Abstract
Respiratory disease and diarrhea are the 2 most common diseases that result in the use of antimicrobial drugs in preweaned calves. Because the use of drugs in food animals, including dairy calves, has the potential for generating cross-resistance to drugs used in human medicine, it is vital to propose farm practices that foster the judicious use of antimicrobials while assuring animal health and productivity. The objective of this study was to use dairy farm calf treatment records to identify antimicrobial drug treatments in calves and to evaluate their effects on the prevalence of antimicrobial-resistant Escherichia coli from rectal swabs of preweaned dairy calves. Eight farms from central New York participated in the study, 3 farms using individual pen housing management and 5 farms using group pen housing management. Eligible study farms could not add antimicrobial drugs to the milk fed to preweaned calves and were required to have farm records documenting antimicrobial drug treatment of calves from birth to weaning. Three fecal E. coli isolates per calf were tested for susceptibility to 12 antimicrobial drugs using a Kirby-Bauer disk diffusion assay. A total of 473 calves were sampled, from which 1,423 commensal E. coli isolates were tested. Of the 9 antimicrobial drugs used on study farms, only enrofloxacin was significantly associated with reduced antimicrobial susceptibility of E. coli isolates, although treatment with ceftiofur was associated with reduced susceptibility to ceftriaxone. The median numbers of days from treatment with ceftiofur and enrofloxacin to rectal swab sampling of calves were 16 d (range: 1–39) and 12 d (range: 6–44), respectively. At the isolate level, treatment with enrofloxacin resulted in odds ratios of 2 [95% confidence interval (CI): 1–4] and 3 (95% CI: 2–6), respectively, for isolation of nonsusceptible E. coli to nalidixic acid and ciprofloxacin compared with calves not treated with enrofloxacin. Treatment with ceftiofur resulted in an odds ratio of 3 (95% CI: 0.9–12) for isolation of nonsusceptible E. coli to ceftriaxone compared with calves not treated with ceftiofur. Treatment with enrofloxacin resulted in selection of isolates that presented phenotypic resistance to both ciprofloxacin and ceftriaxone. Treatment with ceftiofur resulted in a higher prevalence of isolates resistant to ≥3 antimicrobial drugs (97%) compared with no treatment with ceftiofur (73%). These findings reinforce the necessity for continued implementation of practices at the dairy farm that support the sustainable and judicious use of antimicrobial drugs in dairy calves.
Keywords: antimicrobial use, antimicrobial resistance, enrofloxacin, farm records
INTRODUCTION
The most common disease in calves that results in the use of antimicrobial drugs is diarrhea, followed by pneumonia (USDA, 2008). According to the last National Animal Health Monitoring Survey in 2007, the most common drugs used to treat diarrhea belonged to the tetracycline (16%) and β-lactam (9%) classes (USDA, 2008). In the same report, the 2 antimicrobial drugs most commonly used for treatment of respiratory disease were florfenicol (18%) and drugs belonging to the macrolide class (15%). In addition to these drugs, in 2008 enrofloxacin (a fluoroquinolone) was approved for use in nonlactating dairy cattle less than 20 mo of age for the treatment of bovine respiratory disease associated with Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis (FDA, 2008).
Regardless of the benefits of using antimicrobial agents in food-producing animals, concerns from public health, food safety, and regulatory perspectives arise from the potential for development of antimicrobial resistance (Oliver et al., 2011). Because of the paradoxical decrease in the approval of new antimicrobials by the US Food and Drug Administration concomitant with the rise of antimicrobial resistance, there is an urgent need for the sustainable and judicious use of current antimicrobial drugs to extend their effectiveness to treat disease in animals and humans (Spellberg et al., 2011). Approaches that have been shown to be effective focus on farm practices that attempt to limit food animal morbidity and mortality while reducing the use of antimicrobial drugs. Examples include targeted therapy of animals with bovine respiratory disease and diarrhea through the use of appropriate criteria for early and accurate diagnosis (McGuirk, 2008). Such screening programs can allow identification of up to 85% of active disease cases and contribute to reduced morbidity and mortality (McGuirk, 2008). A study by Berge et al. (2009) observed that calves in a conventional therapy group treated as per dairy protocol with sulfamethoxazole-trimethoprim, spectinomycin, penicillin, and bismuth-pectin for diarrhea had 70% more days with diarrhea than calves in the targeted therapy group treated with bismuth-pectin for diarrhea and antimicrobial treatment only in cases of fever or depressed attitude. In addition to monitoring tools, the use of preventive-measures such as adequate feeding of colostrum to calves to prevent failure of passive transfer have been shown to result in lower occurrence of disease, and therefore decreased use of antimicrobial drugs in preweaned calves (Windeyer et al., 2014).
On-farm monitoring of selection of antimicrobial drug resistance is vital to propose alternatives to practices that result in resistance to antimicrobials important in human and animal medicine. The objective of this study was to use dairy farm records to identify and evaluate the effects of antimicrobial drug use on the prevalence of antimicrobial resistant Escherichia coli from rectal swabs of preweaned dairy calves.
MATERIALS AND METHODS
Inclusion Criteria for Farms
The data used for this study was part of a project evaluating calf housing management on antimicrobial drug resistance. Study herds were selected from a convenience sample of commercial dairy farms within a 265-km (165-mile) radius of Cornell University (Ithaca, NY). Farms housed calves in 1 of 2 housing systems: 1) in individual pens, feeding calves milk or milk replacer (IP); or 2) in group pens, feeding calves acidified milk ad libitum (GP). Farms could not add any antimicrobial drugs to the milk fed to preweaned calves. Farms participating in the study used antimicrobial drugs by parenteral route for treatment, control, or prevention of disease in calves. To participate in the current study, eligible farms were required to have antimicrobial drug treatment records of individual calves from birth to weaning for at least 6 mo before the farm visit. Five study farms housed calves using GP and 3 study farms housed calves using IP. A total of 479 preweaned dairy calves were enrolled from 8 farms in central NY (Table 1). Dairy farms were located in the following counties in central New York State: Cayuga, Cortland, Ontario, Tompkins, and Wayne. Farmers were requested to answer a short questionnaire on their calf management practices.
Table 1.
Descriptive data for the total number of calves, fecal Escherichia coli isolates, and average age of calves in the study by calf housing management and age group
| Description | Calves, no. | E. coli isolates,1 no. | Mean age, d (range) |
|---|---|---|---|
| IP2 | |||
| AGE-13 | 37 | 132 | 10 (3–14) |
| AGE-2 | 89 | 217 | 25 (15–34) |
| AGE-3 | 38 | 137 | 49 (37–65) |
| Total | 164 | 486 | |
| GP4 | |||
| AGE-1 | 84 | 285 | 8 (1–14) |
| AGE-2 | 120 | 321 | 25 (15–35) |
| AGE-3 | 111 | 331 | 43 (37–58) |
| Total | 315 | 937 | |
| Overall total | 479 | 1,423 | |
E. coli isolates from rectal swabs.
Calves housed in individual pens without direct contact with neighboring calves and fed milk or milk replacer 2 to 3 times a day.
Age groups refer to calves with ages AGE-1 = 0 to 14 d; AGE-2 = 15 to 35 d; AGE-3 = 36 to 65 d.
Calves housed in group pens with 3 to 25 calves and fed free choice acidified milk.
Study Design and Sample Collection
This study used a cross-sectional design in which rectal swabs were collected from preweaned dairy calves during a single farm visit. At each farm preweaned calves were divided into 3 age groups, and approximately the same numbers of total samples were collected from each age group using a randomization spreadsheet. The randomization spreadsheet was created using the random number function of Excel (Microsoft Corp., Redmond, WA). The age groups were (1) 3 to 14 d of age (AGE-1); (2) 15 to 35 d of age (AGE-2); and (3) 36 to 65 d of age (AGE-3). All calves enrolled in the study were female. Farm sampling was conducted from July to October 2012.
Bacterial Isolation, Culture, and Identification
Transport swabs with Amies medium were used to collect rectal swabs from calves (Becton Dickinson and Company, Franklin Lakes, NJ). Individual fecal swabs were streaked onto MacConkey agar plates on the day of collection and incubated overnight at 37°C. Individual fecal swabs were streaked onto MacConkey agar plates on the day of collection and incubated overnight at 37°C. Up to 3 typical lactose-positive E. coli colonies were picked and stored in Luria-Bertani broth containing 20% glycerol at −80°C (Pereira et al., 2011). Biochemical testing was performed on 20% of the colonies using sulfur-indole-motility medium tubes (Northeast Laboratories, Waterville, ME), confirming E. coli characteristics by the observation of indole production, motility, and no H2S production (Berge et al., 2003).
Antimicrobial susceptibility of E. coli isolates was tested against a modified National Antimicrobial Resistance Monitoring system panel of 12 antimicrobial drugs. The susceptibility testing was done using a Kirby-Bauer disk diffusion agar assay in accordance with the guidelines published by the Clinical and Laboratory Standards Institute (CLSI) and methodology previously described (CLSI, 2008; Hoelzer et al., 2011; Pereira et al., 2011). Internal quality control was performed by inclusion of E. coli ATCC 25922, which was previously determined to be pansusceptible, and a previously characterized in-house E. coli isolate known to have a blaCMY-2 gene and to be resistant to 9 of the antimicrobials tested. Antimicrobial susceptibility for all isolates was assessed using the following panel: ampicillin, 10 μg; cefoxitin, 30 μg; ceftiofur, 30 μg; ceftriaxone, 30 μg; chloramphenicol, 30 μg; ciprofloxacin, 5 μg; gentamicin, 10 μg; nalidixic acid, 30 μg; neomycin, 30 μg; streptomycin, 10 μg; tetracycline, 30 μg; and trimethoprim-sulfamethoxazole, 23.75/1.25 μg. Susceptibility of the isolates to most antimicrobial drugs was categorized (susceptible, intermediate, or resistant) by measuring the inhibition zone according to interpretive criteria adhering to the CLSI guidelines (CLSI, 2008).
Normalized Resistance Interpretation
Inhibition zone diameter histograms for each antimicrobial tested were visually examined for detection of drugs that had an unclear transition between isolates devoid of detectable resistance phenotypes and resistant isolates. For antimicrobial drug histograms where the clinical breakpoints values did not appear consistent with the frequency distribution, epidemiological cut-off values (ECV) were used in place of the CLSI clinical breakpoints to classify isolates as susceptible or non-susceptible (Kronvall, 2003). The ECV are determined on the basis of the distribution of the inhibition zone diameter, where the population that departs from the distribution of the susceptible population is categorized as nonsusceptible (Silley, 2012). The ECV can be used as the most sensitive measure to categorize isolates as susceptible and nonsusceptible and measure the development of resistance (Simjee et al., 2008).
The ECV were calculated using the normalized resistance interpretation (NRI) analysis. The NRI analysis was used with permission from the patent holder, Bioscand AB, Täby, Sweden (European patent number 1383913, US Patent number 7,465,559). An advantage of using the NRI analysis is that it provides a formal method of overcoming situations in which the distribution of zones obtained for nonsusceptible strains manifested low-level resistance (Smith et al., 2012). The NRI uses the fact that zone diameter distributions of the susceptible isolates are Gaussian in shape and that the upper half for zones (the susceptible side of the peaks) should be unaffected by the occurrence of resistance, and might therefore be used for reconstruction of the whole susceptible isolates peak of the distribution (Kronvall et al., 2011). This is achieved by localizing the peak using moving averages, then calculating the fraction of isolates from the highly susceptible side (Kronvall, 2003). After probit transformation of these values, the constants of the straight line between probit values and zone diameters are then calculated using the equation of least-squares and the mean and standard deviation of the ideal normal distribution can be determined. To define the optimal parameter settings for the reconstruction of the real standard distribution of susceptible isolates, a 4-zone moving average with a peak adjustment of 2.5 standard deviation limits from the switch position for the moving average to the proper position of the peak was used (Joneberg et al., 2003).
Statistical Analyses
Escherichia coli isolates were categorized as susceptible or nonsusceptible (intermediate or resistant) in accordance with the interpretive methods and breakpoints previously described. This binomial categorical variable was selected for use in the analysis (instead of the 3-category CLSI classification) for 2 main reasons: to focus the analysis on the presence or absence of isolates that are fully susceptible to the antimicrobial drugs tested, and to reduce erroneous analysis of the data caused by a high number of isolates classified as intermediate for 2 antimicrobial drugs (ceftiofur and ceftriaxone) for which the CLSI breakpoints may not correlate well to acquisition of resistant genes. The records for the dates of antimicrobial drug treatments of sampled preweaned dairy calves were obtained from the farm records and arranged by order of antimicrobial drug treatment after birth using PROC FREQ in SAS (SAS Institute Inc., Cary, NC; Table 2). Additionally, the median, minimum, and maximum age in days of calves when treated with antimicrobial drugs for the first, second, and third time after birth were estimated using PROC MEANS in SAS (Table 2).
Table 2.
Distribution of antimicrobial drug use [% (no.)] of individual calves as documented in farm records and displayed by calf housing type and order of treatment after birth
| Description | IP1
|
GP2
|
||||
|---|---|---|---|---|---|---|
| First treatment | Second treatment | Third treatment | First treatment | Second treatment | Third treatment | |
| Any drug3 | 38 (63) | 18 (31) | 9 (14) | 16 (55) | 6 (19) | 1 (3) |
| Distribution of antimicrobial drug use by treatment order: | ||||||
| β-Lactam, | 79 (50) | 35 (11) | 7 (1) | 12 (7) | ||
| Penicillin G | 17 (11) | 6 (2) | ||||
| Ampicillin | 12 (7) | |||||
| Ceftiofur | 62 (39) | 29 (9) | 7 (1) | |||
| Fluoroquinolones | 3 (2) | 6 (2) | 21 (3) | 49 (27) | 47 (9) | 66 (2) |
| Enrofloxacin | 3 (2) | 6 (2) | 21 (3) | 49 (27) | 47 (9) | 66 (2) |
| Macrolides | 11 (7) | 58 (18) | 43 (6) | 23 (13) | 53 (10) | |
| Tulathromycin | 11 (7) | 55 (17) | 36 (5) | 23 (13) | 53 (10) | |
| Tylosin | 3 (1) | 7 (1) | ||||
| Phenicols | 9 (6) | 44 (1) | ||||
| Florfenicol | 9 (6) | 44 (1) | ||||
| Tetracyclines | 2 (1) | |||||
| Oxytetracycline | 2 (1) | |||||
| Sulfonamide | 30 (4) | |||||
| Sulfamethazine | 30 (4) | |||||
| Median age at treatment, d (range) | 9 (0–57) | 12 (6–46) | 19 (7–46) | 20 (3–37) | 29 (15–49) | 37 (32–43) |
Calves housed in individual pens, fed milk or milk replacer (n = 164).
Calves housed in group pens, fed acidified milk ad libitum (n = 315).
Percent and number of calves receiving any antimicrobial drug treatment.
A multinomial logistic regression was used to determine the calf level effects of antimicrobial drug treatment on the number of nonsusceptible fecal E. coli cultured from each calf. The model was developed using the generalized logit-link function (glogit) in PROC GLIMMIX in SAS. The outcome variable was the number of nonsusceptible fecal E. coli cultured from a calf for an individual antimicrobial drug (range from 0 to 3 isolates). The model was controlled for calf housing type and age group as fixed effects, and herd as a random effect. The independent binomial variables for farm records showing at least one treatment with the referred antimicrobial drug was offered to the model. All possible two-way interactions between the independent variables were added to the model. Variables and interactions were manually excluded from the model if their P-value was >0.05.
The isolate level association between identification of nonsusceptible E. coli and antimicrobial drug treatment of a calf was estimated in a 2-step analysis. First, a bivariate analysis using a chi-squared test in PROC FREQ in SAS was used to determine if identification of a nonsusceptible E. coli was associated with the binomial variable for antimicrobial drug treatment of a calf (treatment at least once with the referred antimicrobial drug or no treatment). Second, antimicrobial drug treatment variables with a P-value <0.05 for the chi-squared tests were added as independent variables to logistic regression models using PROC GLIMMIX in SAS. The models were controlled for calf housing type and age group as fixed effects, and herd as a random effect. All possible two-way interactions between the independent variables were added to the model. Variables and interactions were manually excluded from the model if their P-values was >0.05.
Descriptive analysis of E. coli resistance phenotypes was conducted using PROC FREQ in SAS. CLSI clinical breakpoint values were used for all drugs tested to categorize an isolate as resistant and determine the resistance phenotypes. For all statistical models and tests, variables were considered significant when a P-value <0.05 was observed.
RESULTS
Descriptive Data
Two GP farms and 1 IP farm kept calf antimicrobial treatment records using a notebook; 3 GP farms and 2 IP farms kept records using a computer program. The number of lactating cows at farms ranged from 100 to 199 cows for 1 GP farm, 500 to 999 cows for 2 GP farms and 1 IP farm, 1,000 to 3,000 cows for 1 GP farm and 2 IP farms, and >3,000 cows for 1 GP farm. The total number of calves and E. coli isolates and the average age of calves in the study by calf housing management are shown in Table 1.
Normalized Resistance Interpretation
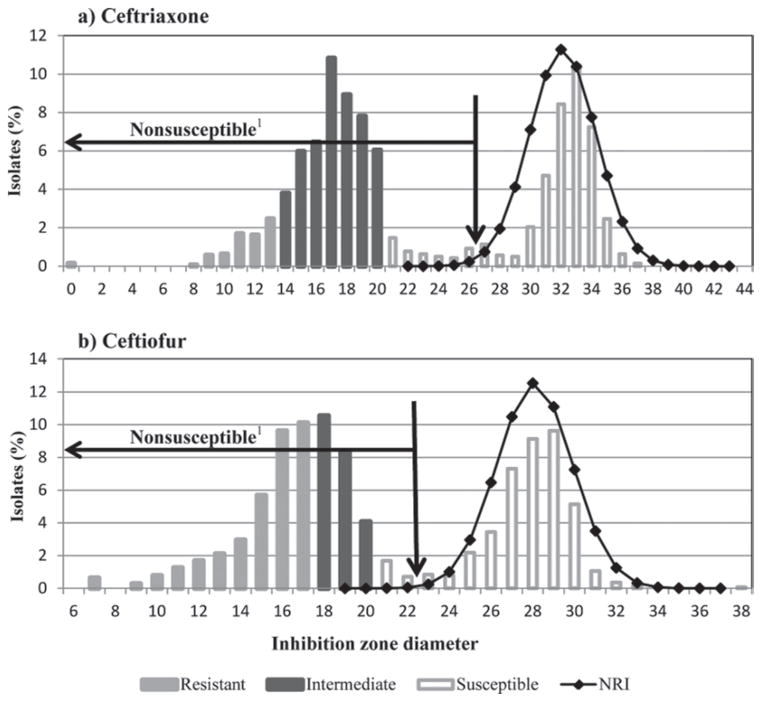
Visual examination of histograms of antimicrobial drugs zone diameter sizes revealed that for ceftiofur and ceftriaxone, the distribution based on clinical breakpoints values did not appear consistent with the frequency distribution (Figure 1). The reconstructed ideal population of susceptible strains using the NRI method identified a mean of 27.6 for ceftiofur and 31.7 for ceftriaxone. Using a 2.5 standard deviation limit below the mean, the new breakpoints for susceptible isolates using these parameters were defined as 23 mm for ceftiofur and 26.5 mm for ceftriaxone.
Figure 1.
Zone diameter histogram with susceptibility classification of Escherichia coli according to Clinical and Laboratory Standards Institute clinical breakpoints. Reconstructed ideal population of susceptible strains shown as a line graph of normalized resistance interpretation (NRI) with a mean of 27.6 for ceftiofur and 31.7 for ceftriaxone. The 2.5 deviation limit below the NRI mean is marked with a vertical arrow (23 for ceftiofur and 26.5 for ceftriaxone) and the isolates below this limit can be considered nonsusceptible for comparative purposes.
Distribution of Antimicrobial Treatment of Preweaned Calves
The on-farm recorded use of antimicrobial drugs in calves is shown in Table 2. A total of 9 antimicrobial drugs belonging to 6 antimicrobial drug classes were used on farms sampled. The most common antimicrobial class used in calves housed in IP was β-lactams, with ceftiofur being the most common drug, whereas for calves housed in GP, it was fluoroquinolones with enrofloxacin being the most common drug. The median number of days from treatment with ceftiofur to rectal swab sampling of calves was 16 d (range: 1–39 d), and the median number of days from treatment with enrofloxacin to rectal swab sampling of calves was 12 d (range: 6–44 d).
Antimicrobial Resistance in E. coli Isolates
Only treatment with enrofloxacin was significantly associated with the number of nonsusceptible fecal E. coli cultured from a calf. Treatment of a calf at least once with enrofloxacin was associated with an increase in the number of fecal E. coli nonsusceptible to ciprofloxacin (P < 0.0001) and nalidixic acid (P < 0.0001; Table 3). Age group and calf housing used for calves were not significantly associated with resistance to ciprofloxacin and nalidixic acid.
Table 3.
Calf-level effect of antimicrobial drug treatment on the number of nonsusceptible fecal Escherichia coli cultured from a calf
| Antimicrobial drug treatment | Nonsusceptible E. coli, no. | Enrofloxacin treatment1
|
P-value | Odds ratio2 | |
|---|---|---|---|---|---|
| Tx, % (n = 42) | NoTx, % (n = 437) | ||||
| Nalidixic acid | 0 | 55 | 94 | <0.0001 | Reference |
| 1 | 2 | 4 | |||
| 2 | 5 | 1 | |||
| 3 | 38 | 1 | 41.0 (10—172) | ||
| Ciprofloxacin | 0 | 62 | 96 | <0.0001 | Reference |
| 1 | 3 | 2 | |||
| 2 | 4 | 1 | |||
| 3 | 31 | 0.5 | 92.0 (14—613) | ||
Calves with farm records with at least one treatment with enrofloxacin from birth to fecal sampling (Tx) or calves with a farm record with no treatment with enrofloxacin from birth to fecal sampling.
Odds ratio in case of 3 nonsusceptible isolates from a calf compared with 0 nonsusceptible E. coli isolates (reference) when a calf received at least one antimicrobial treatment of enrofloxacin. The 95% confidence interval is in parentheses.
The effect of antimicrobial treatment of calves with enrofloxacin was significantly associated with the non-susceptibility of E. coli to ciprofloxacin and nalidixic acid. Treatment of a calf at least once with enrofloxacin was associated with an increase in the odds of isolates nonsusceptible to ciprofloxacin (P = 0.003) and nalidixic acid (P < 0.0001; Table 4). Treatment of a calf at least once with ceftiofur resulted in a 0.5 odds ratio (P = 0.05; 95% CI: 0.3–1) of identification of isolates non-susceptible to neomycin, and a 3 odds ratio (P = 0.08; 95% CI: 0.9–12) of identification of isolates nonsusceptible to ceftriaxone). For all models where antimicrobial drug treatment had a P-value <0.1, calf age group was associated with the nonsusceptibility of E. coli to ceftriaxone (P = 0.0002), neomycin (P < 0.0001), ciprofloxacin (P = 0.0003), and nalidixic acid (P < 0.0001). The highest prevalence of nonsusceptible isolates for ceftriaxone (CRO = 865 isolates) and neomycin (NEO = 464 isolates) was observed at AGE-2 (CRO = 39%, NEO = 39%) compared with AGE-1 (CRO = 27%, NEO = 32%) or AGE-3 (CRO = 34%, NEO = 28%). The highest prevalence of nonsusceptible isolates for ciprofloxacin (CIP = 70 isolates) and nalidixic (NAL = 91 isolates) was at AGE-3 (CIP = 59%, NAL = 59%) compared with AGE-1 (CIP = 3%, NAL = 3%) or AGE-2 (CIP = 39%, NAL = 37%). No interaction between age group and record of antimicrobial drug treatment was significant. Type of calf housing was not significantly associated with resistance to ceftriaxone, neomycin, ciprofloxacin, or nalidixic acid. Models for which at least one drug treatment variable had a P-value <0.10 are shown in Table 4.
Table 4.
Isolate level effect of antimicrobial treatment of calves on prevalence of nonsusceptible Escherichia coli
| Antimicrobial treatment | Age1
|
Class2 | Enrofloxacin treatment3
|
Ceftiofur treatment3
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | Tx, % (n = 152) | NoTx, % (n = 1,271) | P-value | Odds ratio4 | Tx, % (n = 144) | NoTx, % (n = 1,279) | P-value | Odds ratio4 | ||
| Ceftriaxone | 0.0002 | S | 34 | 40 | 0.3 | 3 | 43 | 0.08 | ||
| NS | 66 | 60 | 0.8 (0.5–1) | 97 | 57 | 3 (0.9–12) | ||||
| Neomycin | <0.0001 | S | 65 | 68 | 0.7 | 62 | 68 | 0.05 | ||
| NS | 35 | 32 | 0.9 (0.6–1) | 38 | 32 | 0.5 (0.3–1) | ||||
| Ciprofloxacin | 0.0003 | S | 80 | 97 | 0.005 | 98 | 95 | 0.31 | ||
| NS | 20 | 3 | 2.0 (1–4) | 2 | 5 | 4 (0.3–58) | ||||
| Nalidixic acid | <0.0001 | S | 73 | 96 | <0.0001 | 94 | 94 | 0.08 | ||
| NS | 27 | 4 | 3.0 (2–6) | 6 | 6 | 6 (0.8–42) | ||||
Categorical age group variable with animals grouped by age in days: 3 to 14 d (AGE-1), 15 to 35 d (AGE-2), and 36 to 65 d (AGE-3).
S = susceptible; NS = nonsusceptible.
E. coli from a calf with a farm record of treatment at least once with the referred antimicrobial drug from birth to rectal swab sampling (Tx) or from a calf with a farm record showing no treatment with the referred antimicrobial drug from birth to rectal swab sampling (NoTx).
Odds ratio for isolation of a nonsusceptible E. coli in the drug-treated group compared with the not-treated group. The 95% confidence interval is in parentheses.
Distribution of E. coli Antimicrobial Resistance Phenotypes
Of the 152 E. coli isolates from calves treated with enrofloxacin, 15.8% were pansusceptible and 77% were resistant to 3 or more antimicrobials. The most common resistance phenotype from enrofloxacin-treated calves was ampicillin-cefoxitin-chloramphenicol-streptomycin-tetracycline, which was present in 7.9% of isolates (Table 5).
Table 5.
Ranking of the most common drug-resistant phenotypes among 1,423 Escherichia coli by farm record treatment (Tx) with enrofloxacin or ceftiofur
| Drug-resistant phenotypes1 | TX rank2 | NoTX rank2 | Tx, % (no.) | NoTx, % (no.) |
|---|---|---|---|---|
| Enrofloxacin (Tx, n = 152; NoTx, n = 1,238) | ||||
| AMP-FOX-CHL-STR-TET | 1 | 3 | 7.9 (12) | 4.4 (56) |
| AMP-TIO-CRO-CIP-NA-TET | 2 | 23 | 5.3 (8) | 1.4 (18) |
| AMP-FOX-NEO-STR-TET | 3 | 4 | 4.6 (7) | 4.2 (54) |
| CHL-STR-TET | 27 | 1 | 0.7 (2) | 5.7 (72) |
| TET | 43 | 2 | 0.7 (1) | 5.1 (65) |
| Pansusceptible | 15.8 (24) | 11.8 (150) | ||
| Ceftiofur (Tx, n = 144; NoTx, n = 1,279) | ||||
| AMP-FOX-TIO-CHL-NEO-STR-TET-COT | 1 | 23 | 14.5 (21) | 1.4 (18) |
| AMP-FOX-CHL-GEN-STR-TET | 2 | 24 | 13.2 (19) | 1.1 (15) |
| AMP-TIO-STR-TET-COT | 3 | 77 | 6.9 (10) | 0.2 (2) |
| CHL-STR-TET | None | 1 | 0 (0) | 5.8 (74) |
| TET | None | 2 | 0 (0) | 5.1 (66) |
| AMP-FOX-CHL-STR-TET | 6 | 3 | 4.1 (6) | 4.9 (62) |
| Pansusceptible | 0.3 (1) | 13.5 (173) | ||
AMP = ampicillin; FOX = cefoxitin; TIO = ceftiofur; CRO = ceftriaxone; CIP = ciprofloxacin; CHL = chloraphenicol; GEN = gentamicin; NA = nalidixic acid; NEO = neomycin; STR = streptomycin; COT = trimethoprim sulfamethoxazole; TET = tetracycline.
Ranking of drug-resistant phenotypes for isolates from a calf with a farm record of receiving at least one treatment with the referred drug (Tx) or from a calf with a farm record showing no treatment with the referred drug (NoTx).
Of the 144 E. coli isolates from calves with ceftiofur treatment, 0.3% were pansusceptible and 97.3% were resistant to 3 or more antimicrobials. The most common resistance phenotype from ceftiofur-treated calves was ampicillin-cefoxitin-ceftiofur-chloramphenicol-neomycin-streptomycin-tetracycline-trimethoprim sulfamethoxazole, which was present in 14.5% of isolates (Table 5).
DISCUSSION
Antimicrobial Resistance in E. coli Isolates
Treatment of a calf with enrofloxacin was significantly associated with a higher number of nonsusceptible fecal E. coli. Compared with calves not treated with enrofloxacin, treated animals had an odds ratio of 41 (P < 0.0001; 95% CI: 10–172) and 92 (P < 0.0001; 95% CI: 14–613) for having 3 versus 0 E. coli isolates nonsusceptible to nalidixic acid and ciprofloxacin, respectively (Table 3). Additionally, isolates from enrofloxacin-treated calves had a higher odds ratio for being nonsusceptible to fluoroquinolones (Table 4). These findings are a major concern because fluoroquinolones are regarded as critically important antimicrobial agents for human medicine according to the World Health Organization (Collignon et al., 2009). Similar findings were observed in a study conducted in swine receiving a daily intramuscular injection of enrofloxacin for 14 d at a dose of 5 mg/kg, where quinolone susceptibility of recolonizing fecal E. coli dropped dramatically compared with pre-treatment isolates (P < 0.0001; Beraud et al., 2008).
Resistance to quinolones can limit therapy options for humans with invasive disease caused by Salmonella species, Campylobacter species, and multidrug-resistant Shigella species (Collignon et al., 2009). The most prevalent mechanism for resistance to quinolones is target modification by mutation in topoisomerase DNA gyrase and topoisomerase IV, 2 enzymes that quinolones bind, thus causing DNA replication fork progression arrest which results in cell death (Jacoby, 2005). In gram-negative bacteria, high levels of quinolone resistance are mainly due to mutation of the genes encoding for the gyrase subunits gyrA and gyrB (mainly in gyrA), whereas mutations in parC and parE, which encode for the subunits of topoisomerase IV are secondary. Additionally, quinolone resistance has been linked with other mechanisms of resistance such as efflux pumps (Poole, 2000). In the vast majority of cases, resistance to the fluoroquinolone ciprofloxacin is always accompanied by resistance to the quinolone nalidixic acid, with a few exceptions reported (Cambau et al., 1993). One of the main ways fluoroquinolones differ from the original quinolone compound is the addition of a fluorine atom at position 6 of the quinolone molecule, which can provide fluoroquinolones with more than a 10-fold increase in gyrase inhibition and up to a 100-fold improvement in MIC (Andersson and MacGowan, 2003). In our study, all isolates resistant to ciprofloxacin were also resistant to nalidixic acid.
Enrofloxacin is the only quinolone drug approved for use in commercial dairy cattle in the United States. The dosage for enrofloxacin in cattle is 7.5 to 12.5 mg/kg as a single-dose therapy or 2.5 to 5 mg/kg as a multiple-day therapy repeating treatment every 24 h for 3 d. The extra-label use of fluoroquinolones in animals in the United States is strictly prohibited, as determined by the Animal Medicinal Drug Use Clarification Act (FDA, 2013). Enrofloxacin administered to cattle is partly metabolized to ciprofloxacin, achieving only 25 to 35% of the concentration of enrofloxacin in the blood (Kaartinen et al., 1995; McKellar et al., 1999). A study by Wiuff et al. (2002) conducted in pigs administered a single intramuscular dose of enrofloxacin at 2.5 mg/kg resulted in measurable concentrations of ciprofloxacin and enrofloxacin up to 6 h after the initial dose. They observed that intestinal contents had concentrations of ciprofloxacin corresponding to only 4 to 5% of the enrofloxacin concentration (0.9 to 5.5 μg of ciprofloxacin per gram of feces) (Wiuff et al., 2002). The traditional concept is that selection of resistant bacteria is dependent on several parameters, among which are the concentrations of free and active antimicrobial drugs reaching the enteric microbiota, the MIC of bacterial populations, and barriers exerted in the colonic ecosystem. To our knowledge, no current data has been published showing the concentration of enrofloxacin or ciprofloxacin in the fecal content of calves after a parenteral administration of enrofloxacin. However, if the concentration in calves is similar to that observed in the Wiuff et al. (2002) study with pigs, after treatment of a calf with enrofloxacin, selection for resistance to quinolones could occur from the presence of enrofloxacin in the feces at concentrations above the MIC of 2 to 4 μg/mL for bacteria from the Enterobacteriaceae family (CLSI, 2008). Because all quinolones have a common mechanism of resistance, resistance to one quinolone will usually result in resistance to all other quinolones, and selection pressure from enrofloxacin treatment could result in the selection of resistance to ciprofloxacin and nalidixic acid (Hopkins et al., 2005). Although the concentration of ciprofloxacin within 6 h after a treatment with enrofloxacin would probably be below the 4 μg/mL MIC for bacteria from the Enterobacteriaceae family based on the findings from Wiuff et al. (2002), this nonlethal concentration of ciprofloxacin could still result in the selection of resistance to fluoroquinolones (CLSI, 2006). Studies have shown that exposure of bacteria to nonlethal concentrations of antimicrobial drugs, including fluoroquinolones, can enrich pre-existing resistant mutants with very small fitness costs and select for de novo resistant mutants (Cebrian et al., 2003; Marcusson et al., 2009; Hughes and Andersson, 2012).
As expected, a higher prevalence of isolates nonsusceptible to neomycin and ceftriaxone was observed for calves at AGE-2 because previous studies have associated increased levels of multiple antimicrobial resistances in calves 2 to 4 wk of age (Berge et al., 2006). Commensal microbiota has been shown to protect the intestine from exogenous pathogens and antimicrobial resistant species through microorganism-mediated direct inhibition and by enhancing host immunity in the intestines (Macpherson and Uhr, 2004; Stecher and Hardt, 2008; Chung et al., 2012; Buffie and Pamer, 2013; Lawley and Walker, 2013). The lack of a developed microbiota in young calves, as observed in metagenomics studies, could be a factor allowing the amplification of bacteria that may have antimicrobial resistance mechanisms that result in a high fitness cost (Oikonomou et al., 2013). However, for ciprofloxacin and nalidixic acid, a higher proportion of resistant isolates was observed at AGE-3, when the calf has a more developed enteric microbiota. A study by Marcusson et al. (2009) evaluating the interplay in the selection of fluoroquinolone resistance and bacterial fitness in E. coli observed that some fluoroquinolone-resistant mutations may reduce drug susceptibility without reducing fitness, and that the addition of a resistance mutation to an already low-fitness mutant strain could cause both a reduction in drug susceptibility and an increase in relative fitness (Marcusson et al., 2009). The low fitness cost of resistance to fluoroquinolones would allow these bacteria to compete with susceptible strains. This would explain the persistence of fluoroquinolone-resistant E. coli in the commensal microbiota of calves even with the development and probable increase in resilience of the commensal microbiota to colonization by antimicrobial resistant bacteria.
Although not statistically significant at the 0.05 level, calves treated with ceftiofur displayed reduced susceptibility of fecal E. coli to ceftriaxone (P = 0.08). A study treating calves with ceftiofur hydrochloride at therapeutic doses (2.2 mg/kg per d) for 5 consecutive days observed a significant increase in the fecal excretion of ceftriaxone resistant bacteria (including Salmonella species) for at least 17 d following initial treatment (Jiang et al., 2006). Ceftiofur is the only third-generation cephalosporin approved for use in cattle and is labeled for the treatment of pneumonia, postpartum metritis, necrotizing pododermatitis, and mastitis (Daniels et al., 2009). Additionally, ceftiofur has been recommended as an extra-label treatment for diarrhea in preweaned calves (following approved label dose, frequency, duration, and route of administration; Constable, 2004). Resistance to third-generation cephalosporins is mainly conferred by the plasmid-encoded AmpC-like CMY β-lactamases and by the plasmid-encoded CTX-M β-lactamases (Daniels et al., 2009; Shaheen et al., 2011). An increase in E. coli susceptible to neomycin (P = 0.05) was also observed in calves treated with ceftiofur. This was an unexpected finding that could have resulted from the treatment with ceftiofur shifting the population of commensal E. coli that carried resistant genes to aminoglycosides. Systemic treatment of cows with ceftiofur has been shown to result in significant decrease in the number of fecal E. coli 2 to 7 d posttreatment, with a shift from >6 cfu/g pretreatment to <4 cfu/g 2 d after treatment (Mann et al., 2011). In our, study the selection pressure induced by ceftiofur treatment could have directly affected E. coli resistant to neomycin because of the high fitness cost that has been associated with many mechanisms of resistance to aminoglycosides (Kramer and Matsumura, 2013; Lioy et al., 2014).
Distribution of E. coli Antimicrobial-Resistant Phenotypes
For calves treated with enrofloxacin, the most common drug-resistant phenotypes (DRP) included resistance to 5 drugs in the β-lactam, phenicol, aminoglycoside, and tetracycline classes (Table 5). However, the most worrying finding was the resistance pattern of the second-most common DRP from enrofloxacin-treated calves, which included ampicillin, ceftiofur, ceftriaxone, ciprofloxacin, nalidixic acid, and tetracycline. Ceftriaxone and ciprofloxacin are the current antimicrobials of choice for treatment of important diseases in humans, including clinical Salmonella infections, and pathogens carrying resistance to both of these drugs could dramatically increase treatment failure. Resistance to fluoroquinolones seems to co-select for class-1 integrons and integron-borne extended-spectrum β-lactam (ESBL) genes because quinolone resistance genes (qnr) are situated in class-1 integron structures, which also carry ESBL resistance (Nordmann and Poirel, 2005; Wellington et al., 2013). This could explain why treatment with enrofloxacin would select for resistance to cephalosporins.
The most common DRP from calves with a farm record of treatment with ceftiofur consisted of 8 antimicrobial drugs belonging to the β-lactam (including second- and third-generation cephalosporins), phenicol, aminoglycoside, tetracycline, and sulfonamide classes. Among calves treated with ceftiofur, 97% were resistant to 3 or more antimicrobials versus 73% for calves not treated with ceftiofur. This finding is in accordance with other studies observing that treatment of cattle with ceftiofur results in co-selection of phenotypic antimicrobial resistances besides cephalosporins in E. coli (Singer et al., 2008; Kanwar et al., 2013). A study by Kanwar et al. (2013) observed that the presence of blaCMY-2 in fecal E. coli from cattle, one of the most common third-generation cephalosporin resistance genes in the United States, was associated with selection of multidrug resistance. An explanation for this has been linked to the fact that blaCMY-2 is usually located on a large Inc A/C plasmid that harbors several other resistance genes (Call et al., 2010; Kanwar et al., 2013; Guo et al., 2014).
Normalized Resistance Interpretation
Because the distribution of the inhibition zone diameters for the disk diffusion test for ceftiofur and ceftriaxone had an unclear transition between isolates devoid of detectable resistant phenotype, ECV were determined for these drugs (Kronvall et al., 2011). In our study, we used the NRI analysis to obtain the ECV (Kronvall, 2003). Of the 1,423 E. coli in our study, low-level resistance to ceftriaxone and ceftiofur were respectively associated with 49% and 23% of E. coli isolates which were classified as intermediate using clinical breakpoints (Figure 1). A limitation of using ECV compared with clinical breakpoints is that isolate classification is limited to susceptible or nonsusceptible, and specific detection of clinical resistant isolates that would be of greater interest for everyday clinical laboratory work to advise on therapy in a patient is not possible (Simjee et al., 2008).
CONCLUSIONS
Treatment of preweaned dairy calves with enrofloxacin resulted in significantly reduced susceptibility of fecal E. coli to ciprofloxacin and nalidixic acid. Although not significant at the 0.05 level, treatment with ceftiofur resulted in a reduced susceptibility of E. coli to ceftriaxone and a higher prevalence of multidrug resistant isolates compared with E. coli from calves not treated with ceftiofur. These findings emphasize the need for the implementation of practices that support the sustainable and judicious use of antimicrobial drugs in dairy calves.
Acknowledgments
We thank P. Leids from the New York State Cattle Health Assurance Program (NYSCHAP, Albany, NY) for assisting with the enrollment of farms in this project. We also thank G. Kronvall, Professor emeritus in Department of Microbiology, Karolinska Institute, Tumor and Cell Biology, Stockholm, Sweden, for his assistance and permission to use the NRI analysis. Additionally, we thank P. R. Smith, professor in the Department of Microbiology, School of Natural Sciences, National University of Ireland, Galway, for his assistance with the use of the NRI analysis. Research reported in this publication was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2010-51110-21131 from the USDA National Institute of Food and Agriculture (Washington, DC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the USDA. Research reported in this publication was supported by the Office of the Director, National Institutes of Health (Bethesda, MD) under Award Number T32ODO011000. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
References
- Andersson MI, MacGowan AP. Development of the quinolones. J Antimicrob Chemother. 2003;51(Suppl 1):1–11. doi: 10.1093/jac/dkg212. [DOI] [PubMed] [Google Scholar]
- Beraud R, Huneault L, Bernier D, Beaudry F, Letellier A, del Castillo JR. Comparison of the selection of antimicrobial resistance in fecal Escherichia coli during enrofloxacin administration with a local drug delivery system or with intramuscular injections in a swine model. Can J Vet Res. 2008;72:311–319. [PMC free article] [PubMed] [Google Scholar]
- Berge AC, Atwill ER, Sischo WM. Assessing antibiotic resistance in fecal Escherichia coli in young calves using cluster analysis techniques. Prev Vet Med. 2003;61:91–102. doi: 10.1016/s0167-5877(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Berge AC, Moore DA, Besser TE, Sischo WM. Targeting therapy to minimize antimicrobial use in preweaned calves: Effects on health, growth, and treatment costs. J Dairy Sci. 2009;92:4707–4714. doi: 10.3168/jds.2009-2199. [DOI] [PubMed] [Google Scholar]
- Berge AC, Moore DA, Sischo WM. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl Environ Microbiol. 2006;72:3872–3878. doi: 10.1128/AEM.02239-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother. 2010;54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambau E, Bordon F, Collatz E, Gutmann L. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob Agents Chemother. 1993;37:1247–1252. doi: 10.1128/aac.37.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian L, Sirvent E, Rodriguez Diaz JC, Ruiz M, Royo G. Characterisation of Salmonella spp. mutants produced by exposure to various fluoroquinolones. Int J Antimicrob Agents. 2003;22:134–139. doi: 10.1016/s0924-8579(03)00099-2. [DOI] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Sixteenth Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA: 2006. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Approved Standard. Clinical and Laboratory Standards Institute; Wayne, PA: 2008. [Google Scholar]
- Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis. 2009;49:132–141. doi: 10.1086/599374. [DOI] [PubMed] [Google Scholar]
- Constable PD. Antimicrobial use in the treatment of calf diarrhea. J Vet Intern Med. 2004;18:8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels JB, Call DR, Hancock D, Sischo WM, Baker K, Besser TE. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl Environ Microbiol. 2009;75:3648–3655. doi: 10.1128/AEM.02435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. BAYTRIL 100—Enrofloxacin injectable solution (beef and non-lactating dairy cattle) US Food and Drug Administration; Silver Spring, MD: 2008. Freedom of information summary; Supplemental new animal drug application; NADA 141–068. [Google Scholar]
- FDA. Extra-label drug use in veterinary medicine. US Food and Drug Administration; Silver Spring, MD: 2013. [Google Scholar]
- Guo YF, Zhang WH, Ren SQ, Yang L, Lu DH, Zeng ZL, Liu YH, Jiang HX. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS ONE. 2014;9:e96738. doi: 10.1371/journal.pone.0096738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzer K, Cummings KJ, Warnick LD, Schukken YH, Siler JD, Grohn YT, Davis MA, Besser TE, Wiedmann M. Agar disk diffusion and automated microbroth dilution produce similar antimicrobial susceptibility testing results for Salmonella serotypes Newport, Typhimurium, and 4,5,12:I-, but differ in economic cost. Foodborne Pathog Dis. 2011;8:1281–1288. doi: 10.1089/fpd.2011.0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int J Antimicrob Agents. 2005;25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Hughes D, Andersson DI. Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr Opin Microbiol. 2012;15:555–560. doi: 10.1016/j.mib.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Jiang X, Yang H, Dettman B, Doyle MP. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog Dis. 2006;3:355–365. doi: 10.1089/fpd.2006.3.355. [DOI] [PubMed] [Google Scholar]
- Joneberg J, Rylander M, Galas MF, Carlos C, Kronvall G. Analysis of parameters and validation of method for normalized interpretation of antimicrobial resistance. Int J Antimicrob Agents. 2003;21:525–535. doi: 10.1016/s0924-8579(03)00053-0. [DOI] [PubMed] [Google Scholar]
- Kaartinen L, Salonen M, Alli L, Pyorala S. Pharmacokinetics of enrofloxacin after single intravenous, intramuscular and subcutaneous injections in lactating cows. J Vet Pharmacol Ther. 1995;18:357–362. doi: 10.1111/j.1365-2885.1995.tb00604.x. [DOI] [PubMed] [Google Scholar]
- Kanwar N, Scott HM, Norby B, Loneragan GH, Vinasco J, McGowan M, Cottell JL, Chengappa MM, Bai J, Boerlin P. Effects of ceftiofur and chlortetracycline treatment strategies on antimicrobial susceptibility and on tet(A), tet(B), and blaCMY-2 resistance genes among E. coli isolated from the feces of feedlot cattle. PLoS ONE. 2013;8:e80575. doi: 10.1371/journal.pone.0080575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JR, Matsumura I. Directed evolution of aminoglycoside phosphotransferase (3′) type IIIa variants that inactivate amikacin but impose significant fitness costs. PLoS ONE. 2013;8:e76687. doi: 10.1371/journal.pone.0076687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronvall G. Determination of the real standard distribution of susceptible strains in zone histograms. Int J Antimicrob Agents. 2003;22:7–13. doi: 10.1016/s0924-8579(03)00107-9. [DOI] [PubMed] [Google Scholar]
- Kronvall G, Giske CG, Kahlmeter G. Setting interpretive breakpoints for antimicrobial susceptibility testing using disk diffusion. Int J Antimicrob Agents. 2011;38:281–290. doi: 10.1016/j.ijantimicag.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy VS, Goussard S, Guerineau V, Yoon EJ, Courvalin P, Galimand M, Grillot-Courvalin C. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA. 2014;20:382–391. doi: 10.1261/rna.042572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Mann S, Siler JD, Jordan D, Warnick LD. Antimicrobial susceptibility of fecal Escherichia coli isolates in dairy cows following systemic treatment with ceftiofur or penicillin. Food-borne Pathog Dis. 2011;8:861–867. doi: 10.1089/fpd.2010.0751. [DOI] [PubMed] [Google Scholar]
- Marcusson LL, Frimodt-Moller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuirk SM. Disease management of dairy calves and heifers. Vet Clin North Am Food Anim Pract. 2008;24:139–153. doi: 10.1016/j.cvfa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar Q, Gibson I, Monteiro A, Bregante M. Pharmacokinetics of enrofloxacin and danofloxacin in plasma, inflammatory exudate, and bronchial secretions of calves following subcutaneous administration. Antimicrob Agents Chemother. 1999;43:1988–1992. doi: 10.1128/aac.43.8.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- Oikonomou G, Teixeira AG, Foditsch C, Bicalho ML, Machado VS, Bicalho RC. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. associations of Faecalibacterium species with health and growth. PLoS ONE. 2013;8:e63157. doi: 10.1371/journal.pone.0063157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SP, Murinda SE, Jayarao BM. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: A comprehensive review. Foodborne Pathog Dis. 2011;8:337–355. doi: 10.1089/fpd.2010.0730. [DOI] [PubMed] [Google Scholar]
- Pereira RV, Santos TM, Bicalho ML, Caixeta LS, Machado VS, Bicalho RC. Antimicrobial resistance and prevalence of virulence factor genes in fecal Escherichia coli of Holstein calves fed milk with and without antimicrobials. J Dairy Sci. 2011;94:4556–4565. doi: 10.3168/jds.2011-4337. [DOI] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen BW, Nayak R, Foley SL, Kweon O, Deck J, Park M, Rafii F, Boothe DM. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the united states. Antimicrob Agents Chemother. 2011;55:5666–5675. doi: 10.1128/AAC.00656-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silley P. Susceptibility testing methods, resistance and breakpoints: What do these terms really mean? Rev. Sci Tech. 2012;31:33–41. doi: 10.20506/rst.31.1.2097. [DOI] [PubMed] [Google Scholar]
- Simjee S, Silley P, Werling HO, Bywater R. Potential confusion regarding the term ‘resistance’ in epidemiological surveys. J Antimicrob Chemother. 2008;61:228–229. doi: 10.1093/jac/dkm423. [DOI] [PubMed] [Google Scholar]
- Singer RS, Patterson SK, Wallace RL. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. 2008;74:6956–6962. doi: 10.1128/AEM.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Schwarz T, Verner-Jeffreys DW. Use of normalised resistance analyses to set interpretive criteria for antibiotic disc diffusion data produce by Aeromonas spp. Aquaculture. 2012;326–329:27–35. [Google Scholar]
- Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. Combating antimicrobial resistance: Policy recommendations to save lives. Clin Infect Dis. 2011;52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- USDA. Part III: Reference of dairy cattle health and management practices in the United States. US Department of Agriculture, National Animal Health Monitoring System; Fort Collins, CO: 2008. [Google Scholar]
- Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM, Williams AP. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- Windeyer MC, Leslie KE, Godden SM, Hodgins DC, Lissemore KD, LeBlanc SJ. Factors associated with morbidity, mortality, and growth of dairy heifer calves up to 3 months of age. Prev Vet Med. 2014;113:231–240. doi: 10.1016/j.prevetmed.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Wiuff C, Lykkesfeldt J, Aarestrup FM, Svendsen O. Distribution of enrofloxacin in intestinal tissue and contents of healthy pigs after oral and intramuscular administrations. J Vet Pharmacol Ther. 2002;25:335–342. doi: 10.1046/j.1365-2885.2002.00430.x. [DOI] [PubMed] [Google Scholar]