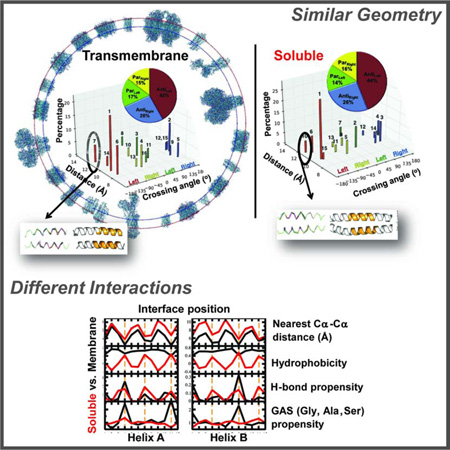
Summary
Alpha-helices are a basic unit of protein secondary structure and therefore the interaction between helices is crucial to understanding tertiary and higher-order folds. Comparing subtle variations in the structural and sequence motifs between membrane and soluble proteins sheds light on the different constraints faced by each environment and elucidates the complex puzzle of membrane protein folding. Here, we demonstrate that membrane and water-soluble helix pairs share a small number of similar folds with various interhelical distances. The composition of the residues that pack at the interface between corresponding motifs shows that hydrophobic residues tend to be more enriched in the water-soluble class of structures and small residues in the transmembrane class. The latter group facilitates packing via sidechain- and backbone-mediated hydrogen bonds within the low-dielectric membrane milieu. The helix-helix interactome space, with its associated sequence preferences and accompanied hydrogen-bonding patterns, should be useful for protein structure engineering, prediction and design.
Keywords: helix-helix association, sequence-structure motif, structural bioinformatics, protein design
Graphical Abstract
Introduction
The α-helix is by far the most common regular secondary structure element. In water-soluble proteins approximately 35% of all protein residues are in the α-helical conformation (Martin et al., 2005). Moreover, membrane proteins are almost exclusively α-helical bundles, with the exception of the β-barrels found in the outer membrane of Gram negative bacteria and mitochondria. Over 30% of the homologous superfamilies described in CATH are comprised mainly or entirely of alpha helices (Greene et al., 2007). These domains are found in both soluble (SOL) and transmembrane (TM) proteins, and carry out a wide range of biological functions.
While SOL domains are well studied, TM domains have only recently begun being elucidated. Since the first TM protein structure was solved in 1984 (Deisenhofer et al., 1984), the folding mechanism of these proteins has gradually become clearer (Bowie, 2005), yet much remains to be discovered. These proteins are estimated to make up 20–30% of open reading frames in known genomes (Wallin and von Heijne, 1998), and are overwhelmingly alpha helical, containing one or multiple membrane-spanning helices. Specific interaction patterns between helices play a critical role in the function, assembly and oligomerization of these proteins (Langosch et al., 2010; Shai, 2001). Likewise, membrane protein misassembly can contribute to a myriad of disease states (Ng et al., 2012). However, due to experimental challenges in crystallization, TM proteins represent only 2% of deposited structures (White, 2009). Despite this shortage, deep computational and bioinformatics-based analyses of helix-helix interactions will accelerate our understanding the folding behavior of helical TM proteins (Nugent and Jones, 2012) and will facilitate their design (Ghirlanda, 2009; Perez-Aguilar and Saven, 2012).
Consequently, the study of basic principles underlying the fold space of the helix-helix interactome, namely understanding the packing of helices, is intrinsic to understanding proteins. For example, in 1977 Chothia, Levitt, and Richardson presented simple helix-helix packing rules as determinants of protein structure (Chothia et al., 1977). An open question is whether helices from TM and SOL proteins are similar in the way they interact with each other and contribute to the overall protein structure. A small subset of SOL helix-helix pairs were shown to be structurally homologous to TM pairs presenting similar properties, even though the overall distributions for SOL dimers are quite different from those of TM dimers (Gimpelev et al., 2004). Here, we investigate the range of currently known SOL helix-helix interactions and compare them to those found in TM proteins focusing on the interplay between sequence and structure. To do this, we extend the approach used previously for characterizing TM dimers (Walters and DeGrado, 2006) to a larger database of TM dimers with stricter criteria and compare the results with dimers from water soluble proteins.
Analysis of sequences derived from helix-helix dimers propels our understanding of helix-helix interactions. The most extensively studied TM helix dimer is Glycophorin A (GpA), a common model system (Lemmon et al., 1992; MacKenzie et al., 1997). Each helix of GpA contains two Gly separated by three amino acids, known as the GxxxG motif (Lemmon et al., 1994), which plays a key role in dimerization. The GxxxG motif is highly overrepresented in the sequences of TM proteins (Senes et al., 2000), and has been well-characterized structurally. GxxxG-containing dimers tend to have a parallel, right-handed geometry, compact helix-helix packing and stabilizing interhelical backbone-mediated hydrogen bonds (MacKenzie et al., 1997; Mueller et al., 2014; Senes et al., 2001).
Comprehensive characterization via a variety of biophysical and biochemical methods has established the GxxxG motif as an important framework of TM helix-helix interaction (Russ and Engelman, 2000). Gly can be commonly replaced by another small residue, such as Ala or Ser in this motif (Mueller et al., 2014; Russ and Engelman, 2000; Senes et al., 2000). The Ala-Coil (Gernert et al., 1995) and GxxxxxxG motif are other prevalent sequence motifs found in membrane protein families (Liu et al., 2002). Additional sequence motifs have been identified, which depend on hydrogen bonds or weak polar interactions, and include derivatives of the small-residue motifs mentioned above (Adamian and Liang, 2002; Bowie, 2005; Gratkowski et al., 2002; Han et al., 2011; Hedin et al., 2011; Herrmann et al., 2009; Langosch and Arkin, 2009; Lawrie et al., 2010; Liang, 2002; Sal-Man et al., 2007; Unterreitmeier et al., 2007; Varriale et al., 2010; Wei et al., 2011; Zhou et al., 2001)
However, a systematic study of sequence-structure relationships on the scale of the whole protein structure database using structural bioinformatics is still lacking. Here we extract helix-helix pairs from high-resolution, non-homologous TM and SOL proteins from the protein data bank (PDB), and cluster them based on sequence-independent geometric similarity. We contrast the relative frequencies of each cluster in both environments and identify specific conformations that are unique to one or the other. Notably, sequence profiles can differ between the TM and SOL datasets, even for geometrically identical clusters. We also analyze the sidechain- and backbone-level interhelical hydrogen bonding interactions of residues in seven clusters of TM helix dimers and in their structural counterparts, namely, SOL dimers, extending an early analysis of Adamian and Liang (Adamian and Liang, 2002). Characterization of these sequence, structural and interaction motifs contribute to our understanding of the folding of helical proteins and aid both in structure prediction (Barth et al., 2009) and de novo design (Samish et al., 2011).
Results
Clustering of TM helical pairs
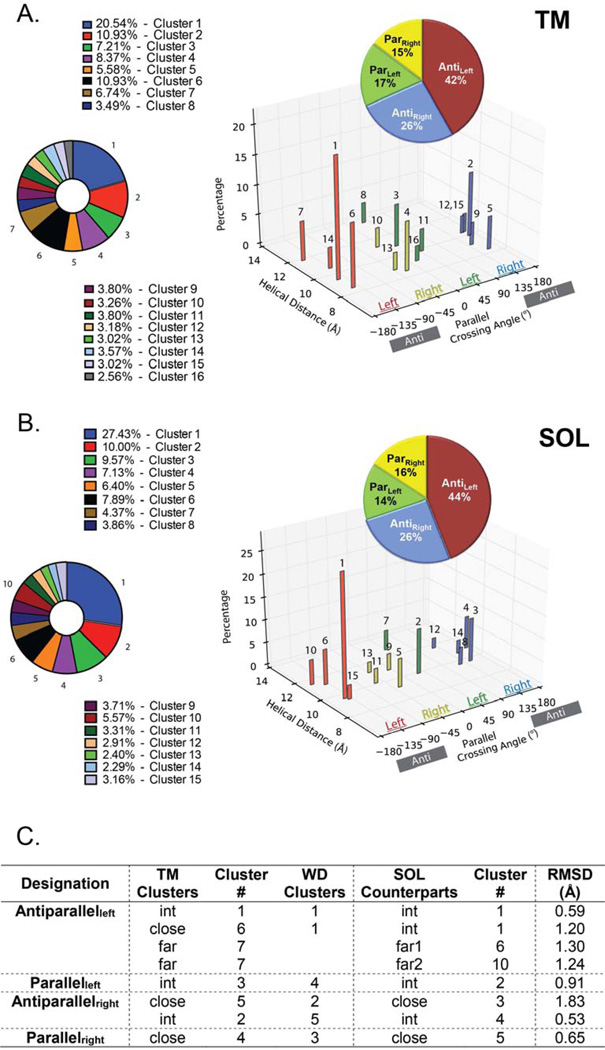
Previously, Walters and DeGrado clustered the helical pairs culled from the existing crystal structures of membrane proteins to define distinct geometries for transmembrane helical pairs, designated here as the WD analysis (Walters and DeGrado, 2006). Since then, the data base has increased roughly 4-fold, allowing us to use more stringent criteria for clustering, and to resolve additional clusters. In our earlier work, we clustered a library of 455 pairs using a greedy clustering algorithm and a 1.5 Å cutoff, and found that 90% fell within geometric clusters. Here, we hoped to find additional geometries, so we used a more generous criterion for inclusion of helical pairs in the database but a more stringent cutoff of 1.25 Å as the clustering criterion. We again used greedy clustering and examined clusters with at least 25 members (representing 1.4% of the pairs, 16 total). Clusters with fewer members are not considered here. Now we find 16 clusters (1290 pairs), which comprise 48% of the pair library of 2694 dimers (Figure 1). This coverage is smaller than the 90% seen in the previous (455 pairs in the library) for several reasons. We increased the minimal size of clusters to 25 members, so rare clusters are now excluded from the analysis. Secondly, the increased geometric stringency (RMSD ≤ 1.25 Å) caused some of the WD clusters (RMSD ≤ 1.5 Å) to split into two clusters that did not separately meet the size threshold for inclusion in the analysis. Finally, and most importantly, we used different geometric criteria to define pairs, allowing large interhelical distances (up to 14 Å) while the previous study required that pairs should have an interhelical distance ≤ 12 Å. In the present study, most of these pairs with large interhelical distances did not fall within well-defined clusters, presumably because their geometries are determined by interactions with other portions of the protein. When we use a cutoff of 0.065 Å−1 for the dimer mean inverse distance (see Materials and Methods) we find that 67% of these more stringently defined pairs are in the 16 clusters. Moreover, 70% of the clustered dimers lie in the first 7 clusters, each of which has more than 70 members. In summary, the geometries of most tightly interacting helices are well represented by the centroids of clusters 1–7 (Figure 2), which we will discuss in detail below. Interestingly, Joo et al. datamined sets of residues that contact each other and computed the crossing angles of the corresponding helices (Joo et al., 2012). Plotting the histogram distribution of these angles results in discrete peaks corresponding to the packing states described here (Figure 2). Similar crossing angle distributions have recently been computed also for membrane proteins (Lo et al., 2011).
Figure 1. Similarities between the TM and SOL helix-helix clusters.
A. Description of the 15 SOL (left panel) and 15 TM (right panel) clusters in respect to their crossing angle, interhelical distance. Helix-helix crossing angle is color coded by 90° segments as in the WD study (Walters and DeGrado, 2006) to Antileft (red), Parright (yellow), Parleft (green) and Antiright (blue) with the percentage of each group (insert pie graph) and B. each cluster (bottom pie graph) shown. C. The RMSD similarity of the top 7 TM clusters relative to their SOL structural counterparts are measured on the 12-residue windows on the centroids with the smallest RMSDs along the most populated 15-residue regions. The corresponding cluster number from the WD study is depicted.
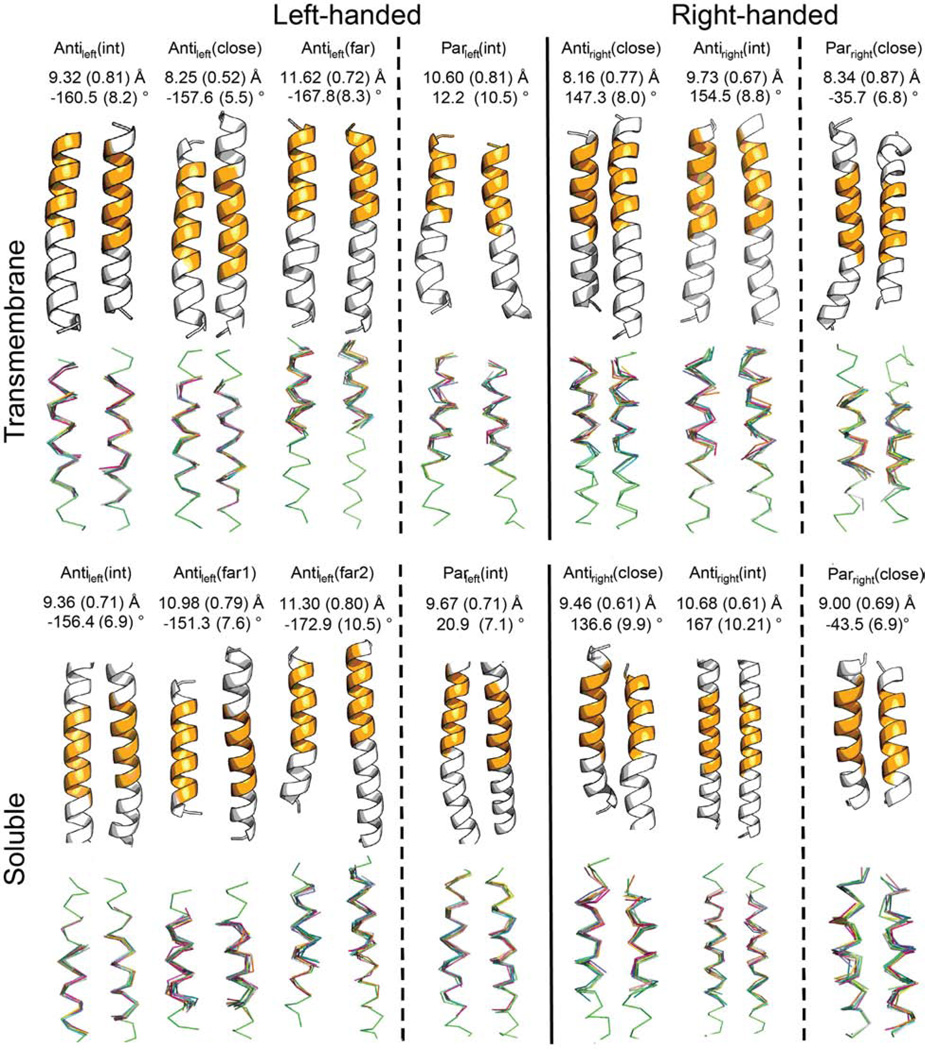
Figure 2. Description of the seven frequent TM and SOL clusters.
Average values of interhelical distance and crossing angle for the clusters are measured on the most populated 12-residue windows of the clusters colored in orange in the centroids and standard deviations are shown in parentheses. The top 10 members in the clusters with the closest RMSD to the centroid are overlapped in the bottom.
Highly populated clusters of 70 members or more have been defined in the present analysis, even though the increased stringency split some of the previously defined clusters into two. The overall division between antiparallel and parallel and left-and right-handed clusters, that is the percentages of members in each class of cluster, is strikingly similar between the water-soluble and transmembrane helix-helix interactome clusters (Figure 1A–B, insert). Yet, the relative weight of helix-helix distances among these clusters displays differences (Figure 1). For example, as seen in Figure 1C, the largest cluster in the previous WD analysis (Walters and DeGrado, 2006) now splits into two clusters (Clusters 1 and 6), which we define as Antileft(int) and Antileft(close), respectively (Figure 1A). In this nomenclature, Antileft(int) refers to an antiparallel dimer with a left-handed crossing angle and an interhelical distance that is intermediate between the other two major antiparallel left-handed clusters with close and far interhelical distances. Other than Antileft, major clusters include Parleft(int), Antiright(close), Antiright(int) and Parright(close). There are other less populated clusters that have, for example, closer and greater interhelical distances than Parleft(int), but they did not reach the criterion of 70 members that we have set for more in depth structural analysis (Table S1).
The most prevalent water-soluble helical pairs have geometries closely related to their membrane counterparts
A total of 5085 water-soluble helical pairs were extracted from a database of predominantly helical proteins, and clustered using the same methods as for the TM pairs, yielding a total of 15 clusters, ranging in size from 754 to 55 members (Figure 1A, Table S1 & S2). Together this set comprises 52% of the total pairs. The TM and SOL helix pair clusters are geometrically highly similar with most being antiparallel (70% and 68% for the SOL and TM datasets, respectively) and left-handed pairs (58.8% and 59.0% for the SOL and TM datasets, respectively). Although, these cluster groups also share similar interhelical distances (Table S1), they differ in the relative abundance of interhelical distances within each cluster (Figure 1).
The top seven SOL clusters (Figure 2B) include 74.0% of the clustered helix pairs. With the exception of the Antiright(close) motif, these are highly similar to the top-seven TM helical clusters (Cα RMSD ≤ 1.3 Å, Figure 1B). Thus, the differences between SOL and TM centroids are generally within the same range as the RMSD between members of a given cluster (up to 1.25 Å). Often, there is a one-to-one relationship between the clusters, although this is not always the case. Three notable exceptions from this rule are: a) the Antileft(close) motif found in TM pairs is not among the top 7 SOL clusters and is rare in the water-soluble database (Cluster 15, see Table S2); b) a motif in the soluble dataset that is relatively close in geometry to the Antileft(int) motif (RMSD = 0.6 Å for the centroids), and somewhat more distant from the Antileft(close) motif (RMSD = 1.2 Å); c) the Antileft(far) motif shows high similarity to two different clusters of related geometry in the water-soluble database (Table S2).
Helices tend to pack more tightly and have shorter interhelical distances in membrane proteins compared to water-soluble proteins (Eilers et al., 2002; Oberai et al., 2009; Senes et al., 2004; Zhang et al., 2009). For example the TM Antiright(close) and Parright(close) motifs have a closer inter-helical distance than the corresponding water-soluble motifs by 0.9 Å and 0.5 Å, respectively. This tightening of the interhelical distance is well documented in previous studies of helix-helix packing of membrane proteins (Cross et al., 2013; Javadpour et al., 1999). Indeed, while the packing energetics of TM and SOL proteins is similar (Joh et al., 2009), TM proteins bury more residues, which are smaller on average, compared to SOL residues (Oberai et al., 2009) thus facilitating this phenomenon. In summary, the SOL and TM helix-helix interactome display similar structural fold space with a small bias towards tighter helix-helix distances in the TM motifs.
Correlations between interhelical distance, hydrophobicity, interhelical hydrogen bonding and residue preferences in aligned sequences of TM and SOL helical pairs
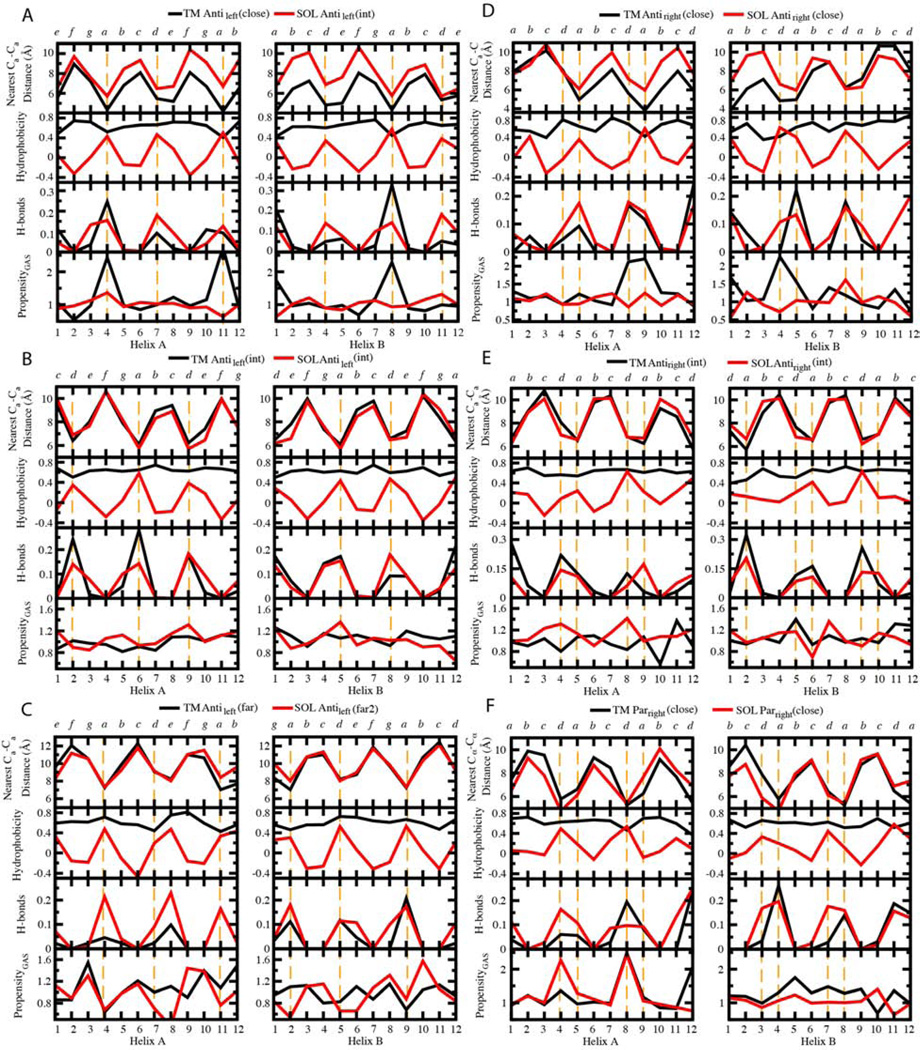
We investigated possible similarities between the nearest Cα-Cα distances, the average hydrophobicity, the hydrogen bonding fraction, and the sequence propensities for each position along the aligned windows of the top 7 TM and SOL clusters (Figures 3, 4 and Figure S1). The structural resemblance of TM and SOL clusters is manifested in the highly similar patterns of the nearest Cα-Cα distance of their centroids. The periodicity of the nearest Cα-Cα distance tends to display the heptad and tetrad repeats for left- and right-handed helix dimers, respectively (Figure 3), confirmed by least squares fitting of a sinusoidal function to the data (Supplementary Table S3). When helices cross with a left-handed crossing angle, the interaction pattern resembles that seen in classically left-handed coiled coils over a limited length of the chain (10–15 residues). We therefore denoted these positions using the classical coiled coil heptad nomenclature, abcdefg (Crick, 1953a, b; Sodek et al., 1972; Talbot and Hodges, 1982). By contrast, the interaction pattern between right-handed helix crossing approximately repeats each four residues, denoted abcd. In both cases, the positions a and d are at the interhelical interface.
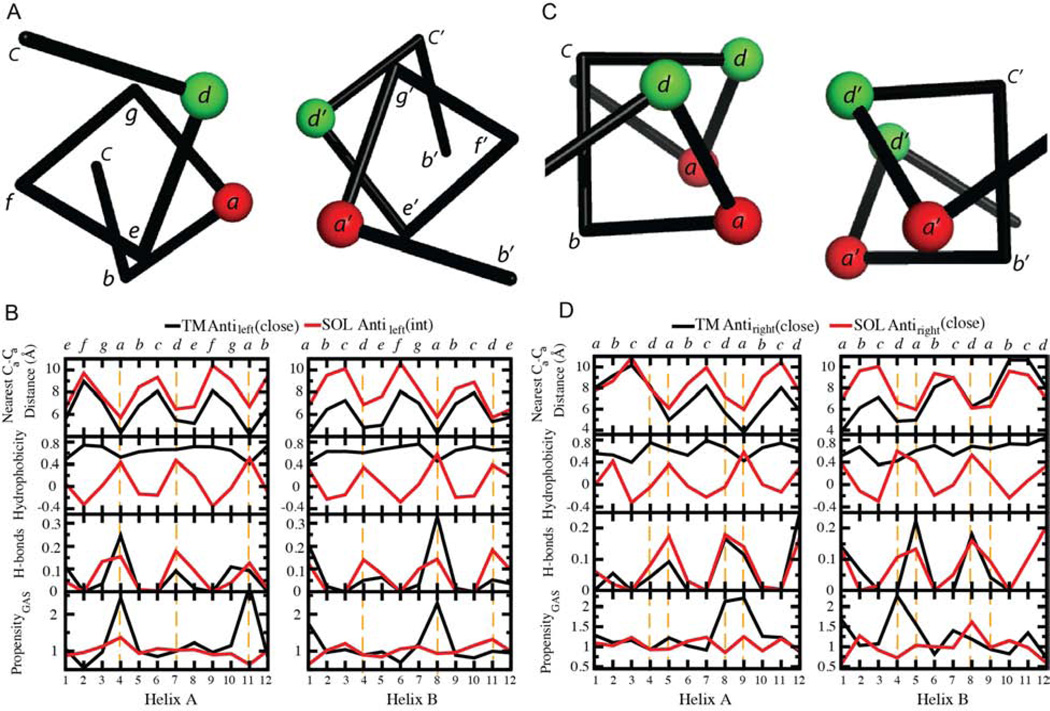
Figure 3. Profiles of the nearest Cα-Cα distance, average hydrophobicity, H-bonding fractions and propensity of small residues GAS on structurally matched windows between TM and SOL clusters.
Residues at the interhelical interface are highlighted by orange dashed lines. The designation of positions in the heptad and tetrad repeats is shown at the top.
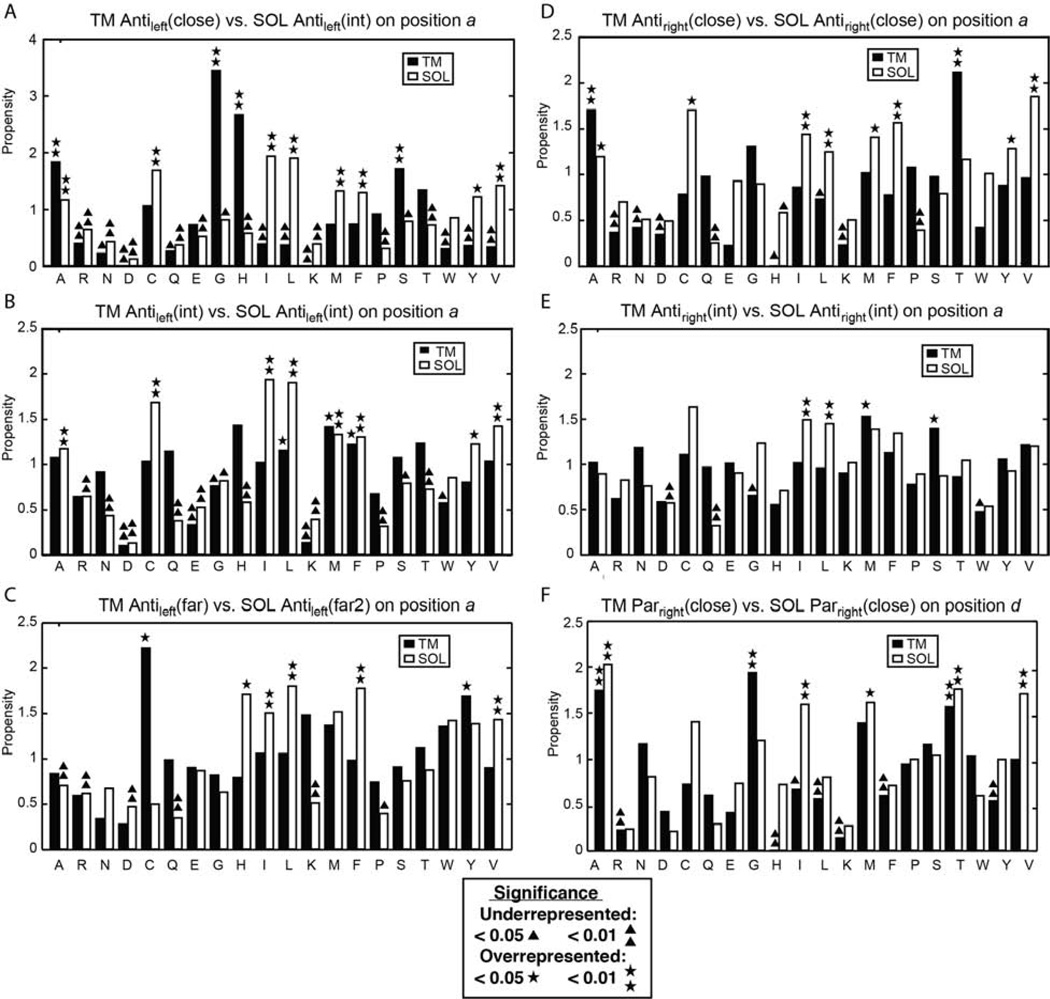
Figure 4. Comparisons of interhelical distances, average hydrophobicity, H-bonding fractions and propensity of small residues GAS for structurally matched TM and SOL motifs.
The 12 residue window of each TM centroid that contains the most cluster members were chosen as a representative sample for analysis. These and the matching windows on each corresponding SOL cluster are analyzed together. Residues at the interhelical interface are highlighted by orange dashed lines. The interhelical distances refer to the closest distance at a given Cα for one helix to a Cα in the neighboring helix. This figure is continued for additional pairs in Figure S1.
Sequence profiles of the interhelical distance, hydrophobicity, interhelical hydrogen bond frequency, and the propensity for a position to be occupied by a small residue, Gly, Ala or Ser (termed here as GAS) provide information concerning the driving force for the assembly of helical pairs in different environments. Figure 3C presents data for the two helices in the TM Antileft(close) motif, and its closest counterpart in the SOL database, the Antileft(int) motif; the profiles for the TM and water-soluble helices are colored black and red, respectively. Focusing first on the interhelical distance profile, one can see that the water-soluble distances tend to be very similar to that of the TM at one end of the bundle, but diverge by about 2–3 Å at the C-terminus of helix A and the N-terminus of helix B in the antiparallel motif. We also see a clear 180° phase shift between the interhelical distance and the mean hydrophobicity at the corresponding position in water-soluble proteins. This relationship reflects the tendency of water-soluble proteins to have apolar residues in buried positions and polar residues at water-accessible positions. This tendency to place hydrophobic residues at the “a” and “d” positions is reflected by different degrees of sinusoidal hydrophobicity propensities in practically all SOL clusters (Figure 4) as well as in propensities of the individual amino acids (Figures 5 and S2). By contrast, the hydrophobicity profile of the TM is uniformly high, reflecting the overall hydrophobic nature of TM helices. Hydrogen bonds are frequently observed along the interfacial “a” and “d” positions of the water-soluble Antileft(int) pair, but they are highly restricted to the “a” positions in the corresponding TM Antileft(close) motif. The difference reflects the closer approach of the helices in the TM motif resulting in shorter interhelical distances at the “a” position. Finally, the TM Antileft(close) motif has a very high propensity for GAS residues at only position “a” of the motif, a tendency that is not present in the water-soluble counterpart. The notable exception is of the significant preference for His at Antileft(close) at “a” and “d” positions (p >0.01, Fig. 4). Upon further investigation, we found this was due to 26 helical pairs (18.4% of Antileft(int)) derived from chlorophyll binding proteins, which use His to coordinate metals (Braun et al., 2011). Meanwhile, a similar TM motif Antileft(int) contains only 3% of pairs from such proteins. Otherwise, we observed a strong tendency to place small residues (Gly, Ala or Ser) at these positions (Figure 5A), a phenomenon seen also for the TM Antiright(close) (Figures 5D, S2F) and Parright(close) (Figures 5F, S2H).
Figure 5. Propensities of amino acids in different positions at the interhelical interface.
Residues labeled by asterisks or triangles are statistically overrepresented or under-represented, respectively as determined by, respectively, the p-value of a binomial test (p <0.05 or <0.01), relative to the expected amino acid frequency as described in Experimental Procedures (Table S4). This figure is continued for additional pairs in Figure S2.
In parallel, bulky and β-branched amino-acids are underrepresented in these close TM motifs yet are more abundant in their water-soluble counterparts, especially with increasing interhelical distance (Figure 5). Thus, the presence of small residues facilitates close helix-helix packing reflected by closer interhelical distances. In summary, the most striking difference in the profiles lies in the strong hydrophobic periodicity seen for the water-soluble pair – reflecting the hydrophobic driving force for assembly in water. In contrast, the TM (close) motifs show a strong periodicity in the GAS propensity – reflecting the strong driving force for folding in membranes associated with the packing of small residues along one face of a TM helix (Eilers et al., 2002; Oberai et al., 2009; Senes et al., 2004; Zhang et al., 2009).
The interhelical distance of related helical pairs is impacted by the composition of the residues at the interface, as reflected in the profiles for the Antileft(close), Antileft(int) and Antileft(far) motifs (Figure 4A–C). A comparison of the interhelical distance profiles for these three left-handed antiparallel motifs shows that the TM and water-soluble motifs are essentially superimposable for the intermediate and far motifs (correlations, all R2 >0.71; periods shown in Table S3). The repeated pattern of hydrophobicity remains strong for all three SOL motifs, while the TM pairs remain uniformly hydrophobic. Conversely, the hydrogen-bonding profiles are only similar between the water-soluble and TM motifs for the Antileft(int) and Antileft(far) motifs (R2 =0.55 for Antileft(far) helix A, but R2 >0.67 for the others, Figure 4B–C). However, for the Antileft(close) motif, the frequency of interhelical hydrogen bonds at interfacial positions is two to three-fold higher for water-soluble helices than TM helices. This finding may reflect the relative paucity of polar residues to form hydrogen bonds in TM helices (Figure 4), rather than the favorability of their formation in an apolar environment (Senes et al., 2004). As the helices become increasingly distant in progressing from the Antileft(close) to Antileft(far) motifs, the propensity for GAS residues decreases, becoming unfavorable for Antileft(far) for both water-soluble and TM motifs.
A comparison of the antiparallel right-handed motifs with the left-handed motifs (Figure 4 left vs. right halves of the figure) shows precisely the same trends, although the periodicity of the profiles is shifted closer to 4-residues from the 3.5-residue period seen for the left-handed motifs. The water-soluble Antiright(close) motif shows a systematic increase in the inter-helical distance at one end of the pair while this divergence is not seen for the corresponding TM motif. The TM Antiright(close) also shows a strong GAS propensity at the “a” and “d” positions where the helices make their closest contact. A strong GAS propensity is not seen in the corresponding SOL motifs possibly reflecting the hydrophobic core (relative to hydrophilic surrounding) found only in the latter motifs (Figure 4). Also, as seen for the Antileft motifs, the geometry of the interacting helices became identical at intermediate interhelical distances for both the water-soluble and TM motifs.
GpA was an early example of a GxxxG motif. Geometrically, the Parright(close) is quite similar to the GpA structure, and the RMSD between GpA and the centroid of the Parright(close) cluster is 1.5 Å (by overlapping a window of 16 residues in the TM helix pairs). The GxxxG motif is rare in this analysis of multispan proteins, representing 11.9% of the top 7 clusters TM clusters. A possible explanation is that GpA is an anchor to a constitutively dimeric glycoprotein rather than a dynamically functioning protein as is the case for most transmembrane proteins. Interestingly, our sequence analysis shows the GAS propensity is stronger at one of the two helices. This finding matches recent results from mutagenesis analysis of the strengths of dimerization of integrin TM helices, which display an asymmetric GxxxG packing motif (Berger et al., 2010). Peaks in the GAS propensity are also seen in one of the two helices in the water-soluble Parright(close) motifs (Fig. 4F).
The clusters have distinct H-bonding connectivity network
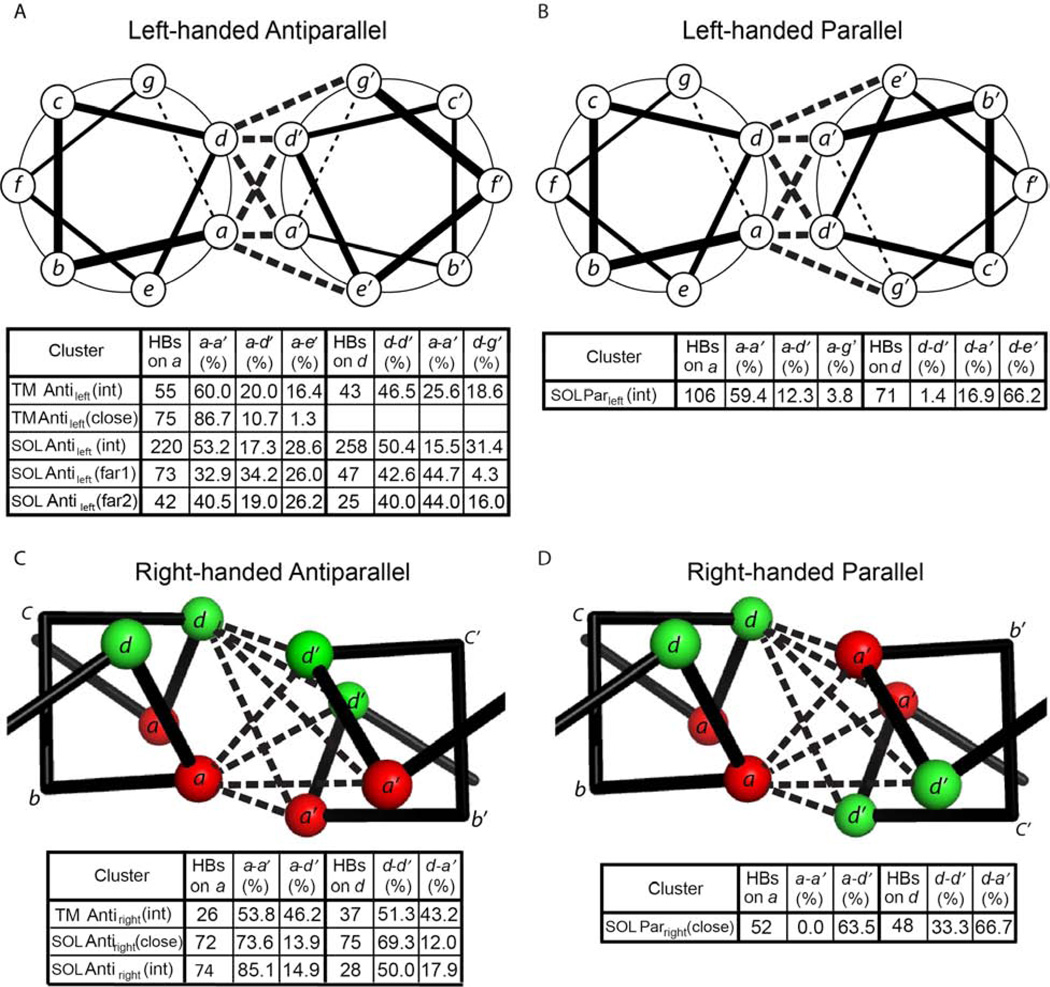
Antiparallel helices can form interhelical hydrogen bonds between residues from interacting helices. Depending on the sidechains and the interhelical geometry, a number of hydrogen-bonding patterns or “connectivities” are possible. For, antiparallel left-handed helical motifs hydrogen-bonding is geometrically feasible between a and a’, d and d’, a and d’, d and g’, or a and e’. However, these do not occur with equal frequencies. Classically, a-to-d’ hydrogen bonding has been extensively studied and used in protein design (McClain et al., 2001; McClain et al., 2002; Oakley and Kim, 1998). However, this interaction pattern is the exception rather than the rule for antiparallel helices. For the motifs for which there are at least 25 observations of hydrogen bonds, a-to-a’ and d-to-d’ hydrogen bonding generally predominates over other hydrogen-bonding connectivities; this is particularly striking for the TM Antileft(close) motif (Figure 4A), in which the proportion of a-a’, a-d’, and a-e’ is 87:11:1 (Figure 6A). As the interhelical distance increases within a motif, the preference for a-a’ and d-d’ becomes less striking (Figure 6A), presumably because the greater interhelical distance provides greater flexibility for sidechain interactions. Interestingly, precisely the same preferences for a-a’ and d-d’ connectivities are seen in the antiparallel right-handed motifs (Figure 6C).
Figure 6. H-bonding connectivity networks for the clusters with different geometry.
The number of hydrogen bonds is the arithmetic summation of those on the most populated position a or d from both chains. The percentage of each contact type, e.g. a-e’, is the fraction of the sum on that position, i.e. sum on an a or d.
The hydrogen bonding connectivities seen in parallel left-handed hydrogen bonding patterns follow the familiar patterns expected from parallel coiled coil motifs (Grigoryan and DeGrado, 2011). The preferred hydrogen bonding at “a” positions involves a-a’ connectivities. By contrast, d-d’ is rare, due to the geometry of the coiled coil. Instead, d residues tend to hydrogen bond to e’ of a neighboring helix (Figure 6B).
The only right-handed parallel cluster with sufficient numbers of interhelical hydrogen bonds to merit analysis was the water-soluble Parright(close) motif (Figure 6D). In this case, a-d’ greatly outnumbered the a-a’ or d-d’ interactions. As mentioned above, the opposite was true for right-handed antiparallel motifs.
The hydrogen bonding connectivity maps also shed light on the conformational specificity of TM and SOL helical bundles. Firstly, in prototypical parallel coiled coils, buried hydrogen bonds typically form between small polar residues in the same register of the heptad repeats (Woolfson, 2005). In antiparallel SOL coiled coils, strong a-d’ and d-a’ interactions are anticipated (Mason and Arndt, 2004), and this was observed in many cases. However, in Antileft(int), there is a strong preference to form a-a’ and d-d’ hydrogen bonds, as well as a tendency to form a-e’ and d-g’ interactions. Hydrogen bonding connectivity maps should help guide the design of complex SOL and TM helical bundles (Tatko et al., 2006).
TM and SOL clusters utilize different residues for H-bonding
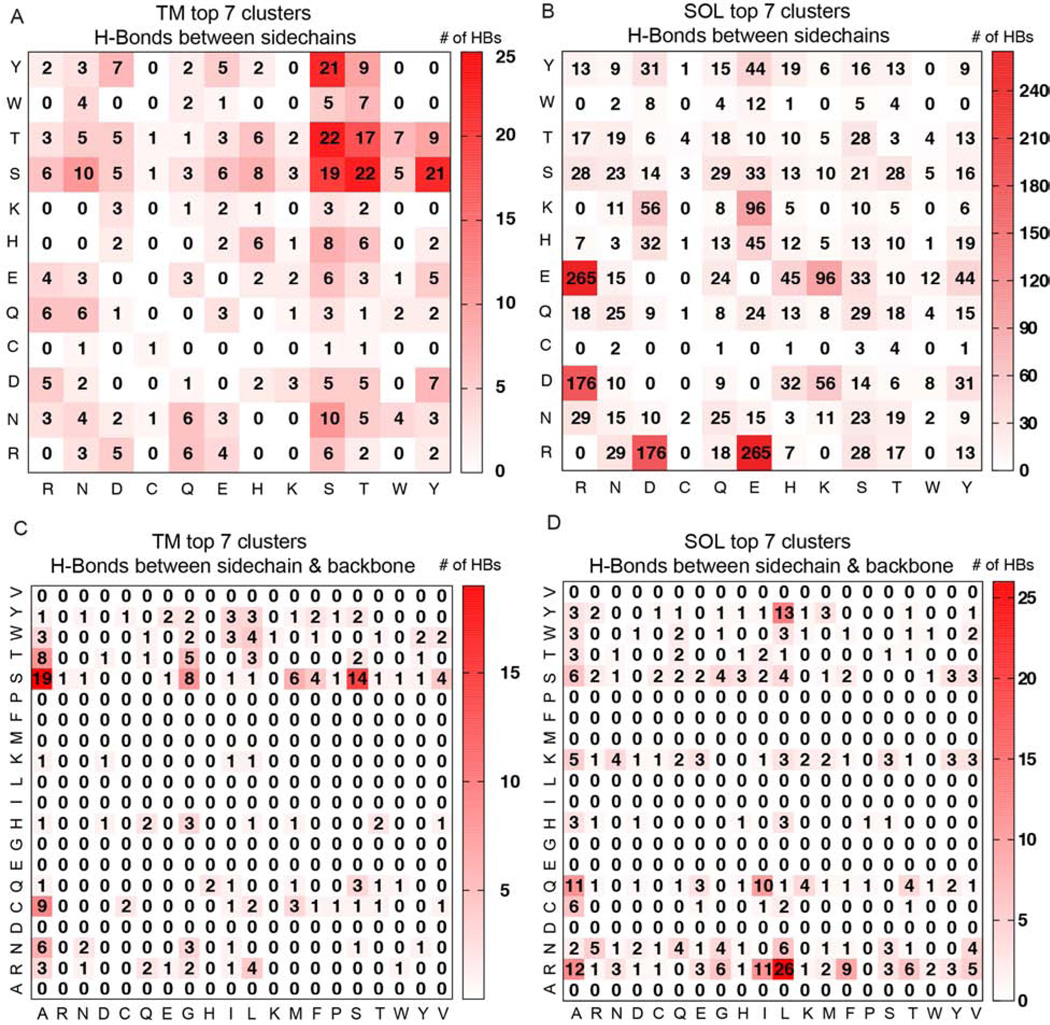
Next, we examined differences and similarities between the inter-helical sidechain-to-sidechain and sidechain-to-backbone interhelical H-bonding in the TM versus the SOL helix dimers. In this nomenclature, e.g. sidechain-to-backbone, the first helix of the pair has a residue in which the sidechain participates in an H-bond and the second helix of the pair has a backbone atom, which participates in the bond. Due to low number of counts for H-bonds in the individual TM clusters, the H-bonds of the top seven TM clusters are summed.
An expected major difference between TM and SOL clusters is the relative abundance of backbone-mediated interhelical hydrogen bonds is expected. In the TM clusters sidechain-to-sidechain and sidechain-to-backbone H-bonds comprise 56% and 44% of the total, respectively, while in the SOL clusters sidechain-to-sidechain and sidechain-to-backbone H-bonds have a population of 80% and 20%, respectively. Consistent with previous surveys of hydrogen bonding (Baker and Hubbard, 1984), the majority of sidechain-backbone H-bonds is from sidechain donors to the backbone carbonyl H-bond acceptors, with a portion of 93% and 94% in the TM and SOL clusters, respectively. Therefore we analyze only sidechain-to-backbone-carbonyl H-bonds below.
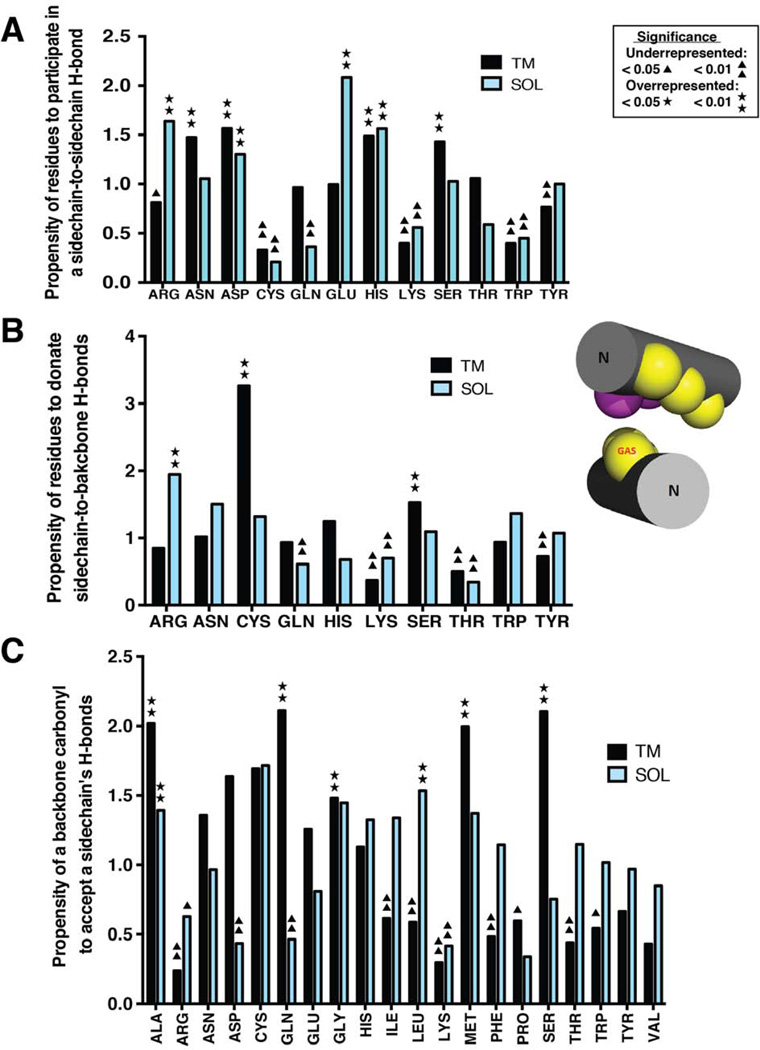
In the sidechain-to-sidechain H-bonding interactions among TM clusters (Figures 7 & 8), Ser is the largest contributor to H-bonding, accounting for 25.4% of occurrences, and showing a significantly high propensity (p-value <0.001) for these interactions even relative to the high abundance of Ser in TM helices (Fig 7A). The other three residues with high propensity (p-value <0.01) are Asn, His, and Asp, which have a much lower frequency in distribution (Table S4). Interestingly, Asn has a 4-fold preference to engage in H-bonds in right-handed crossings, and His has a 4-fold preference in left-handed crossings (data not shown). Each of the other polar residues occurs in less than 12% of H-bonds. The predominance of Ser among sidechain-to-sidechain interactions in the membrane environment is consistent with previous reports by Adamian and Liang (Adamian and Liang, 2002). Ser-Thr, Ser-Tyr, Ser-Ser and Thr-Thr are the most common sidechain-to-sidechain H-bonding contributors, shown in Figure 8.
Figure 7. Propensity of residues in the top 7 TM and SOL clusters to donate or accept an interhelical hydrogen bond of different types.
(A) Interhelical hydrogen bonding propensity of residues participating in sidechain-to-sidechain hydrogen bonds. (B) Interhelical hydrogen bonding propensity of residues that donate a sidechain hydrogen bond to the backbone carbonyl on the helical pair. (C) Interhelical hydrogen bonding propensity of residues that accept a hydrogen bond via the backbone carbonyl to the sidechains of their helical pair. As an example, the TM Parright(close) motif adopts configuration shown in the insert. Positions a and b are represented by yellow and magenta spheres, respectively. The one-sided small residue positions are labelled by GAS. The N-termini of the helices are labeled. Residues labeled by asterisks or triangles are statistically overrepresented or underrepresented as hydrogen bond participants, respectively as determined by the p-value of a binomial test (p <0.05 or <0.01).
Figure 8. Number of interhelical H-Bonds between the sidechains of residues (A, B) and between sidechain and the backbone carbonyl (C, D) in the top 7 TM (A, C) and SOL (B, D) clusters.
In A and B, the numbers in the grids are the arithmetic summations of the numbers of specific sidechain-to-sidechain hydrogen bonds in the top 7 clusters from each category. In C and D, the numbers of H-bonds denote those from the sidechain of the residue on the column to the backbone carbonyl on the residue on the row.
In the top 7 SOL clusters Arg displays a very high sidechain-to-sidechain H-bonding propensity (Figure 7A). The most frequent residues of this H-bonding class are Arg (19.8%), Glu (19.5%) and Asp (12.3%): Arg-Glu (19.0%), Arg-Asp (12.6%) and Lys-Glu (6.9%) are the three most common pairs of H-bonding partners (Figure 8B).
In the sidechain-to-backbone H-bonds of the TM clusters (Figure 7B, Figure 8C), Ser is and Cys are overrepresented as H-bonding donors, with frequencies of 31.8% and (11.0%). Small residues Ala, Gly and Ser are the major backbone carbonyl H-bonding acceptors, with 25.9%, 12.4% and 11.4% of the occurrences, respectively. The small residues may facilitate tight interactions, as found in the case of the Parright(close) model protein GpA (Figure 7B, insert).
In contrast with the TM clusters, the SOL clusters have Arg as the main sidechain-to-backbone H-bonding donor (29.1%), with Gln (13.3%), Ser (11.5%) and Lys (10.9%) next (Figure 8D), but only Arg is overrepresented (Fig.7B). Aliphatic residues without β-branching, namely Leu (18.8%) and Ala (16.4%) are the two major backbone carbonyl H-bonding acceptors (Figure 7C). It is interesting to note the important role of Arg residues in forming both sidechain-to-sidechain as well as sidechain-to-backbone-carbonyl interactions in water-soluble helical pairs. This finding agrees with experimental studies, which showed that this residue is unique among the polar residues in terms of its ability to contribute largely to conformational stability and specificity (Acharya et al., 2006; Borders et al., 1994).
Discussion
This work provides the most extensive analysis of TM and SOL helical interactions, providing a library of helical motifs, and their corresponding sequence preferences. Moreover, the present study provides information concerning the pattern and positions of hydrogen-bonding residues and how they may provide specificity supporting different helical packing interaction motifs. This work also provides the first extensive comparison of geometrically similar TM and water-soluble helical pairs.
Comparing the helix-helix interactome of transmembrane and water soluble proteins leads to key differences. One major difference lies in the greater abundance of tightly interacting helical pairs in TM compared to water-soluble proteins. Water-soluble structures tend to have more interhelical hydrogen-bonds and utilize larger and more charged residues for this task. On one hand, the water soluble helix-helix interactome generally displays a sinusoidal pattern of hydrophobicity. On the other hand, the transmembrane helix-helix interactome displays a significantly more pronounced abundance of small residues at the helix-helix interface, which facilitate backbone-mediated interhelical H-bonding interactions. This contrasts with the old view that membrane proteins are inside-out versions of water-soluble proteins. Instead, the requirements to maintain membrane proteins within a low-dielectric transmembrane environment, or the requirements associated with helix insertion via the translocon, select for TM helices that are highly hydrophobic and don’t necessarily use hydrogen bonds for stability as much as their soluble protein counterparts. Nevertheless, small residue sidechain- and backbone-mediated hydrogen bonds in the membrane milieu may guide helix-helix assembly as well as direct dynamic functionality (Bowie, 2011).
Helix-helix association is also affected by other factors, e.g., hydrophobic mismatch between a TM helix and the membrane (Benjamini and Smit, 2012). Investigation of the clusters will help greatly our understanding of the folding and structure of helical proteins, quantifying broad structural trends which will be useful in structure prediction and design.
Experimental Procedures
Dataset selection
The Orientation of Proteins in Membranes (OPM) database (Lomize et al., 2012) was used as the source for helical TM proteins. We obtained a list of all structures available as of September 26, 2014. To ensure accurate analysis, structures with X-ray resolution lower than 3.2 Å were removed from consideration. From the remaining structures, we used the PISCES server (Wang and Dunbrack, 2003) to cull at the PDB ID level for a maximum sequence homology of 30%. This resulted in a list of 139 representative structures, from which helix-helix pairs were derived. For the soluble database, a query was executed on the PDB as of February 9, 2012 for all structures classified in CATH (Greene et al., 2007) as "mainly alpha" and containing only protein. These were matched against the PDB-TM database (Tusnady et al., 2005) and any TM proteins were removed. This list was also culled using the PISCES server to a maximum of 30% sequence identity. In order to keep the size of the dataset computationally tractable, only structures with a maximum resolution of 2.0 Å were kept, resulting in 765 proteins. For all soluble structures, the biological unit was downloaded from the PDB. The lists of TM and SOL structure covered for analysis are included by a spreadsheet file in Supplemental Data.
We extracted the helical regions from the selected structures using the definitions of the TM segments in the OPM or the HELIX records in the PDB header information for soluble proteins. In order to ensure that these definitions were correct, the annotated regions were filtered to exclude helical breaks or sharp kinks (defined with a loose cutoff: −130° < φ < −20° and −90° < ψ < 30°). They were also extended by up to 4 residues on both the N- and C-terminal sides if the positions meet a stricter definition of helicity (−90° < φ < −35°; −70° < ψ < 0°). This helped to join soluble helices that otherwise might have been counted separately.
Creating the pair library
Two heuristic criteria were used to determine whether a given pair of helices was interacting. First, the minimum distance between the helical axes was required to be no more than 14 Å; second, the mean inverse distance was required to be at least 0.065 Å− over a 12-residue window (see “Window Selection and Alignment” below for a definition of this quantity). Both of these were intended to be generous, as low specificity would merely result in a larger fraction of dimers which cannot be clustered, while low sensitivity would negatively impact our ability to detect and characterize real trends.
Although the overall structural libraries were filtered to reduce sequence homology, individual proteins often contain multiple copies of one or more subunits, resulting in several identical helix pairs. In order to remove this additional source of redundancy polypeptide chains with identical sequences were assigned to a “chain group,” which allowed us to identify and remove duplicate dimers. Two helices can either come from the same chain, different chains, both belonging to the same chain group, or separate chains that also belong to disparate chain groups. The final helix pair library contains 2694 TM dimers and 5085 soluble dimers.
Window Selection and Alignment
To be able to align pairs, we used a distance map representation of each dimer. Briefly, the inverse distance between each Cα atom on one helix and every Cα atom on the other is stored in a matrix. (Residues more than 25 Å apart are given a value of 0.) We selected a twelve-residue segment from each helix, chosen so that we captured the maximum amount of interaction for a given pair. Interaction strength was determined by averaging the interfacial distance map over a 12-residue window on each helix, as calculated using Equation 1:
| (1) |
where M is the “mean inverse distance” or interaction strength, n is the window size (here 12 residues), a and b are the starting residues of the window on each helix, respectively, and xij is the value of the distance map for residues i and j, i.e. the inverse of the distance between the Cα atoms of residues i and j (in Angstroms) or zero if they are more than 25 Å apart. M was maximized by varying a and b over all possible values, from 1 to L-n+1, where L is the length of the particular helix. Since residues that are closer together in three dimensions have a larger entry in the distance map, this picks out the twelve residues on one helix that are closest to twelve residues on the other. Moreover, because of the inverse weighting, this emphasizes each residue’s nearest neighbors, with the distances between the end of one helix and the far end of the other being less important.
We used MaDCaT (Zhang and Grigoryan, 2013) to conduct all-vs.-all searches of the two dimer libraries. Interactions are not always symmetrical along the length of a helix, with six residues on either side of the point of closest approach –some are ‘V’-shaped rather than ‘X’-shaped. Thus had we merely compared the twelve-residue windows to each other directly, we would have missed pairs that otherwise have the same geometry. We therefore searched each query window against the library of whole pairs, as extracted above. We limited the searches to a maximum of 10,000 hits each, which in practice exhausted all possible alignments within our clustering threshold.
Structural Clustering
Examining the alignments calculated by MaDCaT, we chose a 1.25 Å RMSD cutoff for clustering as an appropriate balance between sensitivity and specificity. We used the same 12-residue windows described above; windows which overlapped by six residues or more on either helix were considered identical and clustered together, while windows with smaller overlaps are treated separately. (This allows the total number of alignments to be greater than the number of unique pairs.) To cluster the pairs, we computed all possible sub-threshold alignments to each window. The window with the largest number of alignments from unique, previously unclustered pairs was selected as the next centroid. All matching windows were assigned to that cluster and removed from consideration for further rounds. This process was then repeated until none of the remaining windows matched at least ∼1% of the associated database (25 pairs for TM and 55 pairs for soluble).
We found 16 TM clusters and 15 SOL clusters of helix pairs. Geometrical properties, including crossing angle and interhelical distance of the aligned windows in each cluster are determined by HELANAL (Bansal et al., 2000) implemented by MSL (Kulp et al., 2012). Mean geometric properties (Fig. 2, Tables S1, S2) of each cluster, were determined by the subset of pairs that fall within the most populated 12 residue window on the centroid. These same windows were those used to cluster, and are the subject of sequence, hydrophobicity, and hydrogen bonding analysis (Figures 3–7). The detailed information for TM and SOL clusters about the structural composition, RMSD to the centroids, interhelical distance and crossing angle is provided as two spreadsheet files in Supplemental Data.
Comparing Clusters
For each centroid, we determined the 15-residue window that is most populated by members of that cluster. To compare clusters, we then used MaDCaT to find the best possible alignment of 12 residues between each pair of centroids approximate to those regions. This information allowed us to identify the most closely related clusters from different sets. The centroid of each cluster is fit to a sinusoidal curve using non-linear regression to estimate the cluster’s periodicity. Two-tailed student’s t-test assuming equal variances were performed to confirm periods within the matching windows between TM and SOL were not significantly different.
Sequence Analysis
We used the structural alignments generated by MaDCaT for each cluster to create sequence alignments. Briefly, each centroid pair was renumbered so that the C-terminal residue of the centroid window would be residue 100. Each member of a cluster was then renumbered to match the centroid numbering, such that residues with the same number correspond in the structural alignment. The numbers of observations for every amino acid type were computed for each position in each cluster and normalized to frequencies by dividing by the total number of observations at that position. The frequencies were compared to the expected frequencies of amino acids in helical regions of TM or SOL proteins that form interacting helical pairs using a binomial distribution. We derived the expected frequency of TM amino acids from the percent distribution of amino acids observed at helical, transmembrane residues in the subset of our TM protein data set that formed interacting pairs. Likewise, only alpha-helical residues from the analogous SOL subset, determined by the DSSP Program (Kabsch and Sander, 1983), were observed in deriving the SOL amino acid distribution. These background frequencies are listed in Table S4. The propensity is defined as the ratio between the observed and expected (or background) frequencies. Significant overrepresentation or underrepresentation of an amino acid at a given position, relative to the expected frequency, was determined by the p-value of respective one-tailed directional binomial tests. The counts of observation, frequency and propensity for each amino acid on the positions with at least 25 and 55 total counts of observation for TM and SOL clusters, respectively, are provided as two spreadsheet files in Supplemental Data. Hydrophobicity profiles were calculated based on the normalized consensus scale (Eisenberg et al., 1984).
Hydrogen Bonding Analysis
Hydrogen bonds are determined by HBPLUS program (McDonald and Thornton, 1994) with default parameters. Weak Cα-H—O hydrogen bonds are not included. Two set of hydrogen bond data on positions a and d on the most populated region from each helix are employed to calculate the hydrogen bonding fraction, which is defined as the ratio between the numbers of residues forming interhelical hydrogen bonds and of the population accumulated on the four positions both for a and d. The hydrogen bonding connectivity are calculated by assigning the interhelically hydrogen-bonded residues in the heptad or tetrad repeats from the most populated positions a and d from both chains. The sidechain-to-sidechain interhelical hydrogen bonding propensity is calculated as the ratio between the fraction of Arg, Asn, Asp, Cys, Gln, Glu, His, Lys, Ser, Thr, Trp and Tyr to make sidechain-to-sidechain hydrogen bonds and their fraction in the subset of background distribution (Table S4). Significant overrepresentation or underrepresentation of an amino acid to participate in a hydrogen bond is determined by the binomial test.
Supplementary Material
Acknowledgements
SQZ thanks B. T. Hannigan and G. Gonzales for technical help. MM was supported by NIH T32 GM008284. This work was supported by NIH grant GM54616.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
SQZ, DWK and WFD designed research; SQZ, DWK, MM, and CAS performed research; SQZ, MM, IS and WFD analyzed the data; all authors wrote the paper.
References
- Acharya A, Rishi V, Vinson C. Stability of 100 homo and heterotypic coiled-coil a-a' pairs for ten amino acids (A, L, I, V, N, K, S, T, E, and R) Biochemistry. 2006;45:11324–11332. doi: 10.1021/bi060822u. [DOI] [PubMed] [Google Scholar]
- Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- Baker E, Hubbard R. Hydrogen bonding in globular proteins. Progress in biophysics and molecular biology. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Bansal M, Kumar S, Velavan R. HELANAL: a program to characterize helix geometry in proteins. J Biomol Struct Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini A, Smit B. Robust driving forces for transmembrane helix packing. Biophysical journal. 2012;103:1227–1235. doi: 10.1016/j.bpj.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BW, Kulp DW, Span LM, DeGrado JL, Billings PC, Senes A, Bennett JS, DeGrado WF. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci U S A. 2010;107:703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borders CL, Jr, Broadwater JA, Bekeny PA, Salmon JE, Lee AS, Eldridge AM, Pett VB. A structural role for arginine in proteins: multiple hydrogen bonds to backbone carbonyl oxygens. Protein science : a publication of the Protein Society. 1994;3:541–548. doi: 10.1002/pro.5560030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- Bowie JU. Membrane protein folding: how important are hydrogen bonds? Current opinion in structural biology. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Goldberg E, Negron C, von Jan M, Xu F, Nanda V, Koder RL, Noy D. Design principles for chlorophyll-binding sites in helical proteins. Proteins. 2011;79:463–476. doi: 10.1002/prot.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Levitt M, Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. The fourier transform of a coiled-coil. Acta Cryst. 1953a;6:685–689. [Google Scholar]
- Crick FHC. The Packing of alpha-Helices: Simple Coiled-Coils. Acta Cryst. 1953b;6:689–697. [Google Scholar]
- Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. European Biophysics Journal with Biophysics Letters. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J, Epp O, Miki K, Huber R, Michel H. X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. Journal of molecular biology. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- Eilers M, Patel AB, Liu W, Smith SO. Comparison of Helix Interactions in Membrane and Soluble alpha-Bundle Proteins. Biophys J. 2002;82:2720–2736. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. Journal of molecular biology. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Gernert KM, Surles MC, Labean TH, Richardson JS, Richardson DC. The Alacoil: a very tight, antiparallel coiled-coil of helices. Protein Sci. 1995;4:2252–2260. doi: 10.1002/pro.5560041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlanda G. Design of membrane proteins: toward functional systems. Current opinion in chemical biology. 2009;13:643–651. doi: 10.1016/j.cbpa.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Gimpelev M, Forrest LR, Murray D, Honig B. Helical packing patterns in membrane and soluble proteins. Biophys J. 2004;87:4075–4086. doi: 10.1529/biophysj.104.049288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratkowski H, Dai QH, Wand AJ, DeGrado WF, Lear JD. Cooperativity and specificity of association of a designed transmembrane peptide. Biophys J. 2002;83:1613–1619. doi: 10.1016/S0006-3495(02)73930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LH, Lewis TE, Addou S, Cuff AL, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic acids research. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, DeGrado WF. Probing Designability via a Generalized Model of Helical Bundle Geometry. Journal of molecular biology. 2011;405:1079–1100. doi: 10.1016/j.jmb.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Aligo J, Manna D, Belton K, Chintapalli SV, Hong Y, Patterson RL, van Rossum DB, Konan KV. Conserved GXXXG- and S/T-like motifs in the transmembrane domains of NS4B protein are required for hepatitis C virus replication. Journal of virology. 2011;85:6464–6479. doi: 10.1128/JVI.02298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin LE, Illergard K, Elofsson A. An introduction to membrane proteins. J Proteome Res. 2011;10:3324–3331. doi: 10.1021/pr200145a. [DOI] [PubMed] [Google Scholar]
- Herrmann JR, Panitz JC, Unterreitmeier S, Fuchs A, Frishman D, Langosch D. Complex patterns of histidine, hydroxylated amino acids and the GxxxG motif mediate high-affinity transmembrane domain interactions. J Mol Biol. 2009;385:912–923. doi: 10.1016/j.jmb.2008.10.058. [DOI] [PubMed] [Google Scholar]
- Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophysical journal. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh NH, Oberai A, Yang D, Whitelegge JP, Bowie JU. Similar energetic contributions of packing in the core of membrane and water-soluble proteins. Journal of the American Chemical Society. 2009;131:10846–10847. doi: 10.1021/ja904711k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H, Chavan AG, Phan J, Day R, Tsai J. An amino acid packing code for alpha-helical structure and protein design. Journal of molecular biology. 2012;419:234–254. doi: 10.1016/j.jmb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kulp DW, Subramaniam S, Donald JE, Hannigan BT, Mueller BK, Grigoryan G, Senes A. Structural informatics, modeling, and design with an open-source Molecular Software Library (MSL) Journal of computational chemistry. 2012;33:1645–1661. doi: 10.1002/jcc.22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein science : a publication of the Protein Society. 2009;18:1343–1358. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langosch D, Herrmann JR, Unterreitmeier S, Fuchs A. Helix-helix interaction patterns in membrane proteins. 2010 [Google Scholar]
- Lawrie CM, Sulistijo ES, MacKenzie KR. Intermonomer hydrogen bonds enhance GxxxG-driven dimerization of the BNIP3 transmembrane domain: roles for sequence context in helix-helix association in membranes. Journal of molecular biology. 2010;396:924–936. doi: 10.1016/j.jmb.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- Liang J. Experimental and computational studies of determinants of membrane-protein folding. Current opinion in chemical biology. 2002;6:878–884. doi: 10.1016/s1367-5931(02)00393-9. [DOI] [PubMed] [Google Scholar]
- Liu Y, Engelman DM, Gerstein M. Genomic analysis of membrane protein families: abundance and conserved motifs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0054. presearch0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A, Cheng CW, Chiu YY, Sung TY, Hsu WL. TMPad: an integrated structural database for helix-packing folds in transmembrane proteins. Nucleic Acids Res. 2011;39:D347–D355. doi: 10.1093/nar/gkq1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic acids research. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- Martin J, Letellier G, Marin A, Taly JF, de Brevern AG, Gibrat JF. Protein secondary structure assignment revisited: a detailed analysis of different assignment methods. BMC Struct Biol. 2005;5:17. doi: 10.1186/1472-6807-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem : a European journal of chemical biology. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- McClain DL, Binfet JP, Oakley MG. Evaluation of the energetic contribution of interhelical coulombic interactions for coiled coil helix orientation specificity. Journal of molecular biology. 2001;313:371–383. doi: 10.1006/jmbi.2001.5044. [DOI] [PubMed] [Google Scholar]
- McClain DL, Gurnon DG, Oakley MG. Importance of potential interhelical salt-bridges involving interior residues for coiled-coil stability and quaternary structure. Journal of molecular biology. 2002;324:257–270. doi: 10.1016/s0022-2836(02)01072-0. [DOI] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. Journal of molecular biology. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Mueller BK, Subramaniam S, Senes A. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Calpha-H hydrogen bonds. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E888–E895. doi: 10.1073/pnas.1319944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DP, Poulsen BE, Deber CM. Membrane protein misassembly in disease. Biochimica et biophysica acta. 2012;1818:1115–1122. doi: 10.1016/j.bbamem.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Membrane protein structural bioinformatics. Journal of structural biology. 2012;179:327–337. doi: 10.1016/j.jsb.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Oakley MG, Kim PS. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- Oberai A, Joh NH, Pettit FK, Bowie JU. Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17747–17750. doi: 10.1073/pnas.0906390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Aguilar JM, Saven JG. Computational design of membrane proteins. Structure. 2012;20:5–14. doi: 10.1016/j.str.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sal-Man N, Gerber D, Bloch I, Shai Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. The Journal of biological chemistry. 2007;282:19753–19761. doi: 10.1074/jbc.M610368200. [DOI] [PubMed] [Google Scholar]
- Samish I, Macdermaid CM, Perez-Aguilar JM, Saven JG. Theoretical and Computational Protein Design. Annual review of physical chemistry. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Senes A, Ubarretxena-Belandia I, Engelman DM. The Calpha ---H…O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci U S A. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai Y. Molecular recognition within the membrane milieu: implications for the structure and function of membrane Pproteins. J Membr Biol. 2001;182:91–104. doi: 10.1007/s00232-001-0034-a. [DOI] [PubMed] [Google Scholar]
- Sodek J, Hodges RS, Smillie LB, Jurasek L. Amino-acid sequence of rabbit skeletal tropomyosin and its coiled-coil structure. Proc Natl Acad Sci U S A. 1972;69:3800–3804. doi: 10.1073/pnas.69.12.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JA, Hodges RS. Tropomyosin: A Model Protein for Studying Coiled-Coil and a-Helical Stabilization. Acc Chem Res. 1982;15:224–230. [Google Scholar]
- Tatko CD, Nanda V, Lear JD, Degrado WF. Polar networks control oligomeric assembly in membranes. J Am Chem Soc. 2006;128:4170–4171. doi: 10.1021/ja055561a. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic acids research. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterreitmeier S, Fuchs A, Schaffler T, Heym RG, Frishman D, Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J Mol Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Varriale S, Merlino A, Coscia MR, Mazzarella L, Oreste U. An evolutionary conserved motif is responsible for immunoglobulin heavy chain packing in the B cell membrane. Mol Phylogenet Evol. 2010;57:1238–1244. doi: 10.1016/j.ympev.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RF, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci U S A. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- Wei P, Liu X, Hu M-H, Zuo L-M, Kai M, Wang R, Luo S-Z. The dimerization interface of the glycoprotein Ibβ transmembrane domain corresponds to polar residues within a leucine zipper motif. Protein science : a publication of the Protein Society. 2011;20:1814–1823. doi: 10.1002/pro.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- Woolfson DN. The design of coiled-coil structures and assemblies. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. 2005;70 doi: 10.1016/S0065-3233(05)70004-8. 79-+ [DOI] [PubMed] [Google Scholar]
- Zhang J, Grigoryan G. Mining tertiary structural motifs for assessment of designability. Methods Enzymol. 2013;523:21–40. doi: 10.1016/B978-0-12-394292-0.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kulp DW, Lear JD, DeGrado WF. Experimental and computational evaluation of forces directing the association of transmembrane helices. J Am Chem Soc. 2009;131:11341–11343. doi: 10.1021/ja904625b. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.