Abstract
Non-obese diabetic (NOD) mice are a widely-used model oftype1 diabetes (T1D). However, not all animals develop overt diabetes. This study examined the circulating metabolomic profiles of NOD mice progressing or not progressing to T1D. Total beta-cell mass was quantified in the intact pancreas using transgenic NOD mice expressinggreen fluorescent protein under the control of mouse insulin I promoter.While both progressor and non-progressor animals displayed lymphocyte infiltration and endoplasmic reticulum stress in the pancreas tissue;overt T1D did not develop until animals lost ~70% of the total beta-cell mass.Gas chromatography time of flight mass spectrometry (GC-TOF) was used to measure >470 circulating metabolites in male and female progressor and non-progressor animals (n=76) across a wide range of ages (neonates to >40-wk).Statistical and multivariate analyses were used to identify age and sex independent metabolic markers which best differentiated progressor and non-progressor animals’ metabolic profiles. Key T1D-associated perturbations were related with: (1) increased plasma glucose and reduced 1,5-anhydroglucitol markers of glycemic control; (2) increased allantoin, gluconic acid and nitric oxide-derived saccharic acid markers of oxidative stress; (3) reduced lysine, an insulin secretagogue; (4) increased branched-chain amino acids, isoleucine and valine; (5) reduced unsaturated fatty acids including arachidonic acid; and (6)perturbations in urea cycle intermediates suggesting increased arginine-dependent NO synthesis. Together these findings highlight the strength of the unique approach of comparing progressor and non-progressor NOD mice to identify metabolic perturbations involved in T1D progression.
Keywords: Type 1 diabetes, Pancreatic beta-cells, Beta-cell loss, Metabolomics
INTRODUCTION
Type 1 diabetes (T1D) is characterized by progressive autoimmune destruction of insulin-secreting pancreatic beta-cells. Recent studies have shown that large fractions of T1D patients had insulin-producing cells at autopsy, unrelated to duration of disease or age at death (Meier et al. 2005), suggesting that some beta-cell population is preservedeven in patients with the long-standing T1D. Gianani et al. reported that 30% of patients with long-standing childhood-onset diabetes had numerous insulin-positive cells (Gianani et al. 2010).
Residual functional beta-cell mass affects glycemic control and the risk of developing T1D related complications. Currently, there is no practical means to measure beta-cell mass noninvasively in vivo(Lebastchi and Herold 2012). Hemoglobin A1c is a widely used marker for assessing glycemic control, however, its accuracy is affected by age, ethnicity and co-morbid conditions such as anemia and end-stage renal diseases (Standards of medical care in diabetes--2013 2013; Davidson and Schriger 2010; Ziemer et al. 2010; Robinson and Freedman 2013). Additionally, insulin-deficiency associated with T1D results in metabolic dysregulation beyond hyperglycemia. Dutta et al. showed alterations in amino acids, lipids, immuneand inflammatory factorsin T1D patient during a systemic insulin treatment and following an 8h insulin withdrawal(Dutta et al. 2012). Oresic et al. conducted a prospective metabolomic study in 56 children who progressed to T1D and 73 controls at autoantibody seroconversion and diabetes onset (Oresic et al. 2008). This study reported alterations in branched chain amino acids (BCAAs), glutamate and lipid metabolism which preceded the appearance of autoantibodies in children who progressed to T1D, suggesting that metabolomic dysregulations may have preceded the onset ofautoimmunity.
An organism's metabolomeis closely tied with genotype and environment, making metabolomic profilingan appealing method of characterizing the organism's phenotype and functional state. Recently, Sysi-Aho et al., usedNOD mice to validate the findings of Oresic et al. (Oresic et al. 2008) and characterizedanimal metabolic phenotypesusinglongitudinal lipidomics data(Sysi-Aho et al. 2011). They found that female NOD mice who later progressed to autoimmune diabetes exhibited the same lipidomic pattern as prediabetic children (Sysi-Aho et al. 2011). Similarly, Madsen et al. performed metabolomic analysisin plasma of NOD mice and C57BL/6 female mice (0-15 weeks) and found that NOD mice displayed altered metabolic profiles resembling thoseof children who later progressed to T1D (Madsen et al. 2012). It is of note that all of the aforementioned studies focused on metabolic perturbations occurring prior orduring seroconversion and did not include analysis of animals older than 25 weeksor nonprogressors.
In the present study we characterized the progression of autoimmune diabetes in both male and female NOD mice across a wide range of ages (neonates to >40-wk) and then utilized gas chromatography time of flight mass spectrometry (GC-TOF) to compare>470 circulating metabolites between animals progressing or avoiding T1D. Beta-cell mass in the intact pancreas was quantified using transgenic mice in which beta-cells were genetically labeled with green fluorescent protein (GFP) under the control of mouse insulin I promoter (MIP) (Kilimnik et al. 2009; Miller et al. 2009; Kim et al. 2009; Jo et al. 2011; Kilimnik et al. 2012). Progression of beta-cell destruction by immune cell infiltration was examined in the whole pancreas in relation to the vascular network as well as at the cellular level. Following the initial characterization, we identified simply two groups: non-progressors and progressors to T1D regardless of age or sex, by precisely correlating to the total residual beta-cell mass.It is well known that although NOD mice,an inbred laboratory mouse line, shareall the same genes and environment, only a subset of animals develop overt diabetes (i.e. chronic hyperglycemia). TheseNOD mice that maintain normoglycemia (i.e. non-progressors) have typically beenexcluded in most studies of T1D in the past. We reasoned that there should be a critical threshold for the functional beta-cell mass to maintain normoglycemia, which should be reflected in circulating metabolites that report the functional state. Comparison of the metabolic profiles of progressor and non-progressor NOD mice, which undergo the same autoimmune destruction of beta-cells, but display alterations in glucose control, is suggested to reveal distinct metabolomic pathways involved in T1D disease development.
RESEARCH DESIGN AND METHODS
Mice
Transgenic mice expressing green fluorescent protein (GFP) specifically in beta-cells under the control of mouse insulin I promoter (MIP) were generated on NOD background. All the procedures involving mice were approved by the University of Chicago Institutional Animal Care and Use Committee.
Quantification of beta-cell mass in intact pancreas
The entire distribution of beta-cells was captured using a method previously described (Kilimnik et al. 2012). Briefly, mouse whole pancreata were excised, fixed in 4% PFA and cleared in 100% glycerol. GFP-expressing beta-cells were visualized using an Olympus IX-80 spinning disk confocal microscope (Olympus, Melville, NY) with imaging software StereoInvestigator (MicroBrightField, Williston, VT). Quantification of beta-cell mass was carried out using a macro written for Fiji/ImageJ (http://rsbweb.nih.gov/ij/). MATLAB (MathWorks, Natick, MA) was used for mathematical analyses.
Visualization of the vascular network in the intact pancreas
Pancreatic blood vessels were labeled through cardiac perfusion of DiI (a lipophilic carbocyanine red fluorescent tracer, Invitrogen, Carlsbad, CA).
Histology and immunohistochemistry
Mouse pancreata were excised and frozen or paraffin-embedded. Sections were stained with hematoxylin and eosin (H&E), and immunostained with the following primary antibodies (all 1:500): rat anti-mouse CD4 and CD8 (BD Biosciences Pharmingen, San Diego, CA), polyclonal guinea pig anti-porcine insulin (DAKO, Carpinteria, CA), mouse monoclonal anti-human glucagon (Sigma-Aldrich, St. Louis, MO) and polyclonal goat anti-somatostatin (Santa Cruz, Santa Cruz, CA). Nuclei were stained with DAPI (Sigma-Aldrich). The primary antibodies were detected using ABC staining (Thermo Fisher Scientific, Rockford, IL) or a combination of DyLight 488, 549, and 649-conjugated secondary antibodies (1:200, Jackson ImmunoResearch Laboratory, West Grove, PA).
Transmission electron microscopy
Microdissected pancreas tissues were fixed with 4% PFA and 0.02% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h and embedded in resin. Ultrathin sections (80 nm) were stained with uranyl acetate and lead citrate. Images were taken using a Tecnai F30 microscope (FEI, Hillsboro, Oregon).
Metabolomic analysis
NOD mice (n = 76) were assessed as diabetic or non-diabetic based on their fasting (4 hr) blood glucose levels at sacrifice, which defined 23 hyperglycemic (glucose ≥ 250 mg/dL) and 53 normoglycemic animals (Supplemental Table S1). The MiniX database (Scholz and Fiehn 2007) was used as a Laboratory Information Management System (LIMS) and for sample randomization prior to all analytical procedures. Detailed sample preparation and data acquisition methods are reported in the supplemental methods. Briefly, plasma aliquots (15 μL), stored at −80°C, were thawed, extracted, derivatized and metabolites levels were quantified by GC-TOF mass spectrometry as previously described (O. Fiehn et al. 2008). All samples were analyzed in one batch, throughout which data quality and instrument performance were monitored using quality control and reference plasma samples (National Institute of Standards and Technology; NIST). Quality controls (n=8), comprised of a mixture of standards and analyzed every 10 samples, were monitored for changes in the ratio of analyte peak heights, and used to ensure equivalent instrumental conditions (p>0.05, t-Test comparing observed to expected ratios of analyte response factors) over the duration of the sample acquisition(O. Fiehn et al. 2008). During data processing, to ensure the exclusion of low confidence or background metabolites, measured specieswere reported if positively detected at very high mass spectral confidence in at least 50% of samples within the diabetic or non-diabetic groups of animals. Detailed criteria for peak reporting including: mass spectral matching, spectral purity, signal-to-noise and retention time are discussed in detail elsewhere (Oliver Fiehn et al. 2005). Known artifact peaks such as polysiloxanes or phthalates were excluded from data export in BinBase. Missing values were replaced by investigating the extracted ion traces of the raw data, subtracted by the local background noise. Data, reported as quantitative ion peak heights, was normalized by the sum intensity of all annotated metabolites and used for further statistical analysis. This presents a well-defined metric for qualitative comparisons of differences in metabolite levels between diabetic and non-diabetic animals. However it should be noted that the response factor between instrumental signal and metabolite concentration is undefined, and thus the comparison of percent change between diabetic and non-diabetic animals may not accurately represent a stoichiometric change in metabolite abundances.
Statistical and multivariate data analysis
All statistical data analyses were implemented in R (v 3.0.1) statistical programming language and environment (R Development Core Team. R: A language and environment for statistical computing. 2011). Analysis of covariance (ANCOVA) was performed on base 10 logarithm transformed measurements to test for differences in metabolite levels between diabetic and non-diabetic animals, adjusting for sex and age. The resulting test statics were adjusted for the false discovery rate (FDR) due to multiple hypotheses tested (Benjamini Y 1995), and reported as FDR-adjusted p-values, padj, and direct estimates of FDR, q-values (Strimmer 2008). Compared to the family-wise error rate correction (FWER) as described by Bonferroni, FDR adjustment is less stringent and results higher power (less false negatives) for studies with large numbers of statistical tests.
Multivariate predictive modeling was used to identifykey markers of large-scale metabolic changes between diabetic and non-diabetic animals. Orthogonal signal correction partial least squares discriminant analysis (O-PLS-DA) (Svensson O. 2002) was implemented on sex and age adjusted, base 10 logarithm transformed and autoscaled metabolomic measurements. Model testing and feature selection (Grapov et al. 2012) was used to identify and validate the top 10% metabolic discriminants of diabetic from non-diabetic animals. Briefly, the full sample set (n=76) was split between 2/3 training and 1/3 test sets while conserving the proportion of diabetic and non-diabetic samples in each set. Training data was used to carry out feature selection and model optimization, and final model performance was validated using the test data. A two latent variable (LV) model was calculated using the full variable set (n=476) from the training data, and its loadings and scores were used to identify the top ~10% of all metabolic determinants of the T1DM phenotype. Metabolites were retained in the model based on significant correlation of raw values with model scores (Spearmans, padj ≤ 0.05) (Wiklund et al. 2008) and absolute value of model loadings on LV1 ≥ 90th quantile (Palermo et al. 2009). The identified top metabolic descriptors of T1D were further validated using 100 replications of internal training and testing cross-validation, permutation testing and final model performance was estimated through the prediction of class labels for the initially held out test data (Supplemental Table S5). The training set was further randomly divided into 2/3 pseudo- trainingand 1/3 pseudo-test sets (n=100), which were then used to calculate the cross-validated fit to the training data (Q2) and root mean squared error of prediction (RMSEP) respectively. Permuted model Q2 and RMSEP distributions were calculated using identical procedures to those described above with the addition of randomly permuted sample class labels. Overall model sensitivity and specificity were estimated based on the predictionof classes labels for the initially held out test setusing a model fit to the complete training data (Supplemental Table S5).
Biochemical and partial correlation network analyses were used to evaluate metabolite associations and interpret statistical and multivariate results within a biological and experimental context. A biochemical and chemical similarity network (Barupal et al. 2012) was constructed between all measured metabolites with KEGG (Kanehisa et al. 2012) and PubChem CID (Bolton et al. 2011) identifiers (n = 188). Partial correlations were calculated between the O-PLS-DA identified top 10% feature set and all statistically significant T1D-associated perturbations. This was achieved using reduced order or q-order partial correlations (Castelo and Roverato 2009) (q=1, 19, 38, 56, n=1000), which have been previously implemented for Gaussian graphical models of high-dimensional metabolomic data (Oresic et al. 2012). An average non-rejection rate β=0.4 was used as a criteria for accepting pairwise relationships, for which coefficients of partial correlation, p-values and FDR adjusted p-values(Benjamini Y 1995) were calculated.
The developed networks were visualized using Cytoscape (Shannon et al. 2003), and network mapping was used to encode statistical and multivariate modeling results through the network edge and vertex attributes.
RESULTS
Immune cell infiltration and beta-cell destruction in prediabetic NOD mice
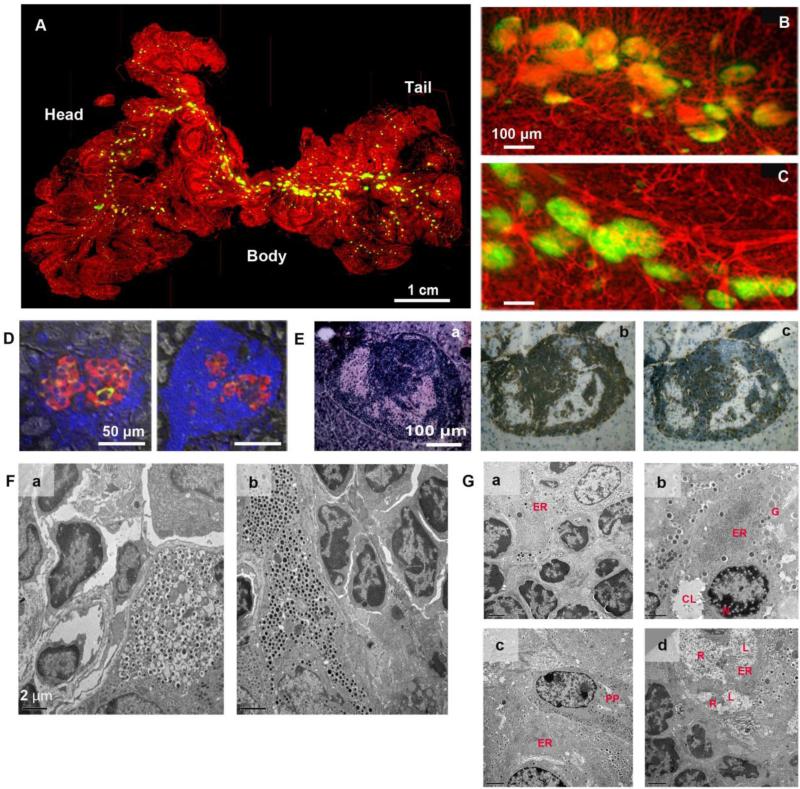
Spatial islet distribution was examined in relation to the vascular network in the intact pancreas. A representative image of a prediabetic NOD mouse shows a cluster of large islets in the body region (Fig. 1A). A close view demonstrates substantial beta-cell destruction where intraislet capillaries were exposed due to a loss of GFP-expressing beta-cells (Fig. 1B) compared to the body region of a wild-type mouse (Fig. 1C). A number of islets in prediabetic NOD mice show only a few beta-cells with marked immune cell infiltration (Fig. 1D). Adjacent sections of a hypertrophic islet (Fig. 1E.a) showed CD4 and CD8 positive T lymphocytes in and around the islet (Fig. 1E. b, c). Ultrastructual analysis of islets in diabetic and non-diabetic littermates (>40-wk old) revealed that neither group of mice escaped from autoimmune attack and individual beta-cells showed different degrees of damages in beta-cells within the same islets/pancreas. In normoglycemic NOD mice, damaged beta-cells with empty vesicles were often surrounded by lymphocytes (Fig. 1F.a). In contrast, some beta-cells in diabetic mice were intact despite the presence of lymphocyte infiltration (Fig. 1F.b), suggesting that an inbred strain of NOD mice undergo the same pathological destruction of beta-cells regardless of the manifestation of chronic hyperglycemia per se. Ultrastructural changes in beta-cells under autoimmune attack were highlighted by progressive degradation of endoplasmic reticulum (ER; Fig. 1G). Well-developed ER, a marker of cells under stress, was observed (Fig. 1G.a). Progressive ER degradation was documented with ribosome detachment from the ER membrane and aggregation as an initial event accompanied with nuclear damage with formation of form-like structures, immature granules with less dense cores and cytoplasmic liquefaction (Fig. 1G.b). An adjacent pancreatic polypeptide-cell to such injured beta-cells appears to be intact (Fig. 1G.c). Beta-cell degradation including ER swelling, ribosome shedding, amorphous cytoplasmic material and cytoplasmic, liquefaction were all observed in the same beta-cell (Fig. 1G.d).
Fig. 1. Immune cell infiltration and beta-cell destruction in prediabetic NOD mice.
A: Visualization of spatial islet distribution in the context of the vascular network in the intact pancreas. A prediabetic NOD mouse at 27-wk. B: The body region of the NOD mouse shown in A. Note that substantial beta-cell destruction is observed in the NOD pancreas (i.e. a loss of GFP-expressing beta-cells). C: Intraislet capillary network in the body region of a wild-type mouse at 21-wk. D: Immunohistochemical staining. Insulin (green), glucagon (red), somatostatin (white) and nuclei (blue). E: Hypertrophic islet with massive infiltration of T-lymphocytes. a. Hematoxylin-Eosin (HE) staining of the islet showing peripheral- and intra-islet infiltrating lymphocytes and remaining endocrine islet cells. b. A serial section stained for CD4-positive lymphocytes by ABC-staining (brown). c. A serial section stained for CD8-positive lymphocytes. F: Ultrastructural analysis of hypertrophic islets in non-diabetic and diabetic littermates. a. Non-diabetic male NOD mouse (41-wk old, 4-hr fasting BG: 136 mg/dL) showing a hyperactive beta-cell with lymphocyte infiltration and vesicles without dense core granules. b. Beta-cells in diabetic female NOD mouse (40-wk old, 4-hr fasting BG: 559 mg/dL) appears to be intact despite the presence of ongoing insulitis. G: Progressive degradation of endoplasmic reticulum (ER).a. Well-developed ER (ER) in a beta-cell undergoing insulitis. b. ER degradation. Ribosomes are detached (shed) from the ER membrane and are aggregated (ER). Nuclear damage is seen with the formation of foam-like structures (N). Immature granules with less dense cores (G) as well as cytoplasmic liquefaction (CL) are observed. c. ER membrane breakdown. ER membrane breakdown resulted in aggregation of shed ribosomes (ER). An adjacent PP-cell (PP) appears to be intact (identified by characteristic moderately dense cores of pancreatic polypeptide-containing secretory granules). d. Beta-cell degradation. ER swelling (ER), ribosome shedding, amorphous cytoplasmic material (R) and cytoplasmic, liquefaction (L) are observed in the same beta-cell.
Progression of autoimmune diabetes in NOD mice
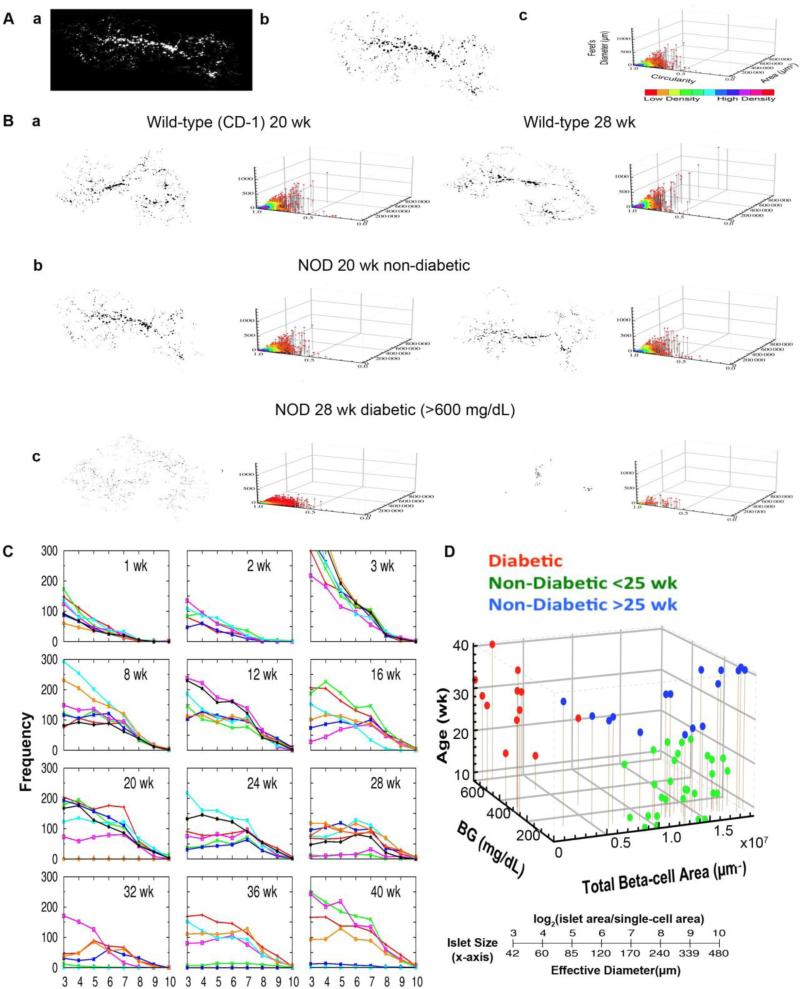
The entire distribution of beta-cells in the intact pancreas was captured and quantified (Fig. 2A.a, b). With area, circularity (a degree of roundness where the number 1.0 depicts a perfect circle) and Feret's diameter (the longest distance within an area), every beta-cell/islet in the whole pancreas is visualized in a 3D scatter plot (Fig. 2A.c). In wild-type mice (on CD-1 background), aging accompanies an increase in large islets with a retained population of small islets including small clusters of beta-cells (Fig. 2B.a). NOD mice preferentially lose large islets before the onset of chronic hyperglycemia and eventually lose most of the islets (Fig. 2B.b). We examined islet number and size distribution from week 1 to 40 (n=5-7 each age group, total 78 mice; Fig. 1C). During neonatal development, there was no difference in beta-cell mass and islet size distribution among littermates. However, marked individual heterogeneity was observed in NOD mice with age (>8-wk). In these mice, we have identified three distinct groups: (1) mice that develop chronic hyperglycemia regardless of age or sex with a loss of ~70% of total beta-cell mass; (2) young mice with normoglycemia (<25 wk); and (3) old mice with normoglycemia (25->40 wk; Fig. 2D). Note that such littermates that escaped developing overt diabetes have been excluded in most studies on NOD mice in the past.
Fig. 2. Progression of autoimmune diabetes in NOD mice.
A:a. Virtual slice capture of a whole mouse pancreas from mouse insulin promoter I (MIP)-green fluorescent protein (GFP) mice on NOD background. b. Measured beta-cell/islet distribution. c. Corresponding 3D scatter plot of islet parameters depicts distribution of islets with various sizes and shapes. Each dot represents a single islet. B:a. Representative data showing islet growth in wild-type mice at 20- and 28-wk of age. b. Examples of beta-cell loss at 20-wk (non-diabetic) and 28-wk (diabetic) in NOD mice. C:Heterogeneous beta-cell loss in NOD mice. Frequency is plotted against islet size.D: Three distinct groups in the development of T1D in NOD mice. 3D scatter plot showing the relationship among blood glucose levels (BG), total beta-cell area and age. Three groups of mice are color-coded as diabetic mice (red), young mice with normoglycemia (<25 wk; green) and old mice with normoglycemia (25-40 wk; blue).
Analysis of plasma metabolites in NOD mice
Unbiased GC-TOF based metabolomics was used to compare plasma profiles of 53 non-diabetic and 23 diabetic animals (Supplemental Table S1). A total of 476 metabolomic peaks were detected, including 188 structurally annotated metabolites. Measured peak heights are reported in normalized units using the diagnostic ‘unique ion’ for each peak.
Comparison of NOD mice physical and biochemical characteristics
Statistical analyses were used to compare physical characteristics and metabolomic measurements between diabetic and non-diabetic NOD mice. Diabetic animals displayed significantly increased fasting glucose, but reduced body weight compared to non-diabetic animals (Supplemental Table S1). ANCOVA in conjunction with FDR correction was used to identify 232 (49%) significantly altered metabolic features (padj≤0.05) between diabetic and non-diabetic animals (Supplemental Table S3). Statistical comparison of the major classes of molecules and biochemical subdomains identified general increases in the majority of circulating metabolites including: carbohydrates, lipids, organic acids, BCAA, ketone bodies and nucleotides in diabetic compared to non-diabetic animals (Supplemental Table S2).
Multivariate classification modeling using O-PLS-DA coupled with feature selection was used to identify the top metabolic descriptors of the T1D plasma phenotype. A subset of 48 metabolites (10%) consisting of 14 known (Table 1) and 34 unknown species (Supplemental Table S4) were selected part of a validated model (Supplemental Table S5) for the discrimination between diabetic and non-diabetic NOD mice.
Table 1.
Top descriptors of type 1 diabetes related metabolic perturbations.
| Rank* | variable | non-diabetic† | diabetic | FC‡ | padj§ | q-value∥ |
|---|---|---|---|---|---|---|
| 1 | 3,6-anhydrogalactose | 2310 ± 1700 | 9920 ± 12000 | 4.3 | 3.56E-08 | 1.07E-08 |
| 2 | 1,5-anhydroglucitol | 78900 ± 21000 | 30900 ± 29000 | 0.4 | 6.74E-08 | 2.02E-08 |
| 3 | glucose¶ | 136000 ± 110000 | 415000 ± 140000 | 3.0 | 4.81E-06 | 1.43E-06 |
| 4 | maltose | 2010 ± 1800 | 6230 ± 3000 | 3.1 | 1.97E-07 | 5.74E-08 |
| 5 | hexuronic acid | 4710 ± 3700 | 12800 ± 4700 | 2.7 | 1.17E-06 | 3.44E-07 |
| 6 | lysine | 121000 ± 49000 | 65700 ± 37000 | 0.5 | 8.69E-04 | 2.58E-04 |
| 7 | gluconic acid | 285 ± 230 | 711 ± 330 | 2.5 | 5.03E-08 | 1.41E-08 |
| 8 | conduritol beta epoxide | 1170 ± 540 | 451 ± 340 | 0.4 | 8.59E-10 | 2.57E-10 |
| 9 | pantothenic acid | 1380 ± 1200 | 4350 ± 2000 | 3.1 | 1.17E-07 | 3.46E-08 |
| 10 | cellobiotol | 479 ± 380 | 1240 ± 1100 | 2.6 | 1.14E-05 | 3.40E-06 |
| 11 | saccharic acid | 444 ± 470 | 1480 ± 880 | 3.3 | 5.03E-08 | 1.47E-08 |
| 12 | methylhexadecanoic acid | 2460 ± 1500 | 4490 ± 1700 | 1.8 | 4.40E-04 | 1.32E-04 |
| 13 | 3-hydroxybutanoic acid | 63500 ± 49000 | 234000 ± 280000 | 3.7 | 6.05E-03 | 1.79E-03 |
| 14 | 2-hydroxyvaleric acid | 802 ± 670 | 2150 ± 1600 | 2.7 | 2.01E-05 | 5.98E-06 |
see Supplemental Table S5 for model performance statistics and Supplemental Table S4 for unknown metabolites
values are reported as the mean ± standard deviation
fold change of means in diabetic compared to non-diabetic animals
false discovery rate adjusted p-value
estimated false discovery rate
measured by GC-TOF
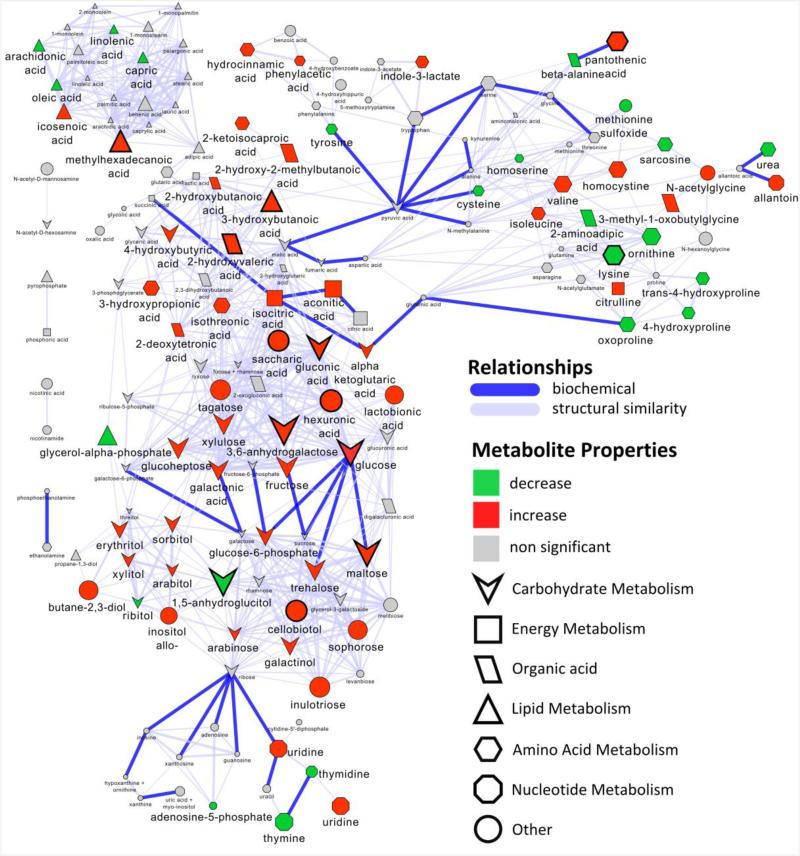
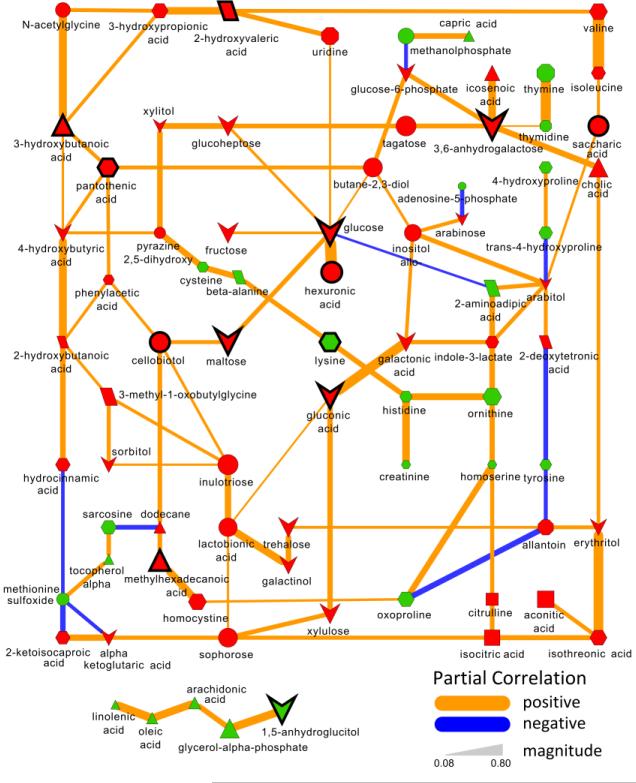
Biochemical and chemical similarity network analysis was used to calculate and displays relationships between precursor and product metabolite reactant pairs and molecules sharing a high degree of structural similarity (Fig. 3). A Gaussian graphical model network was calculated to identify conditionally independent relationships (partial correlation, padj≤0.05) between all significant (padj≤0.05) T1D-associated metabolic perturbations (Fig. 4).
Fig. 3. Biochemical network displaying metabolic differences between diabetic and non-diabetic NOD mice.
Metabolites are connected based on biochemical relationships (blue, KEGG RPAIRS) or structural similarity (violet, Tanimoto coefficient ≥ 0.7). Metabolite size and color represent the importance (O-PLS-DA model loadings, LV 1) and relative change (gray, padj>0.05; green, increase; red, decrease) in diabetic compared non-diabetic NOD mice. Shapes display metabolites’ molecular classes or biochemical sub-domains and top descriptors of T1D-associated metabolic perturbations (Table 1) are highlighted with thick black borders.
Fig. 4. Partial correlation network displaying associations between all type 1 diabetes-dependent metabolomic perturbations.
All significantly altered metabolites (padj≤0.05, Supplemental Table S3) are connected based on partial correlations (padj≤0.05). Edge width displays the absolute magnitude and color the direction (orange, positive; blue, negative) of the partial-coefficient of correlation. Metabolite size and color represent the importance (O-PLS-DA model loadings, LV 1) and relative change (gray, padj>0.05; green, increase; red, decrease) in diabetic compared non-diabetic NOD mice. Shapes display metabolites’ molecular classes or biochemical sub-domains (see Fig. 3 legend), and top descriptors of T1D-associated metabolic perturbations (Table 1) are highlighted with thick black borders.
Diabetic NOD mice indicate altered carbohydrate profiles
Total plasma carbohydrates were increased by 125% in diabetic mice (Supplemental Table S2), and these changes were dominated by a 330% increase in 3,6-anhydrogalactose (Table 1). Interestingly, select carbohydrate species were decreased in diabetics compared to non-diabetic animals including: 1,5-anhydroglucitol, threonic acid and ribitol (60%, 30% and 20%). T1D-dependent alterations in 3,6-anhydrogalactose, 1,5-anhydroglucitol and glucose constituted the top three known metabolic predictors of the diabetic phenotype (Table 1). Elevations in 3,6-anhydrogalactose and glucose were shown to be both biochemically (Fig. 3) and empirically (Fig. 4) related to the general alterations in most carbohydrates, but not to the reduction in 1,5-anhydroglucitol; the change in which paralleled similar reductions in unsaturated fatty acids (Fig. 4). In addition to aforementioned alterations, T1D-dependent increases in maltose, gluconic acid, cellobiotol, saccharic acid and decreases in conduritol beta epoxide were similarly identified as top descriptors of this malady (Table 1).
Diabetic NOD mice display perturbations in lipid profiles
Lipid related metabolites were increased by an average of 171% in diabetic compared to non-diabetic animals, constituting the largest overall magnitudinal shift of any biochemical sub-domain (Supplemental Table S2). Major changes were noted by a 280% and 270% increase cholic acid, and ketone body, 3-hydroxybutanoic acid (Supplemental Table S3). The dominant plasma unsaturated fatty acids: linoleic acid, oleic acid and arachidonic acid were all decreased in diabetic compared to non-diabetic animals, and correlated with the reduction in 1,5-anhydroglucitol (Fig. 4). Overall the T1D-associated increases in methylhexadecanoic acid, 3-hydroxybutanoic acid and 2-hydroxyvaleric acid, were all selected as top predictors for T1D (Table 1).
Diabetic NOD mice display alterations in amino acid profiles
Overall, 28 of 49 measured amino acid metabolites (57%) were significantly perturbed in diabetic mice (Supplemental Table S3). Of these changes, 12 species were significantly decreased including 50% reductions in lysine, ornithine and 4-hydroxyproline in diabetic compared to non-diabetic animals (Supplemental Table S3). Conversely 16 species showed T1D-associated elevations including 210% and 160% increases in pantothenic acid and 3-ureidopropionate. These noted shifts were included in the top predictor set for T1D by 6th and 9th ranked lysine and pantothenic acid (Table 1). The observed reduction in lysine was closely related to similar reductions in cysteine, beta-alanine, histidine, creatinine and ornithine (Fig. 4), while the elevation in pantothenic acid was positively correlated with 3-hydroxybutanoic and indirectly related to increase in the majority of ketone bodies (Fig. 4).
DISCUSSION
The prevalence of T1D in children younger than 20 years of age has increased by 23% in the last decade (Nokoff and Rewers 2013). The fact that the concordance rate for T1D among monozygotic twins is only between 10-40%, and the majority of T1D patients (90%) develop the disease sporadically without family history, suggests that there are non-genetic but environmental pathogenic factors such as diet, viruses and the intestinal microbiome(Dutta et al. 2012). Metabolomics provides complementary information to genomic and proteomic data, capturing the final downstream products that reflect the body's response to lifestyle and environmental factors (McKillop and Flatt 2011). Identifying circulating biomarkers of beta-cell destruction as early in the diabetic state as possible could lead to better understanding of the triggers of autoimmunity and potentially serve as a therapeutic target. Studying these markers of metabolic dysregulation in conjunction with actual pancreatic morphology provides an even more robust phenotype for stages of disease progression.
Since metabolites are conserved across species, studying inbred mice raised in a controlled laboratory setting eliminates variables and enables translation of targeted research findings from animals to humans, a notion which has been demonstrated in previous studies (Sysi-Aho et al. 2011; Madsen et al. 2012). Furthermore, animal studies allow for precise quantification of residual beta-cell mass including islet cell size, number, distribution and shape. Importantly, the cohort of NOD mice used in this study extended beyond 25 weeks and included NOD mice which escaped chronic hyperglycemia, a group which has been largely excluded in previous studies, despite undergoing the same autoimmune destruction of beta-cells. The inclusion of mice which avoid the onset of hyperglycemia is of particular value as this allows for a comparison between progressors and non-progressors undergoing similar beta-cell insult wherein it is the differences between these two groups which allows for a unique insight into T1D pathophysiology and beta-cell viability and function. We have shown that beta-cell loss in NOD mice proceeds through gradual loss of large islet cells before the onset of hyperglycemia followed by an eventual loss of most of the islets including the small clusters. Importantly, we demonstrate that chronic hyperglycemia only occurs in mice with less than 30% remaining total beta-cell mass suggesting that there is a critical threshold of beta-cell loss before onset of severe hyperglycemia and T1D. The onset of T1D may involve a rapidly accelerating cycle of (1) reduction in beta-cell function due to autoimmune, inflammatory and/or metabolic insult; (2)increased insulin demand on functionally compromised beta-cells; (3)worsening lipids and glucose control and increased beta-cell lipo- and glucotoxicity; and (4) beta-cell de-granulation, apoptosis and increasing T1D severity (Dutta et al. 2012; Akirav et al. 2008).
Unbiased metabolomic analysis identified major T1D-associated perturbations in plasma carbohydrates, carbohydrate oxidation products, lipids, amino acids and organic acids. Progressors to T1D also displayed increased markers of oxidative stress, a known etiology of diabetes (Maritim et al. 2003), including: (1) allantoin, a non-enzymatic oxidation product of uric acid and known marker of oxidative stress (Lanza et al. 2010); (2) gluconic acid, a product of glucose oxidation or autoxidation (Bankar et al. 2009); and saccharic acid, a product of glucose oxidation by nitric oxide (NO) (Smith et al. 2012). In light of the T1DM-dependent increases in oxidative metabolites, alpha-tocopherol, a lipophilic antioxidant, was reduced in diabetic compared to non-diabetic animals (Supplemental Table S3), further suggesting a heightened state of oxidative stress. Beta-cells are particularly sensitive to oxidation byproducts (Holohan et al. 2008; Oyadomari et al. 2001; Sampson et al. 2010) and, therefore, it is of particular interest to uncover the connection between changes in carbohydrate oxidation products and global T1DM-associated metabolic perturbations.
Diabetic animals displayed alterations in several urea cycle intermediates suggestive of a T1DM-associated elevationin nitric oxide (NO) synthesis.Urea, ornithine, aspartic acid, and nacetyl glutamate were decreased while citrulline was increased in diabetic compared to non-diabetic animals (Supplemental Table S3). Citrulline can be derived from either ornithine via ornithine transcarbamylase or as a byproduct of arginine-nitric oxide synthase(Mori 2007). The observed T1D-dependent reductions in urea cycle intermediates and concomitant elevations in citrulline may indicate increased arginine-dependent NO synthesis in diabetic compared to non-diabetic animals. Overproduction of NO has been previously implicated in T1D (Kolb and Kolb-Bachofen 1992) and canlead to beta-cell death (Holohan et al. 2008). For example, the cytotoxic effects of cytokine-induced beta cell death are mediated through IL-1ß-induced NOsynthesis through the arginine-nitric oxide synthase(Southern et al. 1990; Kacheva et al. 2011). The liver is the dominant site of both arginine metabolism and the urea cycle, which may suggest impaired liver function in diabetic compared to non-diabetic animals. This is supported by an observed 100% increase in pipecolic acid (Supplemental Table S3), a metabolite of lysine and noted marker of liver disease (Kawasaki et al. 1988), in diabetic compared to non-diabetic animals. However, it remains to be determined if these noted changes are directly linked with beta-cell viability and T1D progression.
Prior metabolomics analyses have been applied separately to T1D and T2D, because of the inherent differences in pathogenesis of these disease states. During the discovery phase of biomarkers in our NOD mouse cohort, initial signals have aligned with other studies previously performed in human T1D trials, human T2D trials as well as other NOD mouse studies, suggesting common mechanisms leading to beta-cell dysfunction as well as compensation (Nakagawa and Ishii 1996; Pflueger et al. 2011; Juraschek et al. 2012). Alterations in plasma free fatty acids have been shown to impact lipid signaling in T2D (Grapov et al. 2012). In our study, T1D-associated reductions in polyunsaturated fatty acid, specifically arachidonic acid, may suggest increased flux into lipid signaling pathways.Of which, both cyclooxygenase-2 and 12-lipoxygenase products of arachidonic acid play critical roles in cytokine-induced human beta-cell destruction (Heitmeier et al. 2004; Tran et al. 2002; Chen et al. 2005; Parazzoli et al. 2012) and remains a focus of ongoing studies. In addition to fatty acids, studies in humans and rodents have indicated that lysine potentiates glucose-stimulated insulin secretion (Kalogeropoulou et al. 2009; Liu et al. 2008). Our analysis indicated a T1D-associated reduction in lysine which may constitute an early marker of beta-cell dysfunction. Elevations in BCAA have been repeatedly reported both in T1D and T2D (Newgard 2012; Adeva et al. 2012; Oresic et al. 2008). The replication of BCAA signatures in our model, as well as others (Oresic et al. 2008; Wang et al. 2011), coupled with the observation that BCAA levels may be early markers for T1D onset (Oresic et al. 2008), highlights the potential interaction between BCAAs and beta-cell destruction and warrants further investigation.
T1DM-associated increases in BCAA weredirectly linked to changes in 2-hydroxyvaleric acid (Fig. 4). This reduced form of a 5 carbon alpha-keto acid, was increased in diabetic compared to non-diabetic animals and constituted thesingle best predictive marker for T1DM (Table 1). Perturbations in 2-hydroxyvaleric acid were also indirectly linked (Fig. 4) to increases in a ketone body, 3-hydroxybutanoic acid (Table 1). Ketone bodies are produced in response to insufficient plasma glucose concentrations or poor glucose utilization, and T1DM-associated inability to maintain ketone body catabolism or renal clearance is observed in cases of diabetic ketoacidosis (Lu et al. 2012). In addition to the T1DM-associated elevation in 3-hydroxybutanoic acid (Stojanovic and Ihle 2011), oxoproline and 4-hydroxyproline were decreased in diabetic compared to non-diabetic animals (Supplemental Table S3), supporting similar observations in diabetic patients exhibiting ketoacidosis (Lu et al. 2012). The noted increases in ketone bodiesare a likely secondary effect of the diabetic animals’ decreased ability to utilize glucose as evident in their hyperglycemia. However the expanded metabolomic analysis may be useful for uncovering the impact of T1DM-associated ketoacidosis on other biochemical domains (e.g. alpha-keto acids, BCAA, proline metabolites) which may constitute improved diagnostic markers of acidosis severity and liver function.
In conclusion, we identified marked differences in the rates of progression of NOD mice to T1D. Metabolomic analysis was used to identify age and sex independent metabolic markers, which may explain this heterogeneity. Future studies combining metabolic end points (as they correlate with beta-cell mass) and genetic risk profiles will ultimately lead to a more complete understanding of disease onset and progression.
Supplementary Material
ACKNOWLEDGEMENTS
The study is supported as a pilot project by the West Coast Metabolomics Center,US Public Health Service Grant DK097154 (to OF); DK-020595 to the University of Chicago Diabetes Research and Training Center (Animal Models Core), DK-072473, AG-042151, and a gift from the Kovler Family Foundation (to MH); and the Intramural research program of the NIH, NIDDK (to VP). The authors thank Mrs. German Kilimnik, Billy Zhao and Mark Zielinski, and Drs. Xioajun Wang and Ryosuke Misawa at the University of Chicagofor the technical assistance.
Footnotes
Author Contributions: JH, AP and MH conducted mouse studies, and JJ, VP and MH analyzed the data. DG, JF and OF designed and performed metabolomic experiments, and DG analyzed the metabolomic data. DG, JF, JH, OF and MH wrote the manuscript. All authors participated in discussion and editing the manuscript. MH is the guarantor of this work and as such had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
No potential conflicts of interest relevant to this article were reported.
REFERENCES
- Adeva MM, Calvino J, Souto G, Donapetry C. Insulin resistance and the metabolism of branched-chain amino acids in humans. Amino Acids. 2012;43:171–181. doi: 10.1007/s00726-011-1088-7. [DOI] [PubMed] [Google Scholar]
- Akirav E, Kushner JA, Herold KC. Beta-cell mass and type 1 diabetes: going, going, gone? Diabetes. 2008;57:2883–2888. doi: 10.2337/db07-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankar SB, Bule MV, Singhal RS, Ananthanarayan L. Glucose oxidase--an overview. BiotechnolAdv. 2009;27:489–501. doi: 10.1016/j.biotechadv.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Barupal DK, Haldiya PK, Wohlgemuth G, Kind T, Kothari SL, Pinkerton KE, et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y HY. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Bolton EE, Chen J, Kim S, Han L, He S, Shi W, et al. PubChem3D: a new resource for scientists. J Cheminform. 2011;3:32. doi: 10.1186/1758-2946-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo R, Roverato A. Reverse engineering molecular regulatory networks from microarray data with qp-graphs. J ComputBiol. 2009;16:213–227. doi: 10.1089/cmb.2008.08TT. [DOI] [PubMed] [Google Scholar]
- Chen M, Yang ZD, Smith KM, Carter JD, Nadler JL. Activation of 12-lipoxygenase in proinflammatory cytokine-mediated beta cell toxicity. Diabetologia. 2005;48:486–495. doi: 10.1007/s00125-005-1673-y. [DOI] [PubMed] [Google Scholar]
- Davidson MB, Schriger DL. Effect of age and race/ethnicity on HbA1c levels in people without known diabetes mellitus: implications for the diagnosis of diabetes. Diabetes Res ClinPract. 2010;87:415–421. doi: 10.1016/j.diabres.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Dutta T, Chai HS, Ward LE, Ghosh A, Persson XM, Ford GC, et al. Concordance of changes in metabolic pathways based on plasma metabolomics and skeletal muscle transcriptomics in type 1 diabetes. Diabetes. 2012;61:1004–1016. doi: 10.2337/db11-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M. Setup and Annotation of Metabolomic Experiments by Integrating Biological and Mass Spectrometric Metadata. In: Ludäscher B, Raschid L, editors. Data Integration in the Life Sciences. Vol. 3615. Springer; Berlin Heidelberg: 2005. pp. 224–239. Lecture Notes in Computer Science. [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, et al. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- Gianani R, Campbell-Thompson M, Sarkar SA, Wasserfall C, Pugliese A, Solis JM, et al. Dimorphic histopathology of long-standing childhood-onset diabetes. Diabetologia. 2010;53:690–698. doi: 10.1007/s00125-009-1642-y. [DOI] [PubMed] [Google Scholar]
- Grapov D, Adams SH, Pedersen TL, Garvey WT, Newman JW. Type 2 diabetes associated changes in the plasma non-esterified fatty acids, oxylipins and endocannabinoids. PLoS One. 2012;7:e48852. doi: 10.1371/journal.pone.0048852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmeier MR, Kelly CB, Ensor NJ, Gibson KA, Mullis KG, Corbett JA, et al. Role of cyclooxygenase-2 in cytokine-induced beta-cell dysfunction and damage by isolated rat and human islets. J BiolChem. 2004;279:53145–53151. doi: 10.1074/jbc.M410978200. [DOI] [PubMed] [Google Scholar]
- Holohan C, Szegezdi E, Ritter T, O'Brien T, Samali A. Cytokine-induced beta-cell apoptosis is NO-dependent, mitochondria-mediated and inhibited by BCL-XL. J Cell Mol Med. 2008;12:591–606. doi: 10.1111/j.1582-4934.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Kilimnik G, Kim A, Guo C, Periwal V, Hara M. Formation of pancreatic islets involves coordinated expansion of small islets and fission of large interconnected islet-like structures. Biophys J. 2011;101:565–574. doi: 10.1016/j.bpj.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraschek SP, Steffes MW, Miller ER, 3rd, Selvin E. Alternative markers of hyperglycemia and risk of diabetes. Diabetes Care. 2012;35:2265–2270. doi: 10.2337/dc12-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacheva S, Lenzen S, Gurgul-Convey E. Differential effects of proinflammatory cytokines on cell death and ER stress in insulin-secreting INS1E cells and the involvement of nitric oxide. Cytokine. 2011;55:195–201. doi: 10.1016/j.cyto.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Kalogeropoulou D, LaFave L, Schweim K, Gannon MC, Nuttall FQ. Lysine ingestion markedly attenuates the glucose response to ingested glucose without a change in insulin response. Am J ClinNutr. 2009;90:314–320. doi: 10.3945/ajcn.2008.27381. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Hori T, Nakajima M, Takeshita K. Plasma levels of pipecolic acid in patients with chronic liver disease. Hepatology. 1988;8:286–289. doi: 10.1002/hep.1840080216. [DOI] [PubMed] [Google Scholar]
- Kilimnik G, Jo J, Periwal V, Zielinski MC, Hara M. Quantification of islet size and architecture. Islets. 2012;4:167–172. doi: 10.4161/isl.19256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilimnik G, Kim A, Jo J, Miller K, Hara M. Quantification of pancreatic islet distribution in situ in mice. Am J PhysiolEndocrinolMetab. 2009;297:E1331–1338. doi: 10.1152/ajpendo.00479.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, Hara M. Islet architecture: A comparative study. Islets. 2009;1:129–136. doi: 10.4161/isl.1.2.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H, Kolb-Bachofen V. Type 1 (insulin-dependent) diabetes mellitus and nitric oxide. Diabetologia. 1992;35:796–797. doi: 10.1007/BF00429103. [DOI] [PubMed] [Google Scholar]
- Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One. 2010;5:e10538. doi: 10.1371/journal.pone.0010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebastchi J, Herold KC. Immunologic and metabolic biomarkers of beta-cell destruction in the diagnosis of type 1 diabetes. Cold Spring HarbPerspect Med. 2012;2:a007708. doi: 10.1101/cshperspect.a007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jeppesen PB, Gregersen S, Chen X, Hermansen K. Dose- and Glucose-Dependent Effects of Amino Acids on Insulin Secretion from Isolated Mouse Islets and Clonal INS-1E Beta-Cells. Rev Diabet Stud. 2008;5:232–244. doi: 10.1900/RDS.2008.5.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhou J, Bao Y, Chen T, Zhang Y, Zhao A, et al. Serum metabolic signatures of fulminant type 1 diabetes. J Proteome Res. 2012;11:4705–4711. doi: 10.1021/pr300523x. [DOI] [PubMed] [Google Scholar]
- Madsen R, Banday VS, Moritz T, Trygg J, Lejon K. Altered metabolic signature in pre-diabetic NOD mice. PLoS One. 2012;7:e35445. doi: 10.1371/journal.pone.0035445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritim AC, Sanders RA, Watkins JB., 3rd Diabetes, oxidative stress, and antioxidants: a review. J BiochemMolToxicol. 2003;17:24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- McKillop AM, Flatt PR. Emerging applications of metabolomic and genomic profiling in diabetic clinical medicine. Diabetes Care. 2011;34:2624–2630. doi: 10.2337/dc11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JJ, Bhushan A, Butler AE, Rizza RA, Butler PC. Sustained beta cell apoptosis in patients with long-standing type 1 diabetes: indirect evidence for islet regeneration? Diabetologia. 2005;48:2221–2228. doi: 10.1007/s00125-005-1949-2. [DOI] [PubMed] [Google Scholar]
- Miller K, Kim A, Kilimnik G, Jo J, Moka U, Periwal V, et al. Islet formation during the neonatal development in mice. PLoS One. 2009;4:e7739. doi: 10.1371/journal.pone.0007739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J Nutr. 2007;137:1616S–1620S. doi: 10.1093/jn/137.6.1616S. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Ishii E. Changes in arachidonic acid metabolism and the aggregation of polymorphonuclear leukocytes in rats with streptozotocin-induced diabetes. BiochimBiophysActa. 1996;1315:145–151. doi: 10.1016/0925-4439(95)00116-6. [DOI] [PubMed] [Google Scholar]
- Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokoff N, Rewers M. Pathogenesis of type 1 diabetes: lessons from natural history studies of high-risk individuals. Ann N Y AcadSci. 2013;1281:1–15. doi: 10.1111/nyas.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oresic M, Seppanen-Laakso T, Sun D, Tang J, Therman S, Viehman R, et al. Phospholipids and insulin resistance in psychosis: a lipidomics study of twin pairs discordant for schizophrenia. Genome Med. 2012;4:1. doi: 10.1186/gm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, et al. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. ProcNatlAcadSci U S A. 2001;98:10845–10850. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G, Piraino P, Zucht HD. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. AdvApplBioinformChem. 2009;2:57–70. doi: 10.2147/aabc.s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parazzoli S, Harmon JS, Vallerie SN, Zhang T, Zhou H, Robertson RP. Cyclooxygenase-2, not microsomal prostaglandin E synthase-1, is the mechanism for interleukin-1beta-induced prostaglandin E2 production and inhibition of insulin secretion in pancreatic islets. J BiolChem. 2012;287:32246–32253. doi: 10.1074/jbc.M112.364612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflueger M, Seppanen-Laakso T, Suortti T, Hyotylainen T, Achenbach P, Bonifacio E, et al. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes. 2011;60:2740–2747. doi: 10.2337/db10-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing. 2011 ISBN 3-900051-900007-900050. [Google Scholar]
- Robinson TW, Freedman BI. Assessing glycemic control in diabetic patients with severe nephropathy. J RenNutr. 2013;23:199–202. doi: 10.1053/j.jrn.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Sampson SR, Bucris E, Horovitz-Fried M, Parnas A, Kahana S, Abitbol G, et al. Insulin increases H2O2-induced pancreatic beta cell death. Apoptosis. 2010;15:1165–1176. doi: 10.1007/s10495-010-0517-5. [DOI] [PubMed] [Google Scholar]
- Scholz M, Fiehn O. SetupX--a public study design database for metabolomic projects. Pac SympBiocomput. 2007:169–180. [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TN, Hash K, Davey CL, Mills H, Williams H, Kiely DE. Modifications in the nitric acid oxidation of D-glucose. Carbohydr Res. 2012;350:6–13. doi: 10.1016/j.carres.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Southern C, Schulster D, Green IC. Inhibition of insulin secretion by interleukin-1 beta and tumour necrosis factor-alpha via an L-arginine-dependent nitric oxide generating mechanism. FEBS Lett. 1990;276:42–44. doi: 10.1016/0014-5793(90)80502-a. [DOI] [PubMed] [Google Scholar]
- Standards of medical care in diabetes--2013 Diabetes Care. 2013;36:S11–66. doi: 10.2337/dc13-S011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic V, Ihle S. Role of beta-hydroxybutyric acid in diabetic ketoacidosis: a review. Can Vet J. 2011;52:426–430. [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Svensson O, K. T., MacGregor JF. An investigation of orthogonal signal correction algorithms and their characteristics. J Chemom. 2002;16:176–188. [Google Scholar]
- Sysi-Aho M, Ermolov A, Gopalacharyulu PV, Tripathi A, Seppanen-Laakso T, Maukonen J, et al. Metabolic regulation in progression to autoimmune diabetes. PLoSComputBiol. 2011;7:e1002257. doi: 10.1371/journal.pcbi.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PO, Gleason CE, Robertson RP. Inhibition of interleukin-1beta-induced COX-2 and EP3 gene expression by sodium salicylate enhances pancreatic islet beta-cell function. Diabetes. 2002;51:1772–1778. doi: 10.2337/diabetes.51.6.1772. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- Ziemer DC, Kolm P, Weintraub WS, Vaccarino V, Rhee MK, Twombly JG, et al. Glucose-independent, black-white differences in hemoglobin A1c levels: a cross-sectional analysis of 2 studies. Ann Intern Med. 2010;152:770–777. doi: 10.7326/0003-4819-152-12-201006150-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.