0 (100.0) |
| Redundancy | 14.1 (12.3) | 41.2 (42.9) |
| Resolution (Å) | 82.36 - 2.65 (2.72 - 2.65) | 55.53 - 3.20 (3.42 - 3.20) |
| No. unique reflections | 34047 | 20932 |
| SAD Phasing | ||
| No. molecules in the a.u. | 2 | |
| No. of Se sites | 16 | |
| Solvent content | 0.63 | |
| FOM a | 0.404 | |
| Refinement Statistics | ||
| Rwork / Rfree b | 0.214 (0.334) / 0.253 (0.331) | |
| No. atoms | 5875 | |
| Protein | 5361 | |
| Zinc | 4 × 1 | |
| PE | 2 × 44 | |
| DC | 7 × 28 | |
| Sulfate | 3 × 5 | |
| Water | 100 | |
| B-factors (Å2) | 76.0 | |
| Protein | 77.2 | |
| Zinc | 62.2 | |
| PE | 70.1 | |
| DC | 70.3 | |
| Sulfate | 60.4 | |
| Water | 59.3 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.014 | |
| Bond angles (deg) | 2.38 | |
| Ramachandran plot c | ||
| preferred (%) | 88.0 | |
| allowed (%) | 7.1 | |
| outliers (%) | 4.9 |
The numbers in parentheses are for the highest resolution shell.
Figure of Merit (experimental phases) = <|ΣP(α)exp(iα)/ΣP(α)|>, where a is the phase and P(α) is the phase probability distribution.
The Rfree was Calculated with a random 5% of the reflections.
Values from Molprobity (http://molprobity.biochem.duke.edu).
RESULTS
Structure of human NAPE-PLD and its membrane association
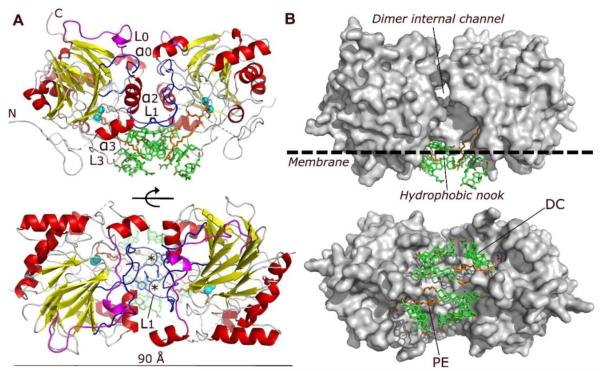
NAPE-PLD was crystallized in lithium sulphate by adding the bile acid DC (0.09%) to the purified protein (Experimental Procedures). The crystal structure was solved by the method of single-wavelength anomalous diffraction (SAD) with phase information derived from the anomalous scattering of 16 selenium atoms in the asymmetric unit (Table 1; see also Experimental Procedures). The enzyme forms a dimer with two interconnected protein subunits, partially separated by an internal channel (Figure 1). Each subunit contains the four-layered αββα core typical of the MβL superfamily (Carfi et al., 1995; De la Sierra-Gallay et al., 2005); however, various structural elements distinguish NAPE-PLD from other MβL proteins and allow this enzyme to interact with membrane phospholipids. These elements include an elongated N-terminal helix α0 and loop L0 (Figure 1A and Figure S1A), which replace the β1 strand present in most MβL proteins, and a second extended loop (L1), which substitutes for the MβL α1 helix (Figure 1A and Figure S1A). The L0 and L1 loops are rigidified by a high frequency of proline residues (12.7% from P68 to P177, compared to 6.6% and 6.2% in NAPE-PLD and human proteome, respectively) (Figure S1A) (Morgan et al., 2013), and mediate most intersubunit interactions (Figure 1A). Together with helices αo and α2, L0 and L1 form at the dimer interface an internal channel with a diameter of ~9 Å (Figure 1B).
Figure 1. Structure of human NAPE-PLD.
(A) Ribbon trace of the NAPE-PLD dimer, front view (top) and bottom view (bottom). Dimer subunits interact mainly through their loops L0 (magenta) and L1 (blue), structural elements that distinguishes NAPE-PLD from other MβL proteins. The subunit orientation produces a symmetric alignment of loops L1, which extend in the dimer the existing monomer binding faces. The active sites contain a binuclear zinc metal center (spheres, color cyan), which faces the lipid bilayer and binds a molecule of PE (carbon atoms in orange). PE derives from the expression system used to produce NAPE-PLD and occupies the binding site reserved to the PE fragment of NAPE. Several molecules of the non-conjugated C24 bile acid DC (carbon atoms in green) bind to the enzyme, creating a lipid substructure at the interface with the membrane. N and C indicate the N-terminus and C-terminus of the protein, respectively. The asterisks indicate the position of L1 residues Q158 and Y159. (B) Surface representation of the NAPE-PLD dimer shown with the same orientation as in (A). An internal channel with a diameter of ~9 Å partly separates the two subunits of NAPE-PLD, and overlooks a hydrophobic nook to which PE and DC are bound. The predicted membrane boundary is shown as dotted line.
A positive electrostatic potential prevails at the subunit interface of the NAPE-PLD dimer (Figure S1B). It is mainly due to several charged residues (K97, K108, R164, R166, R167, K202) that are exposed by both subunits within the internal channel. The presence of the channel and electrostatic repulsion among these residues limits the contact between the two subunits to an area (surface interaction area, ISA/2≈1010 Å2) that is substantially smaller than those generally found in obligate homodimeric proteins (≈1940 Å2) (Bahadur et al., 2003). The area involves the L1 loop and a series of intersubunit H-bonds (residues R153–N194, S157–Q158, Q158–N194, Y159–Y193, and K163–E201) (Figure 1A and Figure S1A). To probe the function of this unusual molecular architecture, we generated a NAPE-PLD in which Q158 and Y159 (Figure 1A and Figure S1A) are replaced by serines. We found that this loop L1-directed double mutation – which does not affect the overall MβL fold and residue polarity of NAPE-PLD monomers (Figure 1A) – disrupts the subunits ability to dimerize (Figure S1C) and hydrolyze NAPE (see below). These findings suggest a critical role for the L1 loop in dimer assembly and catalysis (Figure 1A).
In agreement with its expected membrane association, cryo-electron microscopy studies show that NAPE-PLD is found mainly in proximity of cellular membranes (Figure S2). This is consistent with the presence in the structure of an extended hydrophobic surface that spans both protein dimer subunits and includes residues of the N-terminus, loop L1, helix α3 and loop L3 (Figure 1). At the membrane interface, the NAPE-PLD dimer forms a hydrophobic nook that contains two molecules of PE, one bound to each monomer (Figure 1B). This phospholipid constitutes ~75-80 mol % of the cell membranes of E. coli (Morein et al., 1996) that was used to produce the recombinant protein for crystallization. PE is not a substrate for NAPE-PLD, but its structural similarity with NAPE suggests that it occupies a portion of the site reserved to this molecule (Figure 2A). Consistent with this view, PE inhibits detergent-stimulated NAPE-PLD activity with an IC50 of ≈30 μM (Figure S3A).
Figure 2. Interactions of NAPE-PLD with NAPE and bile acids.
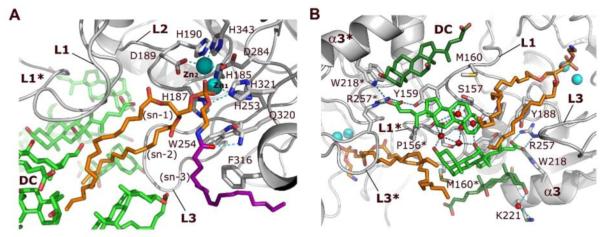
(A) Binding of NAPE to the active site of NAPE-PLD. A molecule of PE (carbon atoms in orange), which derives from the expression system used to produce the enzyme, occupies the binding site reserved to the PE fragment of NAPE. Its phosphate group bidentately bridges the two Zn2+ of the metal center (Zn1 and Zn2). Aspartate D284 bridges the two metal ions while three histidine residues (H185, H187 and H253) fill the coordination sphere of Zn1, and residues D189, H190, and H343 fill the coordination sphere of Zn2. A possible orientation of the N-acyl (oleoyl) substituent of NAPE is shown (carbons drawn in purple). (B) Specific binding sites for steroid acids. Two pairs of DC molecules (carbons colored in green) bind to the L1 loops at the interface of NAPE-PLD with the membrane. The central pair (carbons colored light green) forms a primary micelle of bile acids that exposes its hydrophobic convex side to the membrane interface. These steroid acids expand the hydrophobic surface of the opposite subunit via their steroid A-rings (Supplementary Fig. 1), and interact directly with the fatty acyl chains of PE. A network of H-bonds (dotted lines) for the bile acid carboxylate groups is arranged by the side chains of Y188, W218, and R257, together with that of Y159 of the opposite subunit. Two additional DC molecules (carbons colored dark green) form tight van der Waals contacts with the protein and expose their hydrophilic concave sides. This pairs contributes to orient the steroid carboxylates of the central DC pair (carbons colored light green). Red spheres symbolize water molecules involved in hydrogen bonding. Asterisks indicate residues and loops of the opposite dimer subunit.
A binuclear zinc center orchestrates NAPE hydrolysis
In proximity of the polar head of PE, electron density map shows the presence of two metal ions, which bind the PE phosphate group in a trigonal bipyramidal geometry (Figure 2A). The metal center of NAPE-PLD contains zinc as revealed by X-ray fluorescence analyses (Figure S3B). The L1 loop provides most of the hydrophobic surface involved in the binding of the sn-1 chain of PE, while the L3 loop binds the sn-2 chain and stretches out into the membrane interface (Figure 2A). The arrangement of L1, L3 and helix α3 shapes a broadening of the hydrophobic nook, in which the N-acyl chain of NAPE can be accommodated (Figure 2A). This provides both an entry point for NAPE into the active site and a membrane exit path for products of NAPE hydrolysis. Moreover, the width of this cavity may explain the ability of NAPE-PLD to recognize NAPE species with diverse N-acyl substituents. Residues H185 to D189, which are part of the L2 loop, coordinate the substrate-bound binuclear metal center, while H321 forms a hydrogen-bond with the ethanolamine oxygen of PE (Figure 2A). An adjacent glutamine residue, Q320, is ideally positioned to form a hydrogen bridge with NAPE’s carboxamide oxygen (Figure 2A). These interactions may help the zinc center to orient NAPE and orchestrate catalysis (Figure S3C and Figure S3D).
Bile acids regulate the biosynthesis of FAEs
In addition to PE, the hydrophobic nook of NAPE-PLD contains several molecules of bound DC, the natural bile acid that was used to crystallize the protein (Figure 1). Two of these steroid acids are contiguous to the tail end of the fatty acyl chains of PE and interact with residues in the L1 loops of both enzyme subunits (Figure 2B). They form a primary micelle of bile acids (Dietschy, 1968), exposing their convex hydrophobic sides to the membrane interface (Figure 2B). A tight binding pocket of the protein structure holds in place these bile acids. The side chains of residues W218, and R257, together with that of Y159 of the opposite subunit, form four perfectly oriented H-bonds with the bile acid carboxylate group (Y159 OH – O3 DC = 2.3 Å; R257 NE – O3 DC = 3.1 Å; R257 NE – O4 DC = 2.7 Å; W218 NE1 – O4 DC = 3.2 Å) (Figure 2B). Finally, a dense network of water-mediated hydrogen bridges – through the hydroxyl groups in position [3] and [12] of the steroid ring (Figure S1B) – reinforces the stability of the micelle and its binding to NAPE-PLD (Figure 2B). The specific and symmetric interactions between each bile acid molecule and the residues R257 and Y159 of both subunits, indicate that, upon binding, the bile acid micelle might stabilize the conformation of both the loops L3 and L1 (Figure 2B), which have a critical role in protein dimerization (Figure 1, Figure 2, and Figure S1C). A second pair of DC molecules forms tight van der Waals contacts with helix α3 of the same subunit, and with residue M160 of the opposite subunit (~3.6 Å close to the side chains of W218, C222, and M160) (Figure 2B). Together, the four bile acid molecules create an L1-bound symmetric lipidic substructure at the interface of NAPE-PLD with the membrane (Figure 2B and Figure S1B), which is in a position to influence dimer assembly and admission of NAPE into the active site.
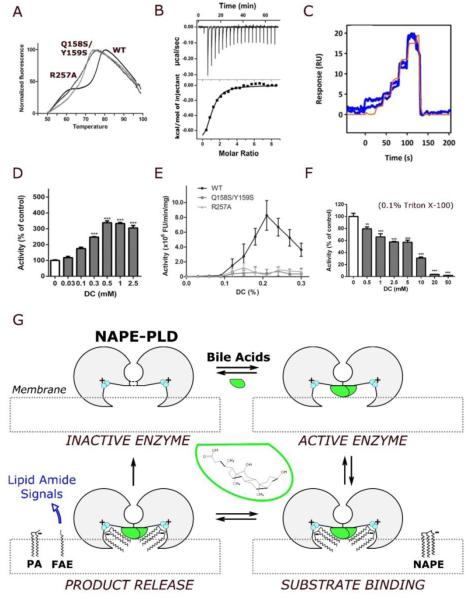
Consistent with this hypothesis, the NAPE-PLD mutant Q158S/Y159S, which cannot form stable dimers (Figure S3), has a substantially lower melting temperature (Tm = 70 °C) than does wild-type NAPE-PLD (Tm = 77°C) (Figure 3A). Furthermore, the R257A mutant, which lacks a key residue implicated in DC binding (Figure 2B), has a Tm value of 71°C (Figure 3A). In addition to stabilizing the NAPE-PLD dimer, DC regulates its activity. Indeed, (i) the bile acid binds to NAPE-PLD with a dissociation constant (KD) of ≈38 μM (Figure 3B), comparable to KD values measured at known bile acid receptors (e.g., FXR) (Makishima et al., 1999); moreover, (ii) this association is rapid and reversible (ka=9.9 ×103 M−1s−1; kd=0.44 s−1) (Figure 3C); and (iii) results in enzyme activation (EC50=186 μM) (Figure 3D). No such activation is seen with mutants that are unable to bind DC (mutant R257A) or form stable homodimers (mutant Q158S/Y159S) (Figure 3E). Noteworthy, DC inhibits the detergent-stimulated activity of NAPE-PLD (Triton X-100, 0.1%) with an IC50 of 8.8 mM (Figure 3F), a concentration that might be reached in vivo (Dietschy, 1968), suggesting a concentration-dependent bimodal regulation of the enzyme. These results indicate that the bile acid DC selectively binds to NAPE-PLD at micromolar concentrations and, by doing so, it promotes protein dimer formation and enzyme activity (Figure 3G). Overall, our findings suggest that the interaction between NAPE-PLD and natural bile acids might have potential biological relevance.
Figure 3. Bile acids regulate dimer formation and catalytic activity.
(A) Representative fluorescence thermal shift profiles for the stabilization of the NAPE-PLD dimer in the presence of DC (0.2% w/v). The analysis yields a melting temperature Tm = 77°C for the wild-type enzyme, and Tm = 71°C and 70°C for the mutants R257A and Q157S/Y158S, respectively (values are means for 5 replicates). (B) Binding thermodynamics profile for the titration of DC against NAPE-PLD obtained by isothermal titration calorimetry (ITC) at 25°C. Each peak corresponds to one injection of DC (top panel, raw titration data; bottom panel, integration of the data, corrected for heat of dilution). Microcalorimetric analysis yields a KD=38.5(±0.8) μM, ΔH=−1.2(±0.2) kcal/mol, and TΔS=−4.9(±0.3) kcal/mol (errors quoted standard deviations of n=3 experiments). (C) Profiles of the binding kinetics for DC on immobilized NAPE-PLD obtained at 25°C by surface plasmon resonance (SPR) with FastStep injections. Analysis (of n=3 replicates) yields a KD=44(2) μM, and dissociation constant Kd=0.44(3) s−1. (D) Concentration-dependent activation of NAPE-PLD by DC. The activity of purified human recombinant NAPE-PLD was measured in the absence of other detergents, using a liquid chromatography/mass spectrometry-based assay (LC/MS). (E) In the absence of synthetic detergent, DC stimulates the activity of wild-type (WT) human recombinant NAPE-PLD (0.1% w/v DC ≈ 2.4 mM DC), but not of NAPE-PLD mutants R257A and Q158S/Y159S. Enzyme activity was measured using a fluorescence-based assay (Methods). (F) In the presence of Triton X-100 (0.1%), DC inhibits the activity of wild-type enzyme using a liquid chromatography/mass spectrometry-based assay (Methods). (G) Hypothetical mechanism of NAPE-PLD activation. According to this model, illustrated clockwise from top left, bile acids (green) promote the assembly of inactive NAPE-PLD subunits (gray) into an active dimer. The resulting lipid-protein complex can recognize NAPE species at the membrane interface. The positively charged binuclear zinc center (cyan) coordinates anionic NAPEs, generated by N-acylation of zwitterionic PE, and orchestrates their hydrolysis to form bioactive FAEs along with phosphatidic acid (PA). A diversity of bioactive FAEs can be generated similarly that activate membrane GPCRs (e.g. cannabinoid receptors, CBs) or nuclear PPARs.
DISCUSSION
The crystal structure of human NAPE-PLD reveals how this enzyme has adapted to perform three key processes needed to initiate FAE signaling in cells. First, an extended lipid-binding surface spanning the NAPE-PLD dimer – unprecedented in the MβL superfamily – allows the enzyme to associate with membranes (Figure 1). Second, a hydrophobic nook facing the lipid bilayer (Figure 1) provides a recognition filter for NAPEs as glycerophospholipid class, while simultaneously accommodating different N-acyl substituents (Figure 2B). The enzyme has a positively charged zinc center that overlooks the membrane, which provides an attraction point for anionic NAPEs and helps coordinate their hydrolysis (Figure S3C and Figure S3D). Finally, specific pockets within this cavity tightly bind four bile acid molecules (Figure 2B), creating a unique protein–lipid molecular architecture that is able to initiate the biosynthesis of bioactive lipid amides (Figure 3G). In this process, the steroid acids form hydrogen bonds with residues of the L1 and L3 loops (Figure 2B), contributing to both stabilize the functional dimer assembly of NAPE-PLD and shape the enzyme hydrophobic nook (Figure 1 and Figure 3G).
The primary physiological role of bile acids is the emulsification and absorption of dietary fat in the small intestine. In mammals, they also serve, however, important signaling functions in cholesterol and glucose homeostasis by activating G protein-coupled membrane receptors (e.g. TGR5) (Thomas et al, 2008; Thomas et al, 2009) and nuclear receptors (e.g. FXR) (Parks et al., 1999; MI et al., 2003). Our findings show that NAPE-PLD is a novel cellular target of bile acids, and that this enzyme might orchestrate a cross-talk between bile acids and lipid amide signals. The interaction of bile acids with NAPE-PLD is likely to be functionally important in the gut, where they might regulate mobilization of oleylethanolamide, a FAE that promotes satiety and lipid absorption. Additionally, bile acids have been identified in the brain (Ogundare et al., 2010), where they might contribute to the production of FAEs, including the endocannabinoid anandamide. Irrespective of these possible physiological roles, the bile acid-binding elements in NAPE-PLD offers targets for the design of small-molecule modulators with potential applications in inflammation, obesity and cancer (Hotamisligil et al., 2006).
EXPERIMENTAL PROCEDURES
Plasmid constructs and mutagenesis
cDNA from a whole human Brain Library (Clonetech) was used to amplify the NAPE-PLD cDNA using conventional polymerase chain reaction (PCR) methodologies. The region encoding a truncated protein form (Δ47) from the amino acid D47 to F393 was amplified and inserted into the expression vector pMAL (NEB). The QuickChange site-directed mutagenesis kit (Agilent) was used to introduce the Q158S/Y159S or R257A mutations in pMAL (New England Biolabs) harboring the NAPE-PLD sequence as template. All the constructs were sequenced and validated. The resulting vector harboring the full length gene was transformed into Escherichia coli RosettagamiB (DE3) pLysS cells (Novagen) to produce the encoded recombinant protein with an N-terminus Maltose binding protein (MBP) tag and a C-terminus hexahistidine tag. For the expression of the protein in HEK293T cells, the coding sequences of the full length or Δ47 NAPE-PLD were inserted into pcDNA 3.1 (Life Technologies).
Crystallization and structure determination
Protein samples from pLysS cells were purified by affinity chromatography and, after MBP-tag cleavage, by size exclusion chromatography (16/600 200pg column, GE Healthcare, elution buffer 20 mM HEPES pH 7.8, 200 mM NaCl, in the absence or presence of DC). NAPE-PLD (0.09% DC) was crystallized at 22 °C by vapour diffusion, using a buffer of 0.9 M LiSO4 and 0.1M Hepes pH 7.8. The phase problem was solved using a selenomethionine-substituted crystal and single-wavelength anomalous dispersion (SAD) data. Supplementary Table 1 reports data collection and final refinement statistics. Full methods are described in the Supplemental Experimental Procedures, and include details for protein expression and purification, structure determination, bile acid binding thermodynamics and kinetics, enzyme activity assays, and cryo-immuno electron microscopy analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank the beam line staff of ELETTRA XRD1 (Trieste, Italy) and ESRF ID23 (Grenoble, France) for help with data collection. We thank Dr Andrea Armirotti for MS analysis. We thank Dr Elisa Romeo, Dr Ilaria Pettinati, Dr Benedetto Grimaldi, and Prof. Silvano Geremia for helpful discussion. The financial support of Marie Curie Action IRG (FP7-PEOPLE-2010-RG, contract no PIRG07-GA-2010-268385 to P.M.) and the National Institute on Drug Abuse (grants DK073955 and DA012413 to D.P.) is gratefully acknowledged.
P.M. performed the biochemical and biophysical analyses, crystallized the protein, and contributed to design the experiments; I.B., M.I., and M.B. carried out the enzymatic assays; R.M. and P.M. performed the electron microscopy experiments; G.G. directed the project, determined the crystal structure, interpreted the results and wrote the manuscript; D.P. co-directed the project, interpreted the results and wrote the manuscript.
Footnotes
SUPPLEMETAL INFORMATION
Supplemental information includes five supplemental figures, supplemental experimental procedures, supplemental references, and can be found with this article online at...
ACESSION NUMBERS
Coordinates and structure factors of human membrane NAPE-PLD have been deposited in the Protein Data Bank under the association code 4QN9.
REFERENCES
- Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- Carfi A, Pares S, Duée E, Galleni M, Duez C, Frère JM, Dideberg O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD. Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog Lipid Res. 2004;43:302–327. doi: 10.1016/j.plipres.2004.03.002. [DOI] [PubMed] [Google Scholar]
- De la Sierra-Gallay IL, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nature Rev. Drug Discov. 2008;7:438–455. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Matias I. Endocannabinoid control of food intake and energy balance. Nature Neurosci. 2005;8:585–589. doi: 10.1038/nn1457. [DOI] [PubMed] [Google Scholar]
- Dietschy JM. Mechanism for the intestinal absorption of bile acids. J. Lipid Res. 1968;9:297–309. [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodríguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Piomelli D. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- Garau G, Bebrone C, Anne C, Galleni M, Frère JM, Dideberg O. A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 2005;345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- Godlewski G, Offertáler L, Wagner JA, Kunos G. Receptors for acylethanolamides-GPR55 and GPR119. Prostaglandins Other Lipid Mediat. 2009;89:105–111. doi: 10.1016/j.prostaglandins.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, Krey JF, Walker JM, Holmes PV, Crystal JD, Duranti A, Tontini A, Mor M, Tarzia G, Piomelli D. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Kang L, Wang YS, Uppalapati SR, Wang K, Tang Y, Vadapalli V, Venables BJ, Chapman KD, Blancaflor EB, Mysore KS. Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 2008;56:336–349. doi: 10.1111/j.1365-313X.2008.03603.x. [DOI] [PubMed] [Google Scholar]
- Kathuria S, Gaetani S, Fegley D, Valiño F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Modulation of anxiety through blockade of anandamide hydrolysis. Nat. Medicine. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- Lafrance CP, Blochet JE, Pézolet M. N-acylphosphatidylethanolamines: effect of the N-acyl chain length on its orientation. Biophys J. 1997;72:2559–2568. doi: 10.1016/S0006-3495(97)78899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, Lithgow GJ, Gill MS. N-acylethanolamine signaling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature. 2011;473:226–229. doi: 10.1038/nature10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Masoodi M, Lee E, Eiden M, Bahlo A, Shi Y, Ceddia RB, Baccei C, Prasit P, Spaner DE. A role for oleoylethanolamide in chronic lymphocytic leukemia. Leukemia. 2014;28:1381–1387. doi: 10.1038/leu.2014.10. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Devarakonda S, Harp JM, Han Q, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Molecular Cell. 2003;11:1093–1100. doi: 10.1016/s1097-2765(03)00112-6. [DOI] [PubMed] [Google Scholar]
- Morein S, Andersson A-S, Rilfors L, Lindblom G. Wild-type Escherichia coli Cells Regulate the Membrane Lipid Composition in a “Window” between Gel and Non-lamellar Structures. J. Biol. Chem. 1996;12:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- Morgan AA, Rubestein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogundare M, Theofilopoulos S, Lockhart A, Hall LJ, Arenas E, Sjövall J, Brenton AG, Wang Y, Griffiths WJ. Cerebrospinal fluid steroidomics: are bioactive bile acids present in brain? J. Biol. Chem. 2010;285:4666–4679. doi: 10.1074/jbc.M109.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 2004;279:5298–5305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Piomelli D. A fatty gut feeling. Trends Endocrinol. Metab. 2013;24:332–341. doi: 10.1016/j.tem.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piomelli D, Sasso O. Peripheral gating of pain signals by endogenous lipid mediators. Nature Neurosci. 2014;17:164–174. doi: 10.1038/nn.3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid HH, Schmid PC, Natarajan V. The N-acylation-phosphodiesterase pathway and cell signalling. Chem. Phys. Lipids. 1996;80:133–142. doi: 10.1016/0009-3084(96)02554-6. [DOI] [PubMed] [Google Scholar]
- Solorzano C, Zhu C, Battista N, Astarita G, Lodola A, Rivara S, Mor M, Russo R, Maccarrone M, Antonietti F, Duranti A, Tontini A, Cuzzocrea S, Tarzia G, Piomelli D. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc Natl Acad Sci U S A. 2009;106:20966–20971. doi: 10.1073/pnas.0907417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature Rev. Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Wang J, Okamoto Y, Morishita J, Tsuboi K, Miyatake A, Ueda N. Functional analysis of the purified anandamide-generating phospholipase D as a member of the metallo-beta-lactamase family. J. Biol. Chem. 2006;281:12325–12335. doi: 10.1074/jbc.M512359200. [DOI] [PubMed] [Google Scholar]
- Wellner N, Diep TA, Janfelt C, Hansen HS. N-acylation of phosphatidylethanolamine and its biological functions in mammals. Biochim. Biophys. Acta. 2013;1831:652–662. doi: 10.1016/j.bbalip.2012.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.