Abstract
The significant consequences of ethanol use during pregnancy are neurobehavioral abnormalities involving hippocampal and neocortex malfunctions that cause learning and memory deficits collectively named fetal alcohol spectrum disorder (FASD). However, the molecular mechanisms underlying these abnormalities are still poorly understood and therefore warrant systematic research. Here, we document novel epigenetic abnormalities in the mouse model of FASD. Ethanol treatment of P7 mice, which induces activation of caspase-3, impaired DNA methylation through reduced DNA methyltransferases (DNMT1 and DNMT3A) levels. Inhibition of caspase-3 activity, before ethanol treatment, rescued DNMT1, DNMT3A proteins as well as DNA methylation levels. Blockade of histone methyltransferase (G9a) activity or cannabinoid receptor type-1 (CB1R), prior to ethanol treatment, which respectively inhibits or prevents activation of caspase-3, rescued the DNMT1 and DNMT3A proteins and DNA methylation. No reduction of DNMT1 and DNMT3A proteins and DNA methylation was found in P7 CB1R null mice, which exhibit no ethanol-induced activation of caspase-3. Together, these data demonstrate that ethanol-induced activation of caspase-3 impairs DNA methylation through DNMT1 and DNMT3A in the neonatal mouse brain, and such impairments are absent in CB1R null mice. Epigenetic events mediated by DNA methylation may be one of the essential mechanisms of ethanol teratogenesis.
Keywords: FASD, Epigenetics, Methyltransferase, Neuronal loss, CB1R, Caspase-3
Alcohol drinking during pregnancy is an intractable health problem worldwide and a leading cause of intellectual disability in Western nations (Mattson et al. 2011). The range of dysfunctions associated with alcohol exposure during development is collectively termed fetal alcohol spectrum disorder (FASD) and is characterized by widespread neuropsychological defects (Mattson & Riley 1998, Mattson et al. 1998) that involve hippocampal (HP) and neocortex (NC) dysfunctions (Bookstein et al. 2001, Clark et al. 2000, Mattson et al. 1996), including deficits in learning and memory (Goodman et al. 1999, Mattson et al. 1999). FASD is a major public health crisis with an estimated incidence rate as high as 2-5% in the United States and several Western European countries (May et al. 2009). Rodents are the most commonly used animal models for FASD research; however, their gestational period is much shorter than that of human beings (18–23 days for mice/rats), and in a significant amount of third trimester equivalents (Bayer et al. 1993) brain development takes place following birth in these species (Cronise et al. 2001, Tran et al. 2000). In rodent models, the brain is particularly sensitive to ethanol between postnatal days 6 and 10 (P6–10) due to the fact that the beginning of the second week is a critical period of synaptic development (Lanore et al. 2010, Marchal & Mulle 2004). A single episode of binge-like ethanol exposure on P7 was shown to induce robust activation of caspase-3 (a marker for neurodegeneration) in several brain regions (Ikonomidou et al. 2000, Sadrian et al. 2012, Saito et al. 2010, Wilson et al. 2011, Subbanna et al. 2013b), perturb local and interregional brain circuit integrity in the olfacto-hippocampal pathway (Sadrian et al. 2012, Wilson et al. 2011) resulting in impaired learning and memory task performance in adulthood (Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2013a) as observed in human FASD (Lebel et al. 2012, Mattson et al. 2011, Norman et al. 2013). So far, there are no effective treatments for FASD because our understanding of the molecular cause of FASD is limited.
Recently, studies from a number of independent laboratories have demonstrated that ethanol has the capacity to bring epigenetic changes to contribute to the development of FASD (Downing et al. 2011, Kaminen-Ahola et al. 2010a, Kaminen-Ahola et al. 2010b, Kim & Shukla 2005, Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2014b, Subbanna et al. 2013b, Zhou et al. 2011a). Epigenetic modifications of genomic DNA and histone proteins are critical in orchestrating the transcriptome of different cell types and their developmental potentials (Ma et al. 2010, Reik 2007, Suzuki & Bird 2008). Abnormal changes in histone modifications and/or DNA methylation play a major role in modulating gene expression and cellular functions that result in long-lasting altered phenotypes (Vaissiere et al. 2008) and several human developmental disorders (Campuzano et al. 1996, Gavin & Sharma 2010, Makedonski et al. 2005, Petronis 2003, Ryu et al. 2006, Warren 2007). Studies from several laboratories have demonstrated that exposure to ethanol at various developmental stages is associated with genome-wide/gene-specific alterations in histone modifications (Kim & Shukla 2005, Pal-Bhadra et al. 2007, Park et al. 2005, Subbanna et al. 2013b, Moonat et al. 2013), changes in DNA methylation (Downing et al. 2011, Garro et al. 1991, Haycock & Ramsay 2009, Liu et al. 2009, Ouko et al. 2009, Zhou et al. 2011b), and long-lasting altered phenotypes reminiscent of fetal alcohol syndrome (Kaminen-Ahola et al. 2010b). Collectively, these observations suggest that ethanol has the ability to act as a potent epigenetic modulator and induce deficits in neuronal differentiation (Veazey et al. 2013) and possibly maturation leading to learning and memory deficits (Izumi et al. 2005, Noel et al. 2011, Sadrian et al. 2012, Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2013a, Wilson et al. 2011) as observed in human FASD (Lebel et al. 2012, Mattson et al. 2011, Norman et al. 2013). Based on these interesting facts, the present study was undertaken to evaluate the mechanisms related to DNA methylation using a mouse model of FASD which induces widespread activation of caspase-3 immediately after ethanol exposure in P7 mice. We document one of the possible novel mechanisms through which DNA methylation was reduced in the mouse model of FASD. In addition, P7 CB1R null mice that exhibit no ethanol-induced activation of caspase-3 are resistant to ethanol-induced impairment of DNMT1 or DNMT3A or DNA methylation.
Methods
Animals
CB1R wild type (WT) and global knock out (KO) (CB1R gene is ablated in all tissues) mice were generated from CB1R heterozygous breeding colony at NKI. C57BL/6J mice or CB1RWT and KO mice on C57BL/6J background were housed in groups under standard laboratory conditions (12 h light / 12 h dark cycle) with food and water available ad libitum. Animal care and handling procedures followed Institutional (NKI IACUC) and National Institutes of Health guidelines. The genotype of CB1RWT and KO mice was determined as described before (Basavarajappa et al. 2003).
Ethanol treatment
An ethanol treatment paradigm, which has been previously shown to induce robust apoptotic neurodegeneration with no lethality in P7 mice (Olney et al. 2002), was used in the current study. Half of the pups (male and female) in each litter were treated subcutaneously (s. c.) with saline and the other half with ethanol at P7 (based on the day of birth) (2.5 g/kg s. c. at 0 h and again at 2 h) as described previously by our laboratory (Sadrian et al. 2012, Wilson et al. 2011, Subbanna et al. 2013a, Subbanna et al. 2013b). Blood ethanol levels (BEL) in pup serum were then determined using a standard alcohol dehydrogenase-based method (Lundquist 1959). In time kinetic studies, saline was injected for 0 h ethanol treatment.
Drugs treatment
In our previous studies, pre-administration of G9a related protein (GLP) inhibitor, Bix-01294 (Bix, 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmethyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride) (Cayman, Michigan, USA)], CB1R antagonist (SR, SR 141716A, gift from RBI) and broad-spectrum caspase inhibitor, and quinoline-Val-Asp(Ome)-CH2-O-phenoxy (Q-VD-OPh) (SM Biochemicals, Anaheim, CA, USA) 30 min before first dose ethanol treatment prevented activation of caspase-3 in P7 mice (Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2014b, Subbanna et al. 2013a, Subbanna et al. 2013b). Therefore, we used Bix or SR or Q-VD-OPh to inhibit caspase-3 activation in the current study. Bix and SR were dissolved separately in 10μl of ethanol followed by a two or three drops of Tween 80 (10 μl) and further volume was made up with sterile saline solution. The optimum dose of Bix or SR (1 mg/kg) solution was administered by s. c. injection at a volume of 5 μ/g body weight 30 min before first ethanol administration. The Bix or SR vehicle solution was injected as a control. The optimum dose of Q-VD-OPh (1 mg/kg) was administered by s. c. injection at a volume of 5 μl/g body weight 30 min before first dose ethanol treatment. Q-VD-OPh was dissolved in sterile saline solution. Sterile saline was used as vehicle in these experiments. Mice were kept with the dams until the pups were killed and their brains removed 4–24 h after the first saline/ethanol injection. Bix or SR or Q-VD-OPh treatment did not alter P7 ethanol induced intoxication (sleeping time) at the time of brain harvest (Subbanna et al. 2013a, Subbanna et al. 2013b). Bix or SR or Q-VD-OPh alone treated P7 mice looked like saline treated mice and did not cause any inflammation or bleeding in any of the organs (Subbanna et al. 2013a, Subbanna et al. 2013b). The brains were processed for several analyses, as described below.
Immunohistochemistry
Eight hours after the first dose ethanol/saline injection, the pups were anesthetized with isoflurane (1-3%) and perfused with a solution containing 4% paraformaldehyde and 4% sucrose in 0.05 M cacodylate buffer (pH 7.2). The free-floating sections containing hippocampus and cortex were immunostained using anti-rabbit cleaved caspase-3 (Asp175) (CC3) (polyclonal, #9661, 1:500, Cell Signaling, Danvers, MA, USA) by the ABC reagents (Vectastain ABC Elite Kit, Vector Labs, Burlingame, CA, USA) with a peroxidase substrate (DAB) kit (Vector Labs) as described previously (Subbanna et al. 2013a, Subbanna et al. 2013b). The primary antibodies were omitted from the reactions as a control for secondary antibody specificity. All photomicrographs were taken through a 2.5× or 40× objective with a Nikon Eclipse TE2000 inverted microscope attached to a digital camera (DXM1200F, Morrell Instrument Company, Melville, NY, USA).
Protein extraction, electrophoresis and immunoblot
For Western blot analysis, 4–24 h after the first saline or ethanol injection, pups were killed by decapitation, hippocampus and neocortex were dissected, flash frozen and stored at −80°C. Homogenates from the hippocampus and neocortex of the pups were processed and subjected to immunoblot as described previously (Basavarajappa et al. 2014, Basavarajappa & Subbanna 2014). Developmental brain samples were prepared by scarifying the P0 to P90 mice according to their date of birth respectively. Blots were Ponceau S stained to confirm equal loading in each lane and were incubated with primary antibody anti-rabbit-DNMT3A (monoclonal, # 3598, 1:1000), anti-rabbit-DNMT1 (monoclonal, # 5032, 1:1000), anti-rabbit CC3 (Asp175) (polyclonal, #9661, 1:1000) and anti-mouse-β-actin (monoclonal, #3700, 1:1,000) (Cell Signaling, Danvers, MA, USA) for 3 h at room temperature (25 degree centigrade) or overnight at 4°C and processed as previously described by our laboratory (Basavarajappa et al. 2008). Incubation of blots with a secondary antibody (goat anti-mouse peroxidase conjugate, #AP 124P, 1:5000, Millipore; goat anti-rabbit, #AP132P, 1:5000, Millipore) alone did not produce any bands.
Quantitative Real-Time Polymerase Chain Reaction (qPCR)
For the qPCR studies, 4–24 h after the first saline or ethanol injection, pups were killed by decapitation, neocortex and hippocampus were dissected, flash frozen and stored at −80°C. The samples were subjected to qPCR as described before (Subbanna et al. 2013a, Subbanna et al. 2013b) using Mm00599763_m1 (dnmt1), Mm00432881_m1 (dnmt3a) and 4352932 (Gapdh)] from Applied Biosystems. GAPDH was used as an endogenous mRNA control. Three independent runs were carried out for each set of samples. For each run, triplicate reactions were carried out for each sample. Data obtained was analyzed with the use of SDS2.4 software (Applied Biosystems). The amount of target (dnmt1 and dnmt3a), normalized to endogenous reference (Gapdh) and relative to a calibrator was given by-2ΔΔCt.
Global DNA methylation assay
For DNA methylation assay, 4–24 h after the first saline or ethanol injection, pups were killed by decapitation, neocortex and hippocampus were dissected, flash frozen and stored at −80°C. Genomic DNA was isolated from hippocampus and neocortex by Qiagen kit (Qiagen Sciences, Maryland, USA). The levels of global DNA methylation were determined by using Epigentek (Farmingdale, NY, USA) methylflash methylated DNA quantification kit (Colorimetric) (Epigentek, Farmingdale, NY, USA) according to the manufacturer’s instructions. 5-methylcytosine (5-mC) fraction of DNA was detected using capture and detection antibodies and then quantified by reading the absorbance at 450 nm. Synthetic unmethylated (50 % of cytosine) and methylated (50 % of 5-mC) DNA oligomers (Epigentek, Farmingdale, NY, USA) were used as a negative and positive control respectively. The percentage of 5-mC was calculated using the formula provided in the kit procedure and were normalized to percentage of control (The graphs represent the global DNA methylation levels multiplied by an arbitrary factor to set the control to 100).
Statistical analysis
Unless indicated otherwise, the experiments involved an equal number of pups per treatment and were performed in triplicate. All of the data are presented as the mean ± SEM. A statistical comparison of the data was performed by either a student’s t test or one-way analysis of variance (ANOVA) or a two-way ANOVA with Bonferroni’s post hoc test. In all of the comparisons, p < 0.05 was considered to indicate statistical significance. The statistical analyses were performed using the Prism software (GraphPad, San Diego, CA).
Results
Ethanol treatment of P7 mice activates caspase-3 followed by loss of DNA methylation
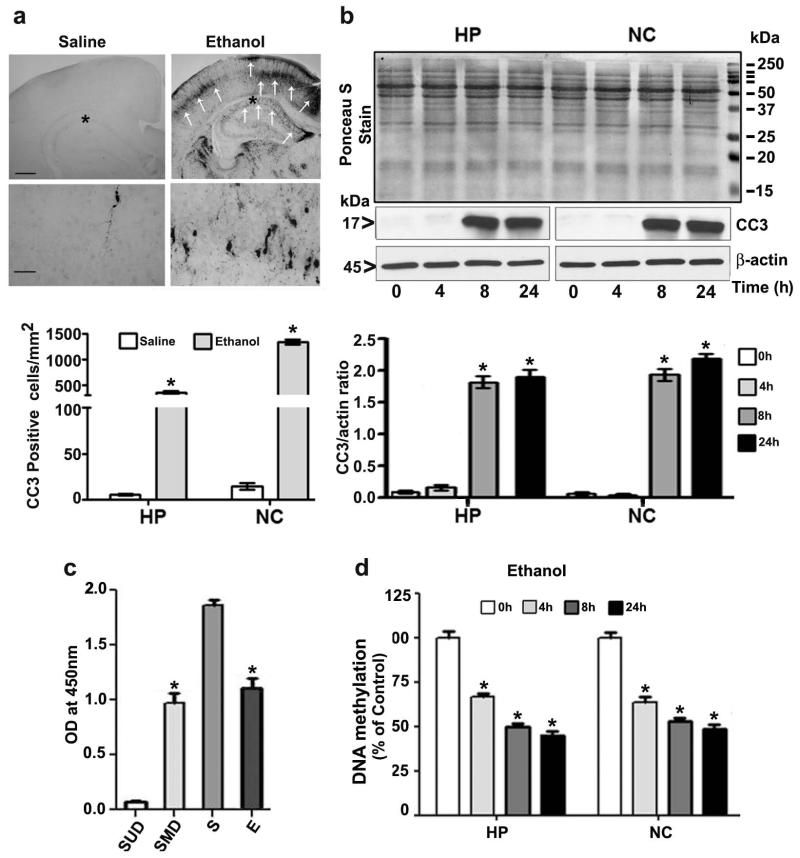
We injected P7 mice with ethanol (2.5 g/kg, sc at 0 h and again at 2h) and measured BELs at two time points. This experimental paradigm resulted in BELs of 0.45 ± 0.4 g/dl at 3 h that was gradually reduced to 0.24 ± 0.07 g/dl at 9 h after the first ethanol injection. We determined neurodegeneration using cleaved caspase-3 antibody (generation of CC3 as a marker for neurodegeneration) in the brains of P7 mice 8 h after first dose of ethanol or saline treatment. As shown in Figure 1a, CC3 immunoreactivity was found throughout the forebrain [hippocampus, (F1, 11 = 68, p<0.05) and cortex (F1, 11 = 180, p<0.05) regions] in the ethanol-exposed P7 brains (One-way ANOVA with Bonferroni’s post hoc test). Using Western blot analysis, we measured CC3 protein levels in hippocampal and neocortical protein cytosolic extracts obtained 4–24 h after the first saline or ethanol injection. Statistical analysis using one-way ANOVA with Bonferroni’s post hoc tests suggest that 8 and 24 h after first dose of postnatal ethanol treatment in P7 mice significantly enhanced CC3 protein levels in both the hippocampus (F3, 28 = 73, p<0.05) and neocortex (F3, 28 = 75, p<0.05) (Fig. 1b) compared to 0 h (saline control). In all our time kinetic studies, saline was injected for 0 h ethanol treatment.
Fig. 1.
Ethanol exposure induces apoptotic neurodegeneration and reduces DNA methylation in the P7 mouse brain. (a) Coronal brain sections (hippocampus and retrosplenial cortex) from saline- and ethanol-treated animals were immunostained with an anti-rabbit CC3 antibody. The white arrows indicate CC3-positive neurons in the hippocampus and retrosplenial cortex, respectively. Scale bars = 200 μm. The respective images were enlarged to show CC3-positive cells (*). The scale bars represent 50 μm. CC3-positive cells were counted in the hippocampus and the retrosplenial cortex (n =10 pups/group). (b) Western blot analysis of CC3 using cytosolic extracts (20 μg) of hippocampal and neocortical samples obtained 4–24 h after the first saline or ethanol injection (n = 15 pups/group). Ponceau S staining confirmed equal loading and β-actin were used as loading controls. *p < 0.05 vs. 0 h or saline control (One-way ANOVA with Bonferroni’s post hoc test). For 0 h ethanol group, saline was injected. (c) Global DNA methylation assay specificity was determined using synthetic unmethylated DNA (contains 50% of cytosine) (negative control) (SUD) and methylated DNA (contains 50% of 5-methylcytosine) (positive control) (SMD). The OD was used with the kit’s included formulas to calculate the global DNA methylation levels. S, saline; E, ethanol. (d) Global DNA methylation quantification in hippocampal (HP) and neocortical (NC) DNA obtained 4–24 h after the first saline or ethanol injection. For 0 h ethanol group, saline was injected. Error bars, SEM. Error bars, SEM. HP, hippocampus; NC, neocortex. (*p < 0.05, n = 10 pups/group) (One-way ANOVA with Bonferroni’s post hoc test).
We then examined the DNA methylation in the nuclear changes experienced by apoptotic neurons in ethanol-treated P7 mice. First, we measured global DNA methylation levels using methylflash methylated DNA quantification kit. Assay specificity was determined using synthetic unmethylated DNA (contains 50% of cytosine) (negative control) (SUD) and methylated DNA (contains 50% of 5-methylcytosine) (positive control) (SMD). The amount of methylated DNA was found to be directly proportional to the optical density (OD) intensity (Fig. 1c). The OD was used with the kit’s included formulas to calculate the global DNA methylation levels. We measured global DNA methylation in hippocampal and neocortical DNA obtained 4–24 h after the first saline or ethanol injection. Global DNA methylation was reduced after ethanol exposure (8 and 24 h) compared to saline-treatment (0 h) in the hippocampus (F1, 20 = 58, p < 0.05) and neocortex (F1, 20 = 62, p < 0.05) (One-way ANOVA with Bonferroni’s post hoc test) (Fig. 1d).
Developmental changes of DNMT1 and DNMT3A proteins in mouse hippocampus and neocortex regions
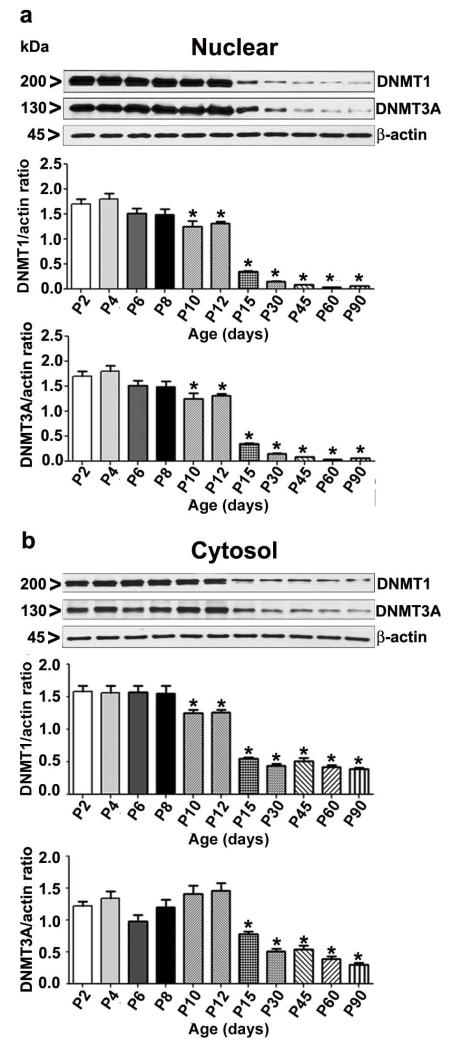
We measured the levels of DNMT1 and DNMT3A, enzymes involved in DNA methylation (Biniszkiewicz et al. 2002, Feng et al. 2010), during various stages of brain development. DNMT1 antibody recognized a major band running at 200kDa (predicted to run at ~183kDa) as observed in several previous studies (Liu et al. 2003, Robertson et al. 2000). DNMT3A antibody recognized a major band running below 140kDa marker and above the 100kDa marker (estimated at ~130kDa) as observed in our previous study (Subbanna et al. 2014b). There is a discrepancy between the observed and predicted molecular weight of DNMT3A (predicted to run at ~101kDa). The post-translational modification can impact the mobility of a protein in SDS-PAGE gels. The post-translational modifications and/or features of DNMT3A that result in the reduced mobility (and thus higher observed molecular weight) have not been well characterized in the literature. Several papers included data demonstrating recombinant and endogenous cellular DNMT3A runs well above the predicted 101kDa (Chang et al. 2011, Subbanna et al. 2014b, Suetake et al. 2011, Takeshima et al. 2006). We then examined the developmental pattern of DNMT1 and DNMT3A protein. Neocortex nuclear and cytosolic protein extracts at several developmental stages were used in Western blot analysis. Nuclear DNMT1 (F8, 45 = 150, p<0.05) and DNMT3A (F8, 45 = 160, p<0.05) protein levels were substantially higher during synaptic development (synaptogenesis) compared to the levels in the adult brain (P90) (Fig. 2a). Similar expression of DNMT1 (F8, 45 = 110, p<0.05) and DNMT3A (F8, 45 = 90, p<0.05) were also found in cytosolic protein extracts (Fig. 2b) (One-way ANOVA with Bonferroni’s post hoc test).
Fig. 2.
Developmental pattern of DNMT1 and DNMT3A proteins associated with nuclear or cytosolic extracts of mouse brain. (a) Western blot analysis of DNMT1 and DNMT3A expression in neocortical nuclear extract during mouse brain development. (b) Western blot analysis of DNMT1 and DNMT3A expression in neocortical cytosolic extract obtained from various mouse brain developmental stages. β-actin was used as a loading control. Representative blot shows developmental expression of DNMT1 and DNMT3A proteins (all data were compared with the P2 group) Error bars, SEM. (*p < 0.05, n = 10 pups/group) (One-way ANOVA with Bonferroni’s post hoc test).
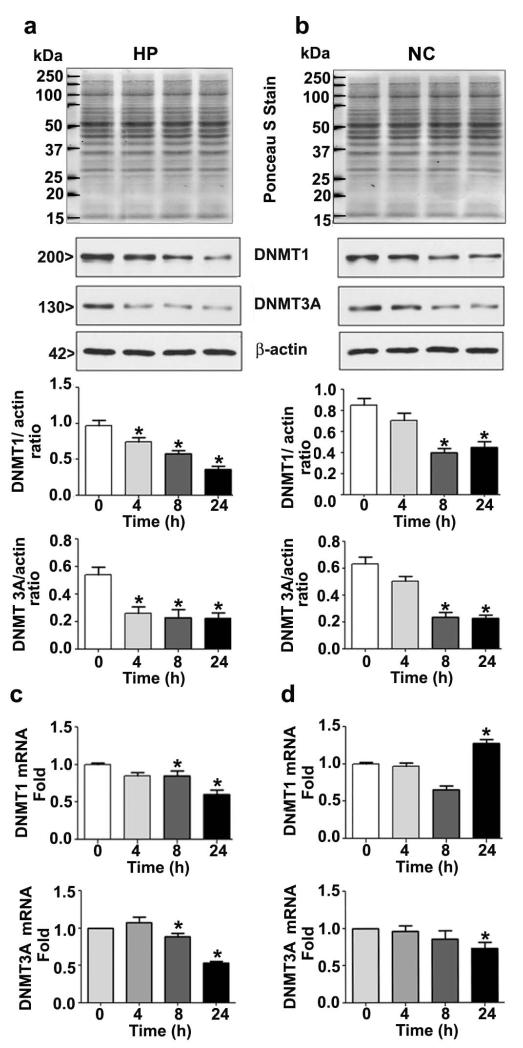
P7 ethanol exposure reduces DNMT1 and DNMT3A protein expression
To examine the mechanism by which ethanol may reduce DNA methylation, we examined the influence of ethanol exposure on DNMT1 and DNMT3A protein expression in the hippocampus and neocortex obtained 4–24 h after the first saline or ethanol injection. Ethanol decreased DNMT1 and DNMT3A protein levels in the hippocampus (Fig. 3a) (DNMT1; F3, 28 = 18, p<0.05; DNMT3A; F3, 28 = 28, p<0.05) and neocortex (Fig. 3b) (DNMT1; F3, 28 = 16, p<0.05; DNMT3A; F3, 28 = 22, p<0.05) at 8-24 h (after first ethanol injection) time points compared to the saline control (0 h) (One-way ANOVA with Bonferroni’s post hoc test). We also measured mRNA levels to understand whether a reduced DNMT1 and DNMT3A protein level is due to suppression of their transcription. The results reveal that decreased DNMT1 mRNA levels by ethanol was found only at the 24 h time point in the hippocampus (Fig. 3c) (F3, 28 = 20, p<0.05). However, in neocortex, decrease in DNMT1 mRNA levels was found at 8 h but it was enhanced at 24 h (F3, 28 = 25, p<0.05) (Fig. 3d). We found reduced DNMT3A mRNA levels in the hippocampus (F3, 28 = 15, p<0.05) at the 8 and 24 h time points. In neocortex, decrease in DNMT3A mRNA levels was found only at 24 h (F3, 28 = 12, p<0.05) (One-way ANOVA with Bonferroni’s post hoc test). Control Gapdh mRNA levels remained unchanged in any of the brain regions examined (data not shown). Over all, ethanol treatment in P7 mice reduced the protein more severely compared to mRNA levels except in neocortex.
Fig. 3.
P7 ethanol treatment reduces the protein levels and differentially regulates mRNA levels of DNMT1 and DNMT3A in mouse brain regions. (a and b) Western blot analysis of DNMT1 and DNMT3A in hippocampus (HP) and neocortex (NC) nuclear extracts obtained 4–24 h after the first saline or ethanol injection. Ponceau S staining confirmed equal loading and β-actin were used as loading controls. (c and d) qPCR analysis of DNMT1 and DNMT3A mRNA in HP and NC extracts obtained 4–24 h after the first saline or ethanol injection groups (n = 10 pups/group). For 0 h ethanol group, saline was injected. [*p < 0.05 vs. saline (0 h) group]. Error bars, SEM. (One-way ANOVA with Bonferroni’s post hoc test).
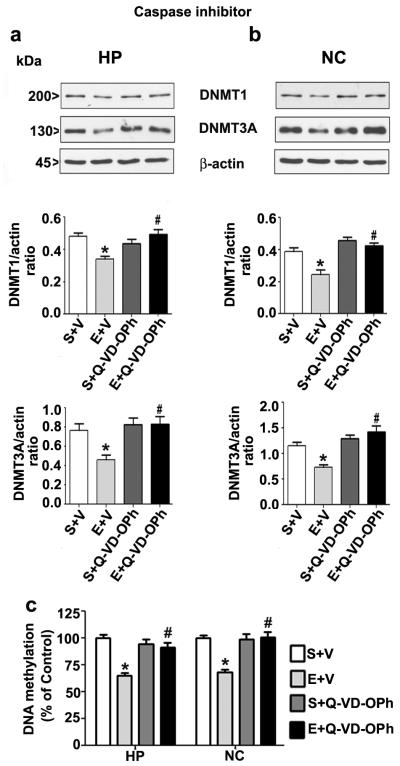
Caspase-3 inhibitor rescues DNMT1 and DNMT3A protein degradation and restores DNA methylation in ethanol treated P7 mice
We hypothesized that the time dependent reduction of DNMT1 and DNMT3A protein may be due to the action of ethanol-activated caspase-3 in P7 mice as activated caspase enzymes degrades many proteins within the cell (Fischer et al. 2003, Kamada et al. 2005, Subbanna & Basavarajappa 2014, Subbanna et al. 2013b). In these studies, we used a third generation dipeptidyl broad-spectrum caspase inhibitor (Q-VD-OPh). Our previous in vivo study have shown that Q-VD-OPh (Renolleau et al. 2007) effectively prevents the generation of CC3 fragment in P7 mice without any detectable toxicity [for references see (Renolleau et al. 2007, Subbanna et al. 2013b)]. It also prevented dimethylated histone H3 lysine 9 (H3K9me2) and total histone H3 protein degradation in ethanol-treated P7 mice (Subbanna et al. 2013b). Administration of optimum dose of Q-VD-OPh (1 mg/kg) before ethanol treatment did not alter the BELs (0.41 ± 0.3 g/dl at 3 h, gradually reduced to 0.23 ± 0.08 g/dl at 9 h after the first ethanol injection), indicating that Q-VD-OPh does not modulate ethanol metabolism as observed in our previous studies (Subbanna et al. 2013b). Pre-administration of Q-VD-OPh before ethanol treatment inhibited the DNMT1 and DNMT3A protein (Fig. 4a and b) degradation in both the brain regions (hippocampus: DNMT1, F1,20 = 23, p < 0.05; DNMT3A, F1,20 = 17, p < 0.05; neocortex: DNMT1, F1,20 = 18, p < 0.05; DNMT3A, F1,20 = 17, p < 0.05). The saline and saline-Q-VD-OPh pre-treatment fail to alter DNMT1 and DNMT3A protein levels (p > 0.05). Pre-administration of Q-VD-OPh before ethanol-treatment rescued impaired DNA methylation (Fig. 4c) in both the brain regions (hippocampus, F1,20 = 25, p < 0.05; neocortex, F1,20 = 28, p < 0.05) (Two-way ANOVA with Bonferroni’s post hoc test).
Fig. 4.
Pharmacological inhibition of caspase-3 rescues P7 ethanol-induced degradation of DNMT1 and DNMT3A proteins as well as DNA methylation in the neonatal mouse brain. (a and b) P7 Mice pre-treated for 30 min with Q-VD-OPh (1 mg/kg) or vehicle were exposed to ethanol and DNMT1 and DNMT3A proteins levels were determined in nuclear extracts of hippocampus (HP) and neocortex (NC) by a Western blot analysis. β-actin was used as a loading control. (c) Global DNA methylation quantification was performed in HP and NC DNA from saline (S), ethanol (E), S + Q-VD-OPh, or E + Q-VD-OPh groups (n= 10 pups/group) (*p < 0.05 vs. S; #p < 0.05 vs. E. Error bars, SEM. (Two-way ANOVA with Bonferroni’s post hoc test).
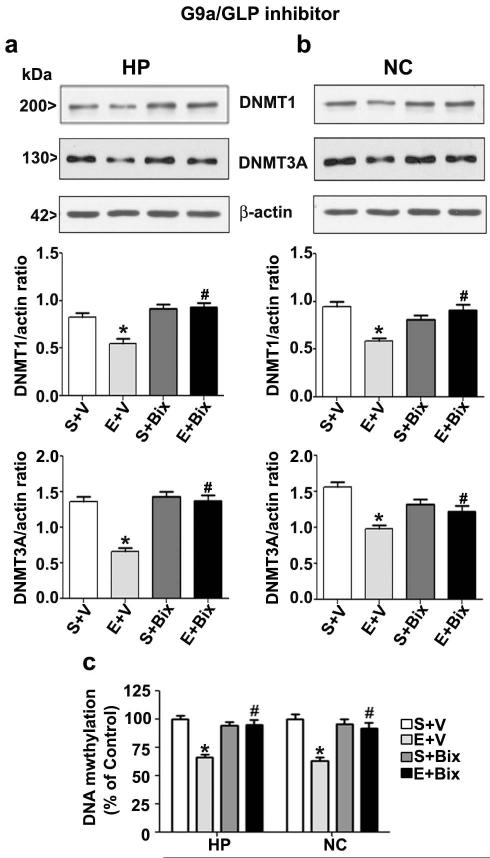
Pre-administration of a G9a/GLP inhibitor prevents ethanol-induced reduction of DNMT1 and DNMT3A protein followed by DNA methylation in P7 mice
In our previous studies, we have shown that pharmacological inhibition of G9a/GLP by Bix before ethanol-treatment prevents activation of caspase-3 in P7 mice without affecting ethanol metabolism (Subbanna & Basavarajappa 2014, Subbanna et al. 2014b, Subbanna et al. 2013b). In this study, we examined whether the reduced levels of DNMT1 and DNMT3A protein observed after ethanol treatment could be rescued by Bix (1mg/kg, optimum dose) pre-treatment. Bix administration before ethanol-treatment prevented DNMT1 and DNMT3A protein loss in both the brain regions (Fig. 5a and b). Statistical analysis by two-way ANOVA with Bonferroni’s post hoc analysis showed a significant effect of ethanol (vs. saline) (hippocampus: DNMT1, F1, 28 = 14, p < 0.05; DNMT3A, F1, 28 = 9, p < 0.05; neocortex: DNMT1, F1, 28 = 16, p < 0.05; F1, 28 = 16, p < 0.05). A significant interaction between ethanol and Bix treatment was also evident (hippocampus: DNMT1, F1, 28 = 11, p < 0.05; DNMT3A, F1, 28 = 17, p < 0.05; neocortex: DNMT1, F1, 28 = 14, p < 0.05; DNMT3A, F1, 28 = 12, p < 0.05). The saline and saline-Bix groups were failed affect significantly (p > 0.05). In our next experiment, we determined whether DNA methylation that was reduced after ethanol treatment could be rescued by Bix-pre-treatment. Comparison of two-way ANOVA with Bonferroni’s post hoc test indicated a significant influence of ethanol treatment (vs. saline) (hippocampus: F1, 28 = 22, p < 0.05; neocortex: F1,28 = 24, p < 0.05). A significant interaction between ethanol and Bix treatments was also evident (hippocampus; F1, 28 = 18, p < 0.05; neocortex; F1, 28 = 20, p < 0.05) (Fig.5c). These findings suggest that Bix prevents the ethanol-induced decrease in DNA methylation by restoring caspase-3 mediated reduction of DNMT1 and DNMT3A protein levels in P7 mice.
Fig. 5.
Pharmacological inhibition of G9a rescues P7 ethanol-induced degradation of DNMT1 and DNMT3A proteins and DNA hypomethylation in the neonatal mouse brain. (a and b) P7 Mice pre-treated for 30 min with Bix (1 mg/kg) or vehicle (V) were exposed to saline (S) ethanol (E) and DNMT1 and DNMT3A protein levels in nuclear extracts were determined by a Western blot analysis in hippocampus and neocortex (n= 6 pups/group). β-actin was used as a loading control. (c) Global DNA methylation quantification was performed in hippocampus and neocortex DNA samples from S + V, E + V, S + Bix or E + Bix groups (n= 10 pups/group) (*p < 0.05 vs. S + V; #p < 0.05 vs. E + V). Error bars, SEM. (Two-way ANOVA with Bonferroni’s post hoc test). HP, hippocampus; NC, neocortex.
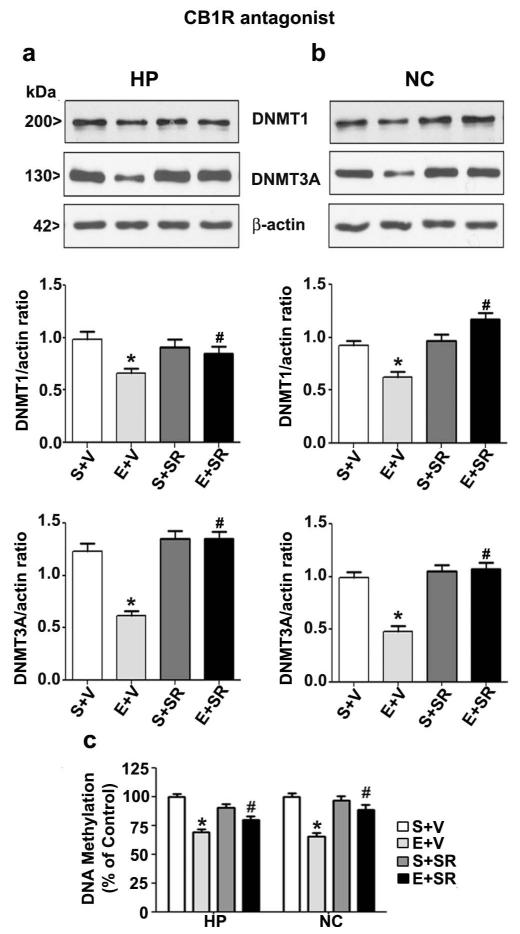
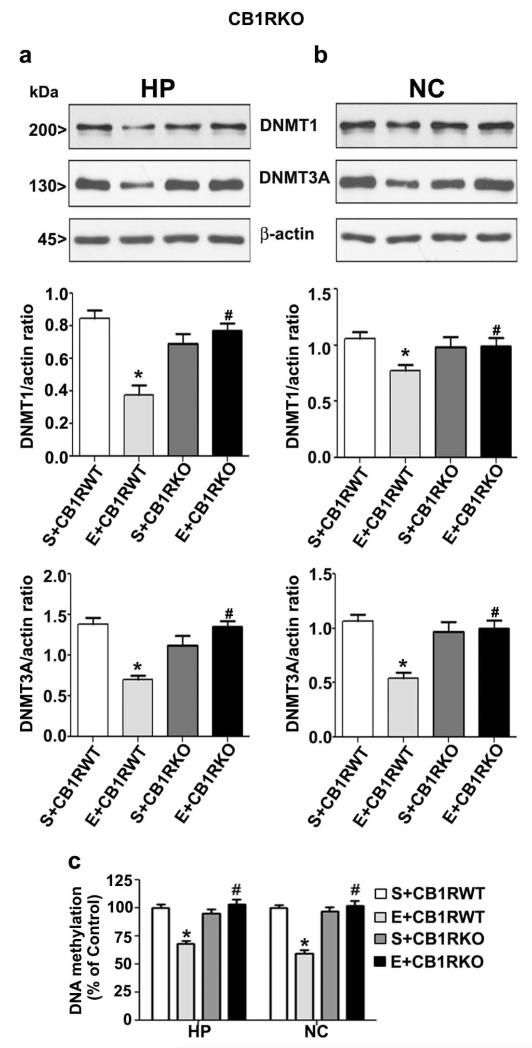
Genetic deletion or pharmacologic blockade of CB1R prevents ethanol-induced reduction in DNA methylation, DNMT1 and DNMT3A levels
In our previous studies, both SR and CB1RKO provided protection against ethanol-induced activation of caspase-3 without altering ethanol metabolism (Subbanna et al. 2014a, Subbanna et al. 2013a). We used a specific CB1R antagonist (SR) or CB1RKO mice in our next experiment to further examine the inhibition of DNA methylation and the levels of the associated proteins DNMT1 and DNMT3A by ethanol is due to wide-spread activation of caspase-3. Our previous findings suggested that maximum inhibition of caspase-3 activation was found at 1 mg/kg (Subbanna et al. 2013a). Thus, we used SR at 1 mg/kg in our current studies. Our findings suggest that SR pre-administration before ethanol-treatment completely rescued the loss of DNMT1 and DNMT3A protein levels in both the brain regions (p < 0.05) (Fig. 6a and b). Statistical analysis using two-way ANOVA with Bonferroni’s post hoc tests indicate significant influence of ethanol (vs. saline) (hippocampus: DNMT1, F1,20 = 20, p < 0.05; DNMT3A, F1,20 = 18, p < 0.05; neocortex: DNMT1, F1,20 = 22, p < 0.05; DNMT3A, F1,20 = 15, p < 0.05) and a significant interaction between ethanol and SR pre-treatment (hippocampus: DNMT1, F1,20 = 14, p < 0.05; DNMT3A, F1,20 = 12, p < 0.05; neocortex: DNMT1, F1,20 = 23, p < 0.05; DNMT3A, F1,20 = 16, p < 0.05). Pre-treatment of SR or vehicle alone fail to alter DNMT1 or DNMT3A protein levels (p > 0.05). Similarly decrease in DNA methylation due to ethanol treatment in P7 mice was also prevented by SR pre-treatment. Statistical analysis using two-way ANOVA with Bonferroni’s post hoc tests confirmed a significant influence of ethanol (vs. saline) (hippocampus: F1,20 = 27, p < 0.05; neocortex: F1,20 = 25, p < 0.05). A significant interaction between ethanol and SR-treatment was also evident (hippocampus: F1,20 = 10, p < 0.05; neocortex: F1,20 = 18, p < 0.05). SR or vehicle pre-treatment failed to reduce DNA methylation (p > 0.05) (Fig. 6c). Statistical analysis of one-way ANOVA with Bonferroni’s post hoc tests suggested that, consistent with SR treatment, the CB1R KO provided protection against the P7 ethanol-induced decrease in DNMT1 and DNMT3A protein levels in both the brain regions (Fig. 7a and b) (hippocampus: DNMT1, F1,20 = 18, p < 0.05; DNMT3A, F1,20 = 13, p < 0.05; neocortex: DNMT1, F1,20 = 23, p < 0.05; DNMT3A, F1,20 = 12, p < 0.05). Similarly, CB1RKO mice provided protection against the P7 ethanol-induced decrease in DNA methylation in the hippocampus (F1,20 = 28, p < 0.05) and neocortex (F1,20 = 26, p < 0.05) (one-way ANOVA with Bonferroni’s post hoc tests) (Fig. 7c).
Fig. 6.
Blocking CB1 receptor with SR rescues ethanol-induced degradation of DNMT1 and DNMT3A proteins and impaired DNA methylation in the neonatal mouse brain. (a and b) Western blot analysis of DNMT1 and DNMT3A proteins and β-actin (loading control) in hippocampal and neocortical nuclear extracts from the four (S+V, E+V, S+SR and E+SR) groups (n = 15 pups/group). P7 mice were treated with ethanol for 8 h, and SR was pretreated for 30 min before the ethanol treatment. (c) Global DNA methylation quantification was performed in hippocampal and neocortical DNA from saline (S), ethanol (E), S + SR, or E + SR groups (n= 10 pups/group) (*p < 0.05 vs. S + V; #p < 0.05 vs. E + V. Error bars, SEM. (Two-way ANOVA with Bonferroni’s post hoc test). HP, hippocampus; NC, neocortex.
Fig. 7.
Genetic deletion of CB1Rs provides protection against ethanol-induced degradation of DNMT1 and DNMT3A proteins and DNA hypomethylation in the neonatal mouse brain. P7 WT and KO mice were treated with ethanol for 8 h (a and b) Western blot analysis of DNMT1 and DNMT3A proteins and β-actin (loading control) in hippocampal and neocortical nuclear extracts from the four treatment (S +CB1R WT, E+CB1R WT, S+CB1R KO + and E+CB1R KO) groups (n = 15 pups/group). S, saline; E, ethanol. (c) Global DNA methylation quantification was performed in hippocampal and neocortical DNA from S +CB1RWT, E +CB1RWT, S+ CB1RKO, or E+CB1RKO groups (n= 10 pups/group) (*p < 0.05 vs. S + CB1RWT; #p < 0.05 vs. E + CB1RWT. Error bars, SEM. S, saline; E, ethanol. (Two-way ANOVA with Bonferroni’s post hoc test). HP, hippocampus; NC, neocortex.
Discussion
In this study, using a mouse model of FASD, we demonstrate for the first time that the mechanism by which postnatal ethanol impairs DNMT1 and DNMT3A followed by DNA methylation in hippocampus and neocortex, two brain regions that are important for learning and memory (Vann & Albasser 2011). This was accomplished through differential transcriptional regulation of both genes and caspase-3-mediated degradation of DNMT1 and DNMT3A, which are produced at significantly higher rate during early brain development compared to that in the mature mouse brain. Overall, our current results are consistent with previous findings demonstrating that DNMT1 and DNMT3A are the most highly expressed early in young age and their levels drop significantly by adulthood in most brain regions (Brown et al. 2008, Feng et al. 2005, Robertson et al. 1999, Simmons et al. 2013). Our results are also supported by one more study in which DNMT1 and DNMT3A protein expression were found in both the nuclear and cytosolic fractions of the mouse brain (Chestnut et al. 2011).
The control of gene transcription, via dynamic modification of DNA methylation, plays an essential function in the regulation of gene expression. The active modification of DNA methylation occurs in a tissue- and time-specific way and appears to be vital for embryonic development (Kakutani et al. 1996, Martin et al. 1999, Matsuda & Yasutomi 1992). Exposure to ethanol during embryonic development has been shown to induce changes in DNA methylation (Downing et al. 2011, Garro et al. 1991, Haycock & Ramsay 2009, Liu et al. 2009, Ouko et al. 2009, Zhou et al. 2011b) and is implicated in the etiology of numerous developmental anomalies (Kaminen-Ahola et al. 2010b, Ouko et al. 2009). Using a mouse model of FASD, our study specifically suggests that ethanol exposure during postnatal development induces wide spread activation of caspase-3 that leads to the loss of DNMT1 and DNMT3A proteins thereby causing DNA hypomethylation. Further, ethanol-activated caspase-3 also cleaves some nuclear proteins such as dimethylated (lysine 9) histone proteins (Subbanna et al. 2013b) in neonatal mice. Our findings are in line with another study in which maternal ethanol consumption (9-11th day of pregnancy) decreases the activity of DNMT in 12th day fetal mice, resulting in DNA hypomethylation (Garro et al. 1991). However, these findings are in contrast with another study in which ethanol-treatment was shown to decrease expression of cell cycle genes through enhanced DNMT activity followed by DNA hypermethylation in neural stem cells (NSCs) (Hicks et al. 2010). In yet another study, repeated postnatal ethanol exposure of P2-10 mice also enhanced DNA methylation in P20 rats (Otero et al. 2012). Despite this observation, the DNMT1 transcription is reduced by ethanol-treatment in both neural stem cells (Hicks et al. 2010) and rat sperm (Bielawski et al. 2002) a finding that supports our data.
In the present study, ethanol took a long time (24 h) to reduce the expression of the methyltransferase genes encoding for DNMT3A and DNMT1 proteins that are important for “de novo” and the “maintenance” of DNA methylation respectively in the hippocampus. But, in the cortex, ethanol even increased the expression of the DNMT1 gene at the time point where ethanol induces the highest activation of caspase-3. This increase may be a compensatory response to the continuous presence of ethanol and suppression of DNMT3A, and this needs to be investigated further in future studies. However, a similar contrasting effect of ethanol exposure in neural stem cells and fetal brain on the expression of DNMT1 mRNA and protein has been reported (Hicks et al. 2010). In an embryonic ethanol model, ethanol enhanced the expression of DNMT3A and 3B, but not DNMT1 (Mukhopadhyay et al. 2013) genes. A recent study reported that ethanol exposure significantly impaired DNA methylation (5-mC staining) in both the dorsal and the ventral neural tube and that led to developmental delay (Zhou et al. 2011c). Together, these data suggest that ethanol, which induces activation of caspase-3 in neonatal mice, can also impact the cellular DNA methylation machinery by modulating DNMT proteins and, thus, play a significant role in the maturation of synaptic circuits during development. Several studies have revealed that DNA methylation is essential to various types of synaptic plasticity and learning and memory behaviors in adult animals (Feng et al. 2010, Miller et al. 2010, Miller & Sweatt 2007, Roth et al. 2009, Weaver et al. 2004). Together, these observations suggest that DNA methylation is important during postnatal development; therefore, its reduction, even for a short period, may cause long-lasting synaptic dysfunction in adult mice, as observed in similar mice models of FASD (Izumi et al. 2005, Sadrian et al. 2012, Subbanna & Basavarajappa 2014, Subbanna et al. 2013a, Wilson et al. 2011).
The cellular proteins that drive biological processes are regulated at different levels, including transcription, translation, post-translational modification, and degradation. In steady-state conditions, cellular proteins are dynamic and most of them are continuously synthesized and degraded. Protein degradation mechanisms adjust protein levels in response to both internal and environmental stimuli. In the nucleus, presence of many caspase-3 substrates were described (Fischer et al. 2003) and were implicated in the nuclear morphological changes that occur in most of the apoptotic cells (Kamada et al. 2005). Consistent with this notion, low concentrations of ethanol induce significantly lower amounts of caspase-3 activation in P7 mice and enhance DNMT3A levels without any proteolytic degradation (Subbanna et al. 2014b). Thus, our study suggests that DNMT1 and DNMT3A proteins may act as substrates for widely activated caspase-3 due to high concentration of ethanol in the developing brain. Accordingly, caspase-3 inhibition rescues loss of DNMT1 and DNMT3A as well as DNA methylation in ethanol-exposed P7 mice. However, our study does not exclude other possible mechanisms of DNMT1 and DNMT3A proteins degradation in ethanol-treated P7 mice. Similar degradation of DNA methyltransferases by ethanol exposure during the embryonic period was observed (Mukhopadhyay et al. 2013). However, this degradation is mediated through the ubiquitin-proteasome pathways (Mason et al. 2012). DNMT1 protein stability was also shown to be regulated by ubiquitin-mediated proteosomal degradation, although the enzymes responsible for the ubiquitination state of DNMT1 have not been reported (Agoston et al. 2005). In addition, DNMT1 stability may also be controlled through other post-translational modification mechanisms such as phosphorylation and methylation (Esteve et al. 2009, Sun et al. 2007, Wang et al. 2009). It is also possible that reduced DNMT1 and DNMT3A protein levels found in the current study may be a result of post-translational modification by ubiquitination followed by degradation through the action of the ubiquitin-26S proteasome system (UPS). Such possibilities warrant future investigation. In our previous study, pre-administration of G9a/GLP inhibitor (Bix) before ethanol-treatment prevented activation of caspase-3 as well as caspase-3-mediated degradation of dimethylated H3K9 (Subbanna et al. 2013b). Furthermore, pre-administration of Bix before ethanol-treatment also prevented long term potentiation LTP as well as learning and memory deficits in adult mice (Subbanna & Basavarajappa 2014). In the current study, pre-administration of Bix also prevented degradation of DNMT1 and DNMT3A proteins and restored DNA methylation. Similarly, pre-administration of CB1R antagonists (SR) which was shown to prevent ethanol-induced activation of caspase-3 in neonatal mice (Subbanna et al. 2013a), also prevented degradation of DNMT1 and DNMT3A proteins and rescued DNA methylation. Interestingly, the ablation of the CB1R, which provides protection against ethanol-induced activation of caspase-3 in neonatal mice (Subbanna et al. 2013a), also restored the levels of DNMT1, DNMT3A proteins and DNA methylation that had been reduced by ethanol. These observations may suggest that observed reduction of DNA methylation for a short period during a critical period of synaptic development (Lanore et al. 2010, Marchal & Mulle 2004) can have a long-lasting impact on genome regulation and synaptic circuit formation leading to neurobehavioral abnormalities as observed in mice and human FASD (Lebel et al. 2012, Mattson et al. 2011, Norman et al. 2013). This is partly because both the pharmacological blockade of G9a (through use of Bix inhibitor) and CB1R (through use of SR) and the use of CB1R null mice led to protection against P7 ethanol-induced activation of caspase-3 but also prevented long-lasting neurobehavioral abnormalities in adult mice (Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2013a, Subbanna et al. 2013b).
Hypomethylation of CpG islands, especially those islands co-localized with gene promoter regions, is generally associated with gene expression. Impaired DNMT activity and the resulting reduction in DNA methylation levels as well as DNA binding proteins was shown to regulate several genes associated with neurodegenerative disorders in adults [for references see (Lagali & Picketts 2011)]. Therefore, it is possible that during early brain development, ethanol-induced DNA hypomethylation may differentially regulate the transcription of genes encoding for survival factor (Kokubo et al. 2009) such as activity-regulated cytoskeleton-associated protein (Arc) (Subbanna et al. 2014a) or CB1R or G9a expression (Subbanna et al. 2013a, Subbanna et al. 2013b), inducing a delay in neuronal development (Chen et al. 2013) and deficits in learning and memory in adult animals (Noel et al. 2011, Sadrian et al. 2012, Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2013a, Wilson et al. 2011). Future studies using chromatin immunoprecipitation coupled with genome-wide analysis will further unveil the impact of impaired DNMT1 and DNMT3A proteins and DNA methylation on specific gene expression and function in ethanol teratogenesis.
In summary, pharmacological inhibition of caspase-3 by Q-VD-OPh or caspase-3 inactivation by means of Bix or SR treatment prior to ethanol treatment prevents impairment of DNMT1 and DNMT3A proteins and DNA methylation. P7 CB1R null mice, which exhibit no ethanol-induced activation of caspase-3, exhibit no impairment of DNMT1, DNMT3A and DNA methylation. This experimental evidence elucidates a novel potential molecular mechanism underlying ethanol teratogenesis, through which ethanol may impair DNA methylation. These impairments, which occur within cells of the developing brain, may have a distinct role in causing long-lasting deficits in learning and memory, as observed in human FASD (Lebel et al. 2012, Mattson et al. 2011, Norman et al. 2013) as well as animal models of FASD (Noel et al. 2011, Sadrian et al. 2012, Subbanna & Basavarajappa 2014, Subbanna et al. 2014a, Subbanna et al. 2013a, Wilson et al. 2011), and should be explored in future studies.
Acknowledgements
This work was supported by NIH/NIAAA grant AA019443 (BSB). DP was supported by NIDA grant F32DA021977. We thank Neha Balapal for editing the final version of the manuscript.
Abbreviations used
- P7
postnatal day 7
- FASD
fetal alcohol spectrum disorder
- BEL
blood ethanol level
- CC3
Cleaved Caspase-3
- HP
Hippocampus
- NC
Neocortex
- CB1R
cannabinoid receptor type 1
- CB1RKO
cannabinoid receptor type 1 knockout
- SUD
synthetic unmethylated DNA
- SMD
synthetic methylated DNA
- DNMT
DNA methyltransferases
- 5-mC
5-methylcytosine
- Bix
Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmethyl)-4-piperidinyl]-4-quinazolinaminetrihydrochloride
- Q-VD-OPh
quinoline-Val-Asp(Ome)-CH2-O-phenoxy
- SR
SR141617A
- H3K9me2
dimethyl histone H3 lysine 9
- Arc
Activity-regulated cytoskeleton-associated protein
Footnotes
Conflict-of-Interest disclosure: The authors declare no competing financial interests.
References
- Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J. Biol. Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- Basavarajappa BS, Nagre NN, Xie S, Subbanna S. Elevation of endogenous anandamide impairs LTP, learning, and memory through CB1 receptor signaling in mice. Hippocampus. 2014;24:808–818. doi: 10.1002/hipo.22272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa BS, Ninan I, Arancio O. Acute Ethanol Suppresses Glutamatergic Neurotransmission through Endocannabinoids in Hippocampal Neurons. J. Neurochem. 2008;107:1001–1013. doi: 10.1111/j.1471-4159.2008.05685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa BS, Saito M, Cooper TB, Hungund BL. Chronic ethanol inhibits the anandamide transport and increases extracellular anandamide levels in cerebellar granule neurons. Eur. J. Pharmacol. 2003;466:73–83. doi: 10.1016/s0014-2999(03)01557-7. [DOI] [PubMed] [Google Scholar]
- Basavarajappa BS, Subbanna S. CB1 Receptor-Mediated Signaling Underlies the Hippocampal Synaptic, Learning and Memory Deficits Following Treatment with JWH-081, a New Component of Spice/K2 Preparations. Hippocampus. 2014;24:178–188. doi: 10.1002/hipo.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Altman J, Russo RJ, Zhang X. Timetables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neurotoxicology. 1993;14:83–144. [PubMed] [Google Scholar]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol. Clin. Exp. Res. 2002;26:347–351. [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, et al. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein FL, Sampson PD, Streissguth AP, Connor PD. Geometric morphometrics of corpus callosum and subcortical structures in the fetal-alcohol-affected brain. Teratology. 2001;64:4–32. doi: 10.1002/tera.1044. [DOI] [PubMed] [Google Scholar]
- Brown SE, Weaver IC, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci. Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Chang Y, Sun L, Kokura K, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nature communications. 2011;2:533. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PloS one. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CM, Li D, Conry J, Conry R, Loock C. Structural and functional brain integrity of fetal alcohol syndrome in nonretarded cases. Pediatrics. 2000;105:1096–1099. doi: 10.1542/peds.105.5.1096. [DOI] [PubMed] [Google Scholar]
- Cronise K, Marino MD, Tran TD, Kelly SJ. Critical periods for the effects of alcohol exposure on learning in rats. Behav. Neurosci. 2001;115:138–145. doi: 10.1037/0735-7044.115.1.138. [DOI] [PubMed] [Google Scholar]
- Downing C, Johnson TE, Larson C, Leakey TI, Siegfried RN, Rafferty TM, Cooney CA. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: effects of a methyl-supplemented diet. Alcohol. 2011;45:65–71. doi: 10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol. Clin. Exp. Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neurosci. Biobehav. Rev. 2010;34:882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AM, Delis DC, Mattson SN. Normative data for 4-year-old children on the California Verbal Learning Test-Children’s Version. The Clinical neuropsychologist. 1999;13:274–282. doi: 10.1076/clin.13.3.274.1748. [DOI] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol. Reprod. 2009;81:618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J. Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Bittigau P, Ishimaru MJ, et al. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science. 2000;287:1056–1060. doi: 10.1126/science.287.5455.1056. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Kitabayashi R, Funatsu M, Izumi M, Yuede C, Hartman RE, Wozniak DF, Zorumski CF. A single day of ethanol exposure during development has persistent effects on bi-directional plasticity, N-methyl-D-aspartate receptor function and ethanol sensitivity. Neuroscience. 2005;136:269–279. doi: 10.1016/j.neuroscience.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J. Biol. Chem. 2005;280:857–860. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Flatscher-Bader T, Wilkins SJ, Anderson GJ, Whitelaw E, Chong S. Postnatal growth restriction and gene expression changes in a mouse model of fetal alcohol syndrome. Birth Defects Res A Clin Mol Teratol. 2010a;88:818–826. doi: 10.1002/bdra.20729. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Whitelaw E, Chong S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010b;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Histone h3 modifications in rat hepatic stellate cells by ethanol. Alcohol Alcohol. 2005;40:367–372. doi: 10.1093/alcalc/agh170. [DOI] [PubMed] [Google Scholar]
- Kokubo M, Nishio M, Ribar TJ, Anderson KA, West AE, Means AR. BDNF-mediated cerebellar granule cell development is impaired in mice null for CaMKK2 or CaMKIV. J. Neurosci. 2009;29:8901–8913. doi: 10.1523/JNEUROSCI.0040-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagali PS, Picketts DJ. Matters of life and death: the role of chromatin remodeling proteins in retinal neuron survival. Journal of ocular biology, diseases, and informatics. 2011;4:111–120. doi: 10.1007/s12177-012-9080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanore F, Blanchet C, Fejtova A, Pinheiro P, Richter K, Balschun D, Gundelfinger E, Mulle C. Impaired development of hippocampal mossy fibre synapses in mouse mutants for the presynaptic scaffold protein Bassoon. The Journal of physiology. 2010;588:2133–2145. doi: 10.1113/jphysiol.2009.184929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Mattson SN, Riley EP, et al. A longitudinal study of the long-term consequences of drinking during pregnancy: heavy in utero alcohol exposure disrupts the normal processes of brain development. J. Neurosci. 2012;32:15243–15251. doi: 10.1523/JNEUROSCI.1161-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell. Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics: official journal of the DNA Methylation Society. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist F. The determination of ethyl alcohol in blood and tissue. Meth Biochem Analy. 1959;7:217–251. [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonski K, Abuhatzira L, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum. Mol. Genet. 2005;14:1049–1058. doi: 10.1093/hmg/ddi097. [DOI] [PubMed] [Google Scholar]
- Marchal C, Mulle C. Postnatal maturation of mossy fibre excitatory transmission in mouse CA3 pyramidal cells: a potential role for kainate receptors. The Journal of physiology. 2004;561:27–37. doi: 10.1113/jphysiol.2004.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev. Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- Mason S, Anthony B, Lai X, Ringham HN, Wang M, Witzmann FA, You JS, Zhou FC. Ethanol exposure alters protein expression in a mouse model of fetal alcohol spectrum disorders. International journal of proteomics. 2012;2012:867141. doi: 10.1155/2012/867141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Yasutomi M. Inhibition of cephalic neural tube closure by 5-azacytidine in neurulating rat embryos in vitro. Anat. Embryol. (Berl) 1992;185:217–223. doi: 10.1007/BF00211820. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Crocker N, Nguyen TT. Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychol. Rev. 2011;21:81–101. doi: 10.1007/s11065-011-9167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson SN, Goodman AM, Caine C, Delis DC, Riley EP. Executive functioning in children with heavy prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 1999;23:1808–1815. [PubMed] [Google Scholar]
- Mattson SN, Riley EP. A review of the neurobehavioral deficits in children with fetal alcohol syndrome or prenatal exposure to alcohol. Alcohol. Clin. Exp. Res. 1998;22:279–294. doi: 10.1111/j.1530-0277.1998.tb03651.x. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Riley EP, Gramling L, Delis DC, Jones KL. Neuropsychological comparison of alcohol-exposed children with or without physical features of fetal alcohol syndrome. Neuropsychology. 1998;12:146–153. doi: 10.1037//0894-4105.12.1.146. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Riley EP, Sowell ER, Jernigan TL, Sobel DF, Jones KL. A decrease in the size of the basal ganglia in children with fetal alcohol syndrome. Alcohol. Clin. Exp. Res. 1996;20:1088–1093. doi: 10.1111/j.1530-0277.1996.tb01951.x. [DOI] [PubMed] [Google Scholar]
- May PA, Gossage JP, Kalberg WO, Robinson LK, Buckley D, Manning M, Hoyme HE. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Developmental disabilities research reviews. 2009;15:176–192. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol. Psychiatry. 2013;73:763–773. doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P, Rezzoug F, Kaikaus J, Greene RM, Pisano MM. Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod. Toxicol. 2013;37:40–48. doi: 10.1016/j.reprotox.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel M, Norris EH, Strickland S. Tissue plasminogen activator is required for the development of fetal alcohol syndrome in mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5069–5074. doi: 10.1073/pnas.1017608108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AL, O’Brien JW, Spadoni AD, Tapert SF, Jones KL, Riley EP, Mattson SN. A functional magnetic resonance imaging study of spatial working memory in children with prenatal alcohol exposure: contribution of familial history of alcohol use disorders. Alcohol. Clin. Exp. Res. 2013;37:132–140. doi: 10.1111/j.1530-0277.2012.01880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney JW, Tenkova T, Dikranian K, Qin YQ, Labruyere J, Ikonomidou C. Ethanol-induced apoptotic neurodegeneration in the developing C57BL/6 mouse brain. Brain Res. Dev. Brain Res. 2002;133:115–126. doi: 10.1016/s0165-3806(02)00279-1. [DOI] [PubMed] [Google Scholar]
- Otero NK, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol. Clin. Exp. Res. 2012;36:1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81:979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. American journal of physiology. Gastrointestinal and liver physiology. 2005;289:G1124–1136. doi: 10.1152/ajpgi.00091.2005. [DOI] [PubMed] [Google Scholar]
- Petronis A. Epigenetics and bipolar disorder: new opportunities and challenges. Am J Med Genet C Semin Med Genet. 2003;123C:65–75. doi: 10.1002/ajmg.c.20015. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Renolleau S, Fau S, Goyenvalle C, Joly LM, Chauvier D, Jacotot E, Mariani J, Charriaut-Marlangue C. Specific caspase inhibitor Q-VD-OPh prevents neonatal stroke in P7 rat: a role for gender. J. Neurochem. 2007;100:1062–1071. doi: 10.1111/j.1471-4159.2006.04269.x. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, Smith KM, Ferrante RJ. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadrian B, Subbanna S, Wilson DA, Basavarajappa BS, Saito M. Lithium prevents long-term neural and behavioral pathology induced by early alcohol exposure. Neuroscience. 2012;206:122–135. doi: 10.1016/j.neuroscience.2011.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Chakraborty G, Hegde M, Ohsie J, Paik SM, Vadasz C. Involvement of ceramide in ethanol-induced apoptotic neurodegeneration in the neonatal mouse brain. J. Neurochem. 2010;115:168–177. doi: 10.1111/j.1471-4159.2010.06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RK, Stringfellow SA, Glover ME, Wagle AA, Clinton SM. DNA methylation markers in the postnatal developing rat brain. Brain Res. 2013 doi: 10.1016/j.brainres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Basavarajappa BS. Pre-administration of G9a/GLP inhibitor during Synaptogenesis Prevents Postnatal Ethanol-induced LTP Deficits and Neurobehavioral Abnormalities in Adult Mice. Exp. Neurol. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Nagaraja NN, Umapathy NS, Pace BS, Basavarajappa BS. Ethanol Exposure Induces Neonatal Neurodegeneration by Enhancing CB1R Exon1 Histone H4K8 Acetylation and Up-regulating CB1R Function causing Neurobehavioral Abnormalities in Adult Mice. International Journal of Neuropsychopharmacology. 2014a doi: 10.1093/ijnp/pyu028. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS. Ethanol Induced Acetylation of Histone at G9a Exon1 and G9a-Mediated Histone H3 Dimethylation leads to Neurodegeneration in Neonatal Mice. Neuroscience. 2014b;258C:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Psychoyos D, Xie S, Basavarajappa BS. Anandamide-CB1 Receptor Signaling Contributes to Postnatal Ethanol-Induced Neonatal Neurodegeneration, Adult Synaptic and Memory Deficits. Journal of neuoscience. 2013a;33:6350–6366. doi: 10.1523/JNEUROSCI.3786-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Umapathy NS, et al. G9a-Mediated Histone Methylation Regulates Ethanol-Induced Neurodegeneration in the Neonatal Mouse Brain. Neurobiol. Dis. 2013b;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake I, Mishima Y, Kimura H, Lee YH, Goto Y, Takeshima H, Ikegami T, Tajima S. Characterization of DNA-binding activity in the N-terminal domain of the DNA methyltransferase Dnmt3a. Biochem. J. 2011;437:141–148. doi: 10.1042/BJ20110241. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhao H, Xu Z, et al. Phosphatidylinositol 3-kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell. Signal. 2007;19:2255–2263. doi: 10.1016/j.cellsig.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews. Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Suetake I, Shimahara H, Ura K, Tate S, Tajima S. Distinct DNA methylation activity of Dnmt3a and Dnmt3b towards naked and nucleosomal DNA. J Biochem. 2006;139:503–515. doi: 10.1093/jb/mvj044. [DOI] [PubMed] [Google Scholar]
- Tran TD, Cronise K, Marino MD, Jenkins WJ, Kelly SJ. Critical periods for the effects of alcohol exposure on brain weight, body weight, activity and investigation. Behav. Brain Res. 2000;116:99–110. doi: 10.1016/s0166-4328(00)00263-1. [DOI] [PubMed] [Google Scholar]
- Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Vann SD, Albasser MM. Hippocampus and neocortex: recognition and spatial memory. Curr. Opin. Neurobiol. 2011;21:440–445. doi: 10.1016/j.conb.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Veazey KJ, Carnahan MN, Muller D, Miranda RC, Golding MC. Alcohol-induced epigenetic alterations to developmentally crucial genes regulating neural stemness and differentiation. Alcohol. Clin. Exp. Res. 2013;37:1111–1122. doi: 10.1111/acer.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Warren ST. The Epigenetics of Fragile X Syndrome. Cell Stem Cell. 2007;1:488–489. doi: 10.1016/j.stem.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wilson DA, Peterson J, Basavaraj BS, Saito M. Local and regional network function in behaviorally relevant cortical circuits of adult mice following postnatal alcohol exposure. Alcoholism Clin and Exp Res. 2011;35:1974–1984. doi: 10.1111/j.1530-0277.2011.01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Zhao Q, Liu Y, Goodlett C, Liang T, McClintick J, Edenberg H, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011a;12:124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol. Clin. Exp. Res. 2011b;35:735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Chen Y, Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Res A Clin Mol Teratol. 2011c;91:703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]