Abstract
Platinum nanoparticles (PtNPs) are able to efficiently catalyze H2O2 to generate oxygen gas. However, due to the lack of an efficient approach or device that is able to measure the produced oxygen gas, the catalytic reaction has never been used for diagnostic applications. Microfluidics technology provides a platform that meets these requirements. The volumetric bar chart chip (V-Chip) volumetrically measures the production of oxygen gas by PtNPs and can be integrated with ELISA technology to provide visible and quantitative readouts without assistance from expensive instrumentation or complicated data processing. Herein we show that PtNPs outperform catalase with respect to stability at high H2O2 concentrations, high temperatures, or long-term reactions, and are resistant to most catalase inhibitors. In addition, we show that the catalase-like activity of PtNPs in concert with the V-Chip can be used to sensitively and specifically detect cancer biomarkers both in serum and on the cell surface.
Keywords: microfluidics, platinum nanoparticles, ELISA, catalytic activity, biomarker assay
Nanomaterials with applicable optical, electronic, magnetic, catalytic, and thermal properties play an important role in clinical diagnosis and medical management.[1] New emerging diagnostic technologies based on nanomaterials show great advantages over traditional methods, particularly in sensitivity, selectivity, and stability. These detection platforms rely on different types of colorimetric,[2] fluorescent,[3] electrochemical,[1b,1c] Raman[4] and magnetic[5] nanoparticles as transducers to convert molecular recognition events into measurable outputs. Catalytic activity is another feature that has recently attracted great interest, as it can amplify signal and increase detection specificity.[2b, 6] Various nanoparticles have intrinsic catalytic activity, and have been designed as catalytic labels for sensitive and selective detection of proteins, nucleic acids, and other molecules.[6c] As highly efficient catalysts, platinum nanoparticles (PtNPs) have been used in medical applications, primarily diagnostics, for detection of biomolecules using electrochemical or colorimetric methods.[7] However, for quantitative detection, expensive instruments are still required, which limited their applications. It has been shown that PtNPs are able to efficiently catalyze the reaction of H2O2 to generate oxygen gas.[7b] Due to the lack of simple approaches or devices able to measure the end product (oxygen gas), the reaction has not been widely adopted for diagnostic applications.
Microfluidic chips allow portability, considerable throughput, and the capacity to integrate with other diagnostic techniques for a complete, point-of-care device.[8] Developed with microfluidics technology, a volumetric bar chart chip (V-Chip) has been developed to volumetrically measure the production of oxygen gas. This can be integrated with an ELISA reaction for quantitative detection of biomarkers, and the output consists of visible bar charts on the V-Chip, without any assistance from instruments, data processing, or graphic plotting.[9] In a previous iteration of this V-Chip, catalase was used as the ELISA probe to generate oxygen gas through catalysis of hydrogen peroxide. However, several problems were encountered in this catalase-propelled microfluidic device. The drawbacks included the high cost of preparation and purification of catalase, its low operational stability due to digestion and denaturation, and the dependence of catalytic activity on environmental conditions. The low catalytic stability leads to unsatisfactory sensitivity for V-Chip applications: catalase is destroyed in the reaction with hydrogen peroxide[10] and its activity is inhibited in the presence of high concentrations of H2O2.[9a]
Herein, we introduce PtNPs to the V-Chip (PtV-Chip) platform as a nanoparticle substitute for catalase (Figure 1). Our results indicate that PtNPs possess the requisite features including: excellent catalytic stability at high H2O2 concentration and over long reaction periods, and maintenance of activity over a broad temperature range and in the presence of catalase inhibitors. In this work, the PtV-Chip was used for quantitative and sensitive detection of the lung cancer biomarker CYFRA 21-1 in buffer and serum based on standard sandwich ELISA. To demonstrate the breadth of potential applications, on-chip cell culture and cell-based ELISA were performed to detect human epidermal growth factor receptor 2 (HER2) and phosphorylated HER2 (pHER2) expression on the surface of breast cancer cells.
Figure 1.
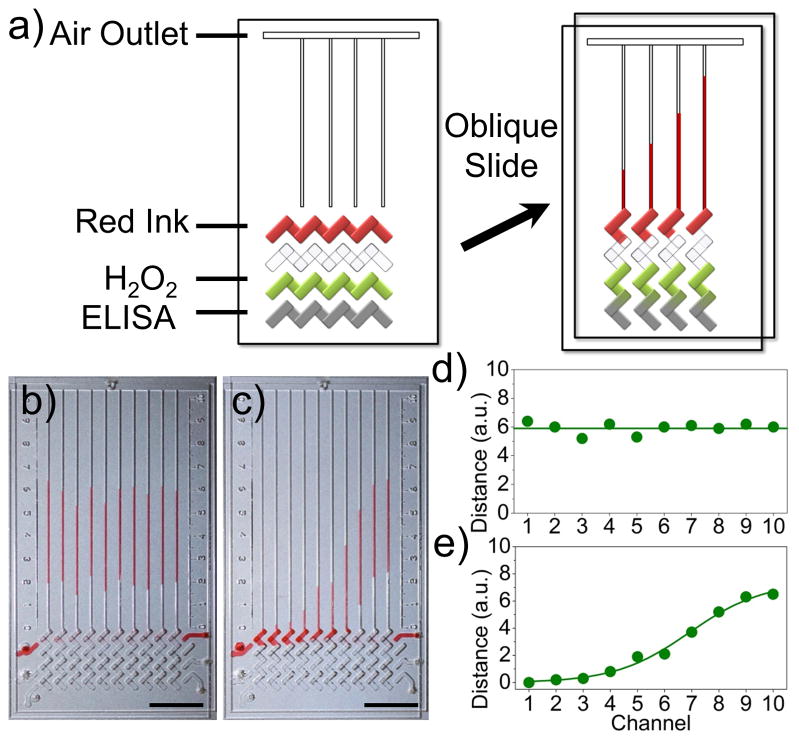
PtNPs have intrinsic catalase activity. a) Schematic view of a typical V-Chip for ELISA application. Ink and H2O2 were preloaded and the ELISA assay was performed in the designated lanes. An oblique slide breaks the flow path and forms the structure on the right, causing PtNPs and H2O2 to react and push the inked bars. b), d) A uniform concentration of PtNPs generates uniform readout distance in the presence of 10 μg·mL−1 of PtNPs and 35 % H2O2. c), e) Bar chart images of diffusing 20 μg·mL−1 of PtNPs in the presence of 35 % H2O2. Scale bar, 1 cm for b and c.
PtNPs with an average diameter of 30 nm were prepared as previously described (Figure S1a, b).[11] We first tested the catalase and peroxidase-like activity by simply mixing PtNPs and H2O2. When PtNPs were added to H2O2, the characteristic “fizzing” was observed due to the production of oxygen gas (Figure S1c). In addition to catalase-like activity, PtNPs also possess intrinsic peroxidase-like activity (Figure S2).[7b] In the presence of H2O2, PtNPs can catalyze the oxidation of the peroxidase substrates to give a color reaction. The V-Chip provides a platform for quantitatively evaluating the production of oxygen gas. As shown in Figure 1b and c, when the PtNP solution was loaded in the V-Chip through the inlet holes, the ink bar chart movements correlate with PtNP concentration. When a uniform PtNP concentration was used, the V-Chip showed uniform ink bar chart advancements (Figure 1b, d). When we created a diffusion gradient of PtNPs, the bar chart resembled a sigmoidal shape (Figure 1c, e). The progressive increments demonstrate the relationship between the height of the V-Chip bars and PtNP concentration, indicating that the PtV-Chip can be used for quantitative biomarker detection.
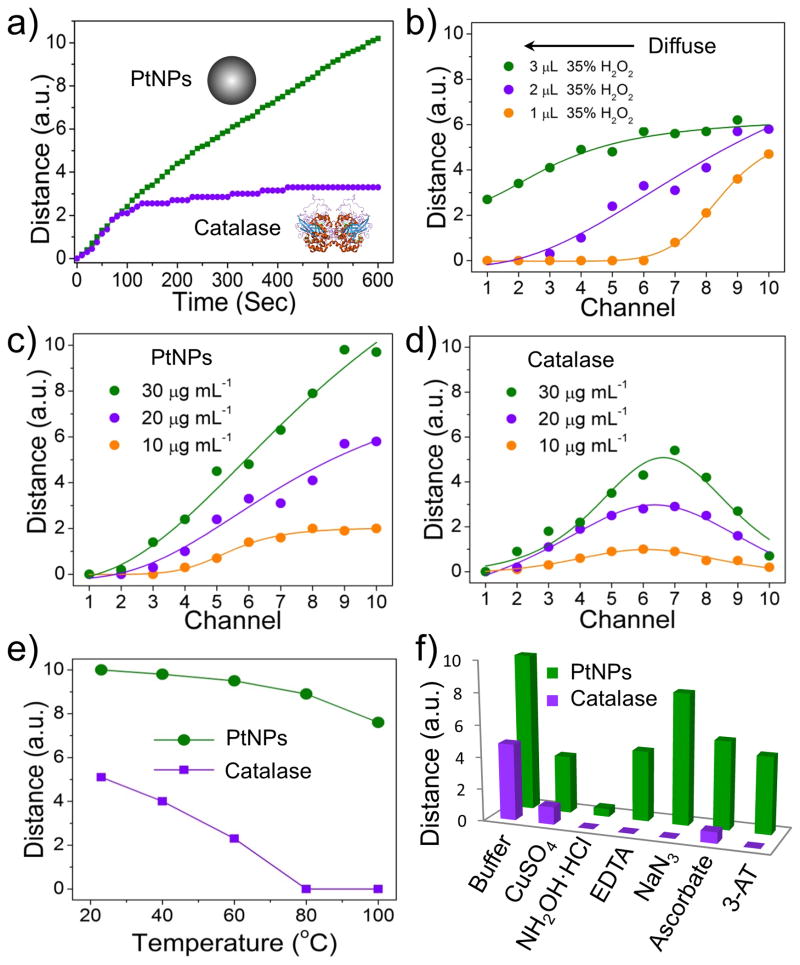
To further examine the suitability of PtNPs to substitute for catalase in the V-chip, we compared their performances in the 10-plex V-Chip. We used PtNPs or catalase diffusion in PBS buffer to study the kinetics of ink bar chart movements in each channel. As shown in Figure S3, loading 2 μL of PtNPs or protein into the preloaded PBS buffer in the bottom lane allows generation of a concentration gradient. The movement of the ink bars was recorded using a camera, and the time-dependent distance of each channel in 10 min intervals are shown in Figure S4. The images clearly show that the ink bar in the chip loaded with PtNPs moved much faster than the chip loaded with same concentration of catalase. In 10 min, ten channels in the PtNP-loaded V-Chip form obvious ink bar charts, but only four channels formed ink bar charts in the catalase-loaded V-Chip. This indicates that lower concentrations of PtNPs are required to produce readout from the V-Chip, and that using PtNPs improves the sensitivity of detection. In addition, the kinetics of ink bar chart movement clearly shows that PtNPs exhibit catalytic stability over a longer reaction period. Figure 2a shows the time-dependent movements of the ink bar in the tenth channel: PtNPs exhibited a continuous increase at 10 min, while catalase quickly reached a plateau in about 2 min, presumably due to degradation.[10]
Figure 2.
Comparison of PtNPs and catalase for catalyze oxygen gas production in 10-plex V-Chip. a) Time-dependent ink advancements with the application of 20 μg·mL−1 PtNPs and catalase. H2O2 concentration is 35% or 4% respectively. b) Bar chart advancements after diffusing different volumes of 35% H2O2 in the presence of 20 μg mL−1 PtNPs. c, d) Bar chart advancements of diffusing 2 μL of 35 % H2O2 in the presence of different concentration of PtNPs (c) or catalase (d). e) Catalytic activity of PtNPs and catalase after treatment at different temperatures for 30 min. f) Catalytic activity of PtNPs (5×) and catalase (1×) in the absence or presence of 5 mM CuSO4, NH2OH·HCl, EDTA, NaN3, ascorbate and 3-amino-1, 2, 4-triazole (3-AT).
An additional attractive feature of PtNPs is that they are catalytically stable over a broad H2O2 concentration range, which can be generated by allowing H2O2 diffuse in preloaded water (Figure 2b). When 2 μL 35% H2O2 was added to preloaded water to generate a concentration gradient, and the same concentration of PtNPs or catalase were then added, the ink bar chart patterns were different (Figure S5). The pattern of the 10-plex V-Chip loaded with PtNPs shows a sigmoidal shape, while the catalase-loaded chip resulted in a Gaussian shape, presumably due to the inhibition of catalase activity at high concentrations of H2O2 (Figure 2c, d). It can be concluded that the optimal H2O2 concentration for the PtNP-mediated reaction is 35%, which provides more substrate for prolonged ink bar chart movement than catalase. In addition, PtNPs also show thermal stability at high temperature and resistance to most catalase inhibitors. As shown in Figure 2e, PtNPs retained 80% activity at 100 °C, while catalase completely lost its activity at 80 °C. We also studied the activity of PtNPs in the presence of six catalase inhibitors.[12] When a 5-fold concentration of inhibitor was used with PtNPs, only NH2OH·HCl showed nearly complete inhibition of PtNPs activity (Figure 2f). It should be noted that, in contrast to catalase, PtNP activity only slightly decreased in the presence of NaN3, which is a bacteriostatic agent widely used to preserve biological samples, such as commercialized antibodies.[13] Therefore, PtNP-conjugated antibodies do not need laborious or costly purification steps to remove NaN3, which is another advantage over catalase.
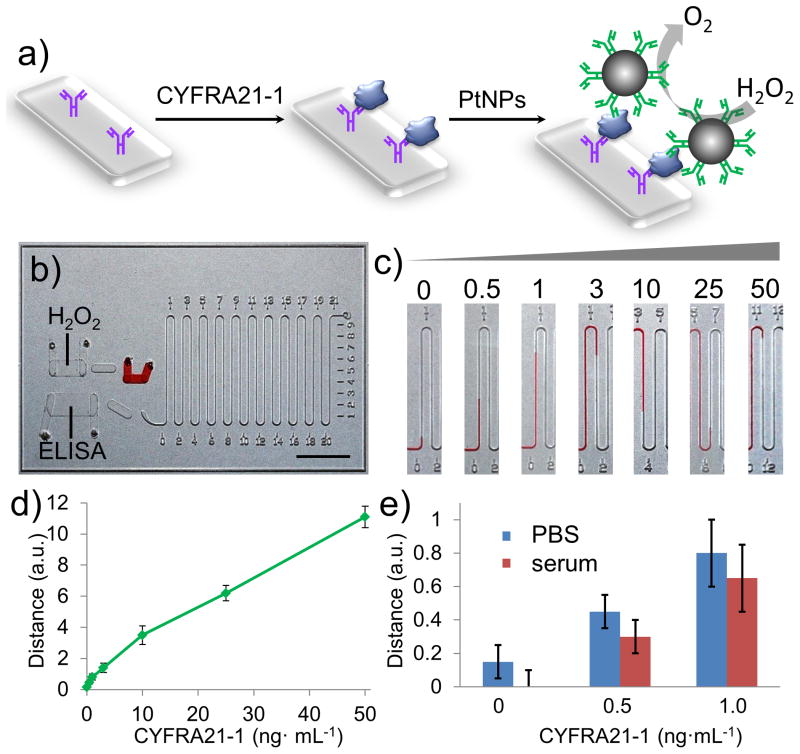
Cytokeratin 19 fragments (CYFRA 21-1) are circulating tumor biomarkers with high specificity and sensitivity for non-small-cell lung cancer (NSCLC), particularly squamous cell carcinoma.[14] As an independent prognostic role, the serum level of CYFRA 21-1 can reflect the disease stage and treatment efficacy in NSCLC.[15] PtNPs were introduced in a single-channel V-Chip for quantitative detection of human CYFRA 21-1. Fabrication and the surface modification of V-Chip are performed according to our previous method.[9] To detect CYFRA 21-1, a typical three-component sandwich ELISA was employed (Figure 3), in which PtNP was conjugated with the probe antibody to react with the hydrogen peroxide and produce oxygen gas. We tested a range of concentrations of CYFRA 21-1 (0.5 to 50 ng·mL−1) using the V-Chip (Figure 3c). Figure 3d shows the 10-min assay results from the test, in which the corresponding ink bar charts provide direct quantitative results of these concentrations. As the concentration of CYFRA 21-1 increased, the bar charts of the V-Chip exhibited a nearly-linear corresponding increase. In each case, the results obtained with solutions containing 0.5 ng·mL−1 were greater by a factor of at least three standard deviations (SDs) than background signals, indicating that the limit of detection resides at or below this concentration. We also studied the ability of the PtV-Chip to detect CYFRA 21-1 under physiological conditions. When the same concentration of CYFRA 21-1 was spiked in serum, the bar chart results exhibit a similar detection limit as that obtained with buffer, indicating that serum matrices do not interfere significantly with PtV-Chip sensitivity (Figure 3e). The cutoff concentration of CYFRA 21-1 for diagnosing NSCLC in serum is 3.3 ng·mL−1,[14] indicating that the detection ability of the PtV-Chip is well within the range of clinical relevance for diagnosing NSCLC.
Figure 3.
PtV-Chip for detection of CYFRA21-1. a) A standard sandwich ELISA was employed in PtV-Chip for biomarker detection. CYFRA21-1 (blue) was added in the assay wells in the bottom plate and bound to the capture antibodies (purple). The PtNP-antibody complexes were loaded into the assay chamber to form sandwich structures. To detect the target biomarkers, PtNP was conjugated with the probe antibody to react with the hydrogen peroxide and produce oxygen gas. b) Image of PtV-Chip after loading reagents and samples. Scale bar, 1 cm. c) Images for single-plex V-Chip results in the absence or presence of different concentration of CYFRA21-1 (0.5, 1, 3, 10, 25, 50 ng·mL−1). d) The CYFRA21-1 calibration curve corresponding to ink advancement at 10 min, with the CYFRA21-1 concentration varying from 0.5 to 50 ng·mL−1. e) Detection results of spiked CYFRA21-1 in PBS buffer and serum. The error bars in d and e represent the SD of three measurements.
The rapid and sensitive detection of biomarker expression on cancer cells or tissues is particularly critical, as it not only can be used for the identification and classification of human tumors, but also provides approaches for selecting the most effective therapeutic targets to improve clinical outcomes. The traditional method for detection of biomarker expression on cancer cells is based primarily on immunohistochemistry and fluorescence in situ hybridization.[16] Cell-based ELISA is an alternatively rapid, convenient, and sensitive assay that is frequently used for measuring the relative amount of biomarkers and evaluating the effect of inhibitors, activators, or various treatments on cancer cells.[17] HER2 is a receptor tyrosine kinase that is detectable in a subset of breast or other adenocarcinomas and transitional cell carcinomas.[18] In the case of breast cancer, HER2 overexpression has been shown to be associated with poor prognosis, and is also a target for therapy.[18a]
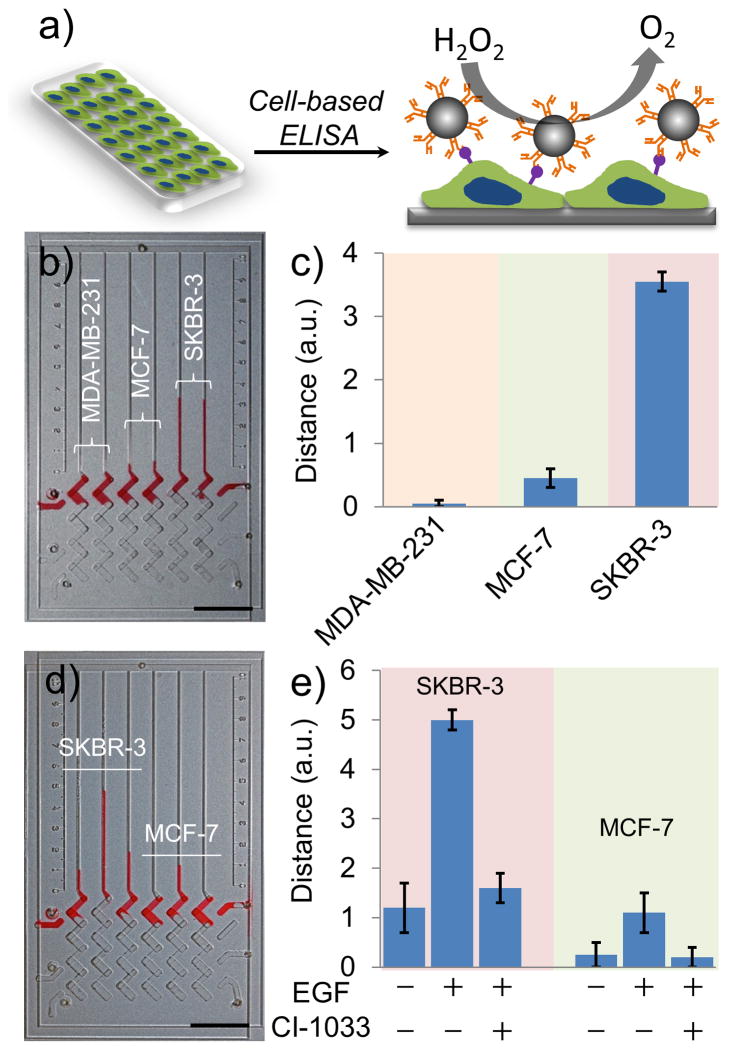
To assess whether the PtV-Chip can be used for biomarker profiling, cells from three breast cancer lines known to variably express HER2 (MDA-MB-231, MCF-7, and SKBR-3) were cultured in the assay wells of the bottom plate, and HER2 expression was assessed (Figure 4a). The rough surface of the assay wells modified with amino groups showed high adhesion for the cancer cells (Figure S6, S7, S8). After overnight incubation, cells were fixed and blocked with BSA, and probe antibody-conjugated PtNPs were loaded. Binding was assessed by the addition of H2O2. To demonstrate binding specificity, we used the peroxidase activity of PtNPs to catalyze the color reaction of DAB in the presence of H2O2, which would result in the staining of bound cells. As shown in Figure S9, MDA-MB-231 showed very little staining, MCF-7 was slightly stained, and SKBR3 showed strong staining, which is consistent with previous reports.[9a] When approximately 1,000 cells were seeded in each ELISA well, the quantitative results can be observed in PtV-Chip results. Considering the distance values of HER2 on SKBR cells to be 100%, and assuming that the distance values are proportional to HER2 expression, MCF-7 cells expressed 13% HER2, while MDA-MB-231 shows almost no expression (Figure 4b, c). To calculate the sensitivity of V-Chip-based cell detection, different numbers of SKBR-3 cells were used. As the number of SKBR-3 cells increased, the V-Chip ink bar increased proportionally, suggesting an increased concentration of bound PtNPs. Using the PtNP-incorporated V-Chip, we could detect as low as 400 SKBR-3 cells (Figure S10).
Figure 4.
Cell-based ELISA for detection of HER2 and pHER2 using PtV-Chip. a) Schematic mechanism for cell-based ELISA for cell surface biomarker detection using PtV-Chip. Cells were seeded in the assay wells in the bottom plate. Antibody conjugated PtNPs were used to target the cell surface biomarkers and to react with the hydrogen peroxide to produce oxygen gas. b), c) PtV-Chip detection of HER2 expression on 1,000 MDA-MB-231, MCF-7, or SKBR-3 cells. d), e) PtV-Chip detection of pHER2 expression on SKBR-3 and MCF-7 cells treated with or without 100 ng·mL−1 EGF and 5 mM CI-1033. Scale bar, 1 cm for (b) and (d). The error bars in b and d represent the SD of three measurements.
In addition to biomarker profiling, the PtV-Chip can also be used to detect protein phosphorylation. Protein phosphorylation plays important roles in the regulation of many cellular processes, including cell cycle, growth, apoptosis, and signal transduction pathways.[19] Because of its central importance to many biological processes, protein phosphorylation has been widely studied to understand its role in human disease.[20] Tyrosine phosphorylation on HER2 at position Y877 was investigated using the cell-based ELISA on the V-Chip. When SKBR-3 and MCF-7 cells were treated with 100 ng·mL−1 epidermal growth factor (EGF) for 1 h, HER2 phosphorylation is increased in both cell types (Figure 4d, e).[21] To further investigate the phosphorylation state of HER2, we treated the cells using EGF and a small molecule inhibitor, CI-1033. This molecule has been shown to irreversibly bind to the kinase domain of HER2.[21a] As showing in Figure 4e, the tyrosine residue at 877 was significantly dephoshyorylated in the presence of 5 mM CI-1033.
In summary, incorporating PtNPs into the V-Chip allows for highly-sensitive, quantitative, and instrument-free detection of protein biomarkers. As a V-Chip-based ELISA probe, PtNPs outperform catalase with respect to catalytic stability at high H2O2 concentrations over long reaction times, thermal stability at high temperatures, and resistance to most catalase inhibitors. In addition, PtNPs are a promising candidate as a catalase biomimetic and offer advantages in ease of preparation and conjugation, low-cost, and biostability. Based on the high sensitivity of PtNP-integrated V-Chip, disease-related biomarkers in serum or on the cell surface can be quantitatively and selectively detected.
Currently, diagnostic technologies are driven by the need to meet the requirements for point-of-care (POC) use in resource-limited settings.[22] Devices need to be sensitive, low cost, portable, and user-friendly to be adopted for wide-spread use. Although several microdevices with integrated microsensors for signal readout have been developed for biomarker quantitation via the miniaturization of conventional bulky instruments, most of these devices remain laboratory prototypes after a decade of development. Their commercial potential is limited because they are too costly, complicated to use, or are not sensitive or specific enough to work in complex biospecimens (blood, serum).[22a, 23] The work presented here has laid the foundation for PtNP-integrated V-Chips to be used in clinical diagnostics, drug screening, and environmental monitoring.
Supplementary Material
Acknowledgments
We are grateful for the support from NIH/NIDA 1R01DA035868-01, NIH/NCI 1R01CA180083-01, the Cancer Prevention and Research Institute of Texas (CPRIT-R1007), Emily Herman Research Fund, and Golfers Against Cancer Foundation. We thank the HMRI scanning electron microscopy (SEM) core facility for instrumental assistance.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201404349.
Contributor Information
Dr. Yujun Song, Department of nanomedicine, Houston Methodist Research Institute, Department of Pathology and Genomic Medicine, Houston Methodist Hospital, Department of Cell and Developmental Biology, Weill Medical College of Cornell University, 6670 Bertener Ave, Houston, TX, 77030, USA
Prof. Xuefeng Xia, Department of nanomedicine, Houston Methodist Research Institute, Department of Pathology and Genomic Medicine, Houston Methodist Hospital, Department of Cell and Developmental Biology, Weill Medical College of Cornell University, 6670 Bertener Ave, Houston, TX, 77030, USA
Prof. Xifeng Wu, Department of Epidemiology, The University of Texas M. D. Anderson Cancer Center, 1155 Herman P. Pressler Blvd, Houston, TX, 77030, USA
Prof. Ping Wang, Department of nanomedicine, Houston Methodist Research Institute, Department of Pathology and Genomic Medicine, Houston Methodist Hospital, Department of Cell and Developmental Biology, Weill Medical College of Cornell University, 6670 Bertener Ave, Houston, TX, 77030, USA
Prof. Lidong Qin, Email: LQin@HoustonMethodist.org, Department of nanomedicine, Houston Methodist Research Institute, Department of Pathology and Genomic Medicine, Houston Methodist Hospital, Department of Cell and Developmental Biology, Weill Medical College of Cornell University, 6670 Bertener Ave, Houston, TX, 77030, USA
References
- 1.a) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; b) Cui Y, Wei Q, Park H, Lieber CM. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]; c) Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]; d) Laurent S, Forge D, Port M, Roch A, Robic C, Vander Elst L, Muller RN. Chem Rev. 2008;108:2064–2110. doi: 10.1021/cr068445e. [DOI] [PubMed] [Google Scholar]; e) Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Lal S, Clare SE, Halas NJ. Acc Chem Res. 2008;41:1842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 2.a) Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]; b) Song Y, Wei W, Qu X. Adv Mater. 2011;23:4215–4236. doi: 10.1002/adma.201101853. [DOI] [PubMed] [Google Scholar]
- 3.a) Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]; b) Nie S, Xing Y, Kim GJ, Simons JW. Annu Rev Biomed Eng. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 4.a) Chen Z, Tabakman SM, Goodwin AP, Kattah MG, Daranciang D, Wang X, Zhang G, Li X, Liu Z, Utz PJ, Jiang K, Fan S, Dai H. Nat Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]; b) Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 5.Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS, Wang SX. Nat Med. 2009;15:1327–U1130. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Katz E, Willner I, Wang J. Electroanalysis. 2004;16:19–44. [Google Scholar]; b) Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N, Wang T, Feng J, Yang D, Perrett S, Yan X. Nat Nanotechnol. 2007;2:577–583. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]; c) Willner I, Willner B. Nano Lett. 2010;10:3805–3815. doi: 10.1021/nl102083j. [DOI] [PubMed] [Google Scholar]; d) Song Y, Qu K, Zhao C, Ren J, Qu X. Adv Mater. 2010;22:2206–2210. doi: 10.1002/adma.200903783. [DOI] [PubMed] [Google Scholar]
- 7.a) Polsky R, Gill R, Kaganovsky L, Willner I. Anal Chem. 2006;78:2268–2271. doi: 10.1021/ac0519864. [DOI] [PubMed] [Google Scholar]; b) Fan J, Yin JJ, Ning B, Wu X, Hu Y, Ferrari M, Anderson GJ, Wei J, Zhao Y, Nie G. Biomaterials. 2011;32:1611–1618. doi: 10.1016/j.biomaterials.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 8.a) Fan R, Vermesh O, Srivastava A, Yen BKH, Qin LD, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang Y, Zhang W, Qin L. Angew Chem Int Ed. 2014;53:2344–2348. doi: 10.1002/anie.201309885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nat Commun. 2012;3:1283. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Song Y, Wang Y, Qin L. J Am Chem Soc. 2013;135:16785–16788. doi: 10.1021/ja4085397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George P. Nature. 1947;160:41–43. doi: 10.1038/160041a0. [DOI] [PubMed] [Google Scholar]
- 11.Huang M, Jin Y, Jiang H, Sun X, Chen H, Liu B, Wang E, Dong S. J Phys Chem B. 2005;109:15264–15271. doi: 10.1021/jp051612e. [DOI] [PubMed] [Google Scholar]
- 12.a) Keilin D, Hartree E. Nature. 1934;134:933. [Google Scholar]; b) Arabaci G, Usluoglu A. J Chem. 2013:6. [Google Scholar]; c) Davison AJ, Kettle AJ, Fatur DJ. J Biol Chem. 1986;261:1193–1200. [PubMed] [Google Scholar]; d) Cohen G, Somerson NL. J Bacteriol. 1969;98:543–546. doi: 10.1128/jb.98.2.543-546.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichstein HC, Soule MH. J Bacteriol. 1944;47:221. doi: 10.1128/jb.47.3.221-230.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieskopf B, Demangeat C, Purohit A, Stenger R, Gries P, Kreisman H, Quoix E. Chest. 1995;108:163–169. doi: 10.1378/chest.108.1.163. [DOI] [PubMed] [Google Scholar]
- 15.a) Pujol JL, Molinier O, Ebert W, Daures JP, Barlesi F, Buccheri G, Paesmans M, Quoix E, Moro-Sibilot D, Szturmowicz M, Brechot JM, Muley T, Grenier J. Br J Cancer. 2004;90:2097–2105. doi: 10.1038/sj.bjc.6601851. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vollmer RT, Govindan R, Graziano SL, Gamble G, Garst J, Kelley MJ, Christenson RH. Clin Cancer Res. 2003;9:1728–1733. [PubMed] [Google Scholar]
- 16.Yeh IT. Am J Clin Pathol. 2002;117(Suppl):S26–35. doi: 10.1309/D76M-QEGU-62BT-HKER. [DOI] [PubMed] [Google Scholar]
- 17.a) Foreman KE, Vaporciyan AA, Bonish BK, Jones ML, Johnson KJ, Glovsky MM, Eddy SM, Ward PA. J Clin Invest. 1994;94:1147–1155. doi: 10.1172/JCI117430. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wu CY, Jan JT, Ma SH, Kuo CJ, Juan HF, Cheng YS, Hsu HH, Huang HC, Wu D, Brik A, Liang FS, Liu RS, Fang JM, Chen ST, Liang PH, Wong CH. Proc Natl Acad Sci U S A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Mitri Z, Constantine T, O’Regan R. Chemother Res Pract. 2012;2012:743193. doi: 10.1155/2012/743193. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ding Y, Liu Z, Desai S, Zhao Y, Liu H, Pannell LK, Yi H, Wright ER, Owen LB, Dean-Colomb W, Fodstad O, Lu J, LeDoux SP, Wilson GL, Tan M. Nat Commun. 2012;3:1271. doi: 10.1038/ncomms2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 20.Cohen P. Eur J Biochem. 2001;268:5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 21.a) Hughes DP, Thomas DG, Giordano TJ, McDonagh KT, Baker LH. Pediatr Blood Cancer. 2006;46:614–623. doi: 10.1002/pbc.20454. [DOI] [PubMed] [Google Scholar]; b) Gijsen M, King P, Perera T, Parker PJ, Harris AL, Larijani B, Kong A. PLoS Biol. 2010;8:e1000563. doi: 10.1371/journal.pbio.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Song Y, Huang YY, Liu X, Zhang X, Ferrari M, Qin L. Trends biotechnol. 2014;32:132–139. doi: 10.1016/j.tibtech.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yager P, Domingo GJ, Gerdes J. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 23.Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Anal Chem. 2012;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.