Abstract
Oesophageal exposure to duodenogastroesophageal refluxate is implicated in the development of Barrett's metaplasia (BM), with increased risk of progression to oesophageal adenocarcinoma. The literature proposes that reflux exposure activates NF-κB, driving the aberrant expression of intestine-specific caudal-related homeobox (CDX) genes. However, early events in the pathogenesis of BM from normal epithelium are poorly understood. To investigate this, our study subjected a 3D model of the normal human oesophageal mucosa to repeated, pulsatile exposure to specific bile components and examined changes in gene expression. Initial 2D experiments with a range of bile salts observed that taurochenodeoxycholate (TCDC) impacted upon NF-κB activation without causing cell death. Informed by this, the 3D oesophageal model was repeatedly exposed to TCDC in the presence and absence of acid, and the epithelial cells underwent gene expression profiling. We identified ∼300 differentially expressed genes following each treatment, with a large and significant overlap between treatments. Enrichment analysis (Broad GSEA, DAVID and Metacore™; GeneGo Inc) identified multiple gene sets related to cell signalling, inflammation, proliferation, differentiation and cell adhesion. Specifically NF-κB activation, Wnt signalling, cell adhesion and targets for the transcription factors PTF1A and HNF4α were highlighted. Our data suggest that HNF4α isoform switching may be an early event in Barrett's pathogenesis. CDX1/2 targets were, however, not enriched, suggesting that although CDX1/2 activation reportedly plays a role in BM development, it may not be an initial event. Our findings highlight new areas for investigation in the earliest stages of BM pathogenesis of oesophageal diseases and new potential therapeutic targets.
Keywords: acid, Barrett's metaplasia, HNF4alpha, oesophageal adenocarcinoma, reflux, taurochenodeoxycholate, tissue engineering
Exposure of the lower oesophagus to gastric refluxate [gastro-oesophageal reflux disease (GORD)] is a critical factor in the pathogenesis of the metaplastic columnar phenotype Barrett's metaplasia (BM). The development of BM places an individual at risk of developing oesophageal adenocarcinoma (OAC), which typically has a poor prognosis with little improvement in survival rates in recent years. Understanding BM pathogenesis is essential if we are to develop scientifically rational strategies to reduce OAC mortality. The development of dysplasia and progression to carcinoma reflects the acquisition of increasing genetic damage, but the mechanism and precise refluxate components which lead to BM are less well understood. Studies suggest that reflux exposure ultimately leads to transcriptional activation of members of the caudal-related homeobox (CDX) gene family, resulting in inappropriate differentiation towards a columnar, intestinal phenotype (Wong et al. 2005). NF-κB activation is observed in BM tissue (Konturek et al. 2004), increasing with progression from normal tissue to OAC (O'Riordan et al. 2005), and is implicated in the aberrant expression of CDX2 (Debruyne et al. 2006).
The gene expression profiles of BM and OAC tissue samples have produced diverse results, potentially reflecting differences in sample production, data analysis and the presence of contaminating tissue types. Nevertheless, it is possible to discriminate between squamous (normal) and columnar (BM/OAC) epithelia on the basis of their transcriptomes (Fox et al. 2005; Gomes et al. 2005) although discriminating between BM and OAC is less successful (Wang et al. 2006; Nancarrow et al. 2011).
Other studies have evaluated gene expression changes in cell lines exposed to acid or bile. However, the relevance of data from immortalized or established OAC cell lines to the initial development of BM is questionable; conventional cell culture systems omit important cell–cell and cell matrix interactions, and there is little published data using primary cells or models that more closely reflect the native human oesophagus. In vivo animal models, incorporating the artificial induction of GORD, can provide a more relevant microenvironment, but the manipulation of specific reflux components becomes technically challenging. There is also species variation in dietary impact on refluxate composition. Therefore, animal experiments may not reflect the events occurring within the human oesophagus. A barrier to progress has been the lack of appropriate experimental models.
We recently described a 3D oesophageal model with an anatomically realistic squamous epithelium, incorporating primary human oesophageal squamous (HOS) cells, oesophageal fibroblasts (HOFs) and a porcine oesophagus-derived matrix (Green et al. 2010). We now report the use of this model to investigate early events in BM development, providing data that are difficult to obtain in other ways.
The model is, however, labour intensive and less suitable for screening multiple compounds. Consequently, we first investigated the impact of the bile salt components of reflux using immortalized oesophageal squamous (HET-1A) cells. As NF-κB activation appears to play a critical role in early BM development, we studied the effect of individual bile salts at neutral pH and pH 4, to identify the bile salts that impact upon NF-κB activation without causing cell death. The bile salts used reflected those observed in GORD (Kauer et al. 1997). Informed by these results, we repeatedly exposed our oesophageal model to taurochenodeoxycholate (TCDC) with and without acid and examined changes in the epithelial gene expression profile.
Materials and methods
All materials were purchased from Sigma-Aldrich (Dorset, UK) unless otherwise stated.
HET-1A cell culture
HET-1A cells (ATCC-LGC, Middlesex, UK) were cultured in serum-free BRFF-EPM2 media (Axxora, Nottingham, UK). The medium was replaced every 3–4 days, and cells were passaged at 80% confluency.
Effect of pulsatile exposure to bile salts on HET-1A cells
HET-1A cells were seeded in the inner 60 wells of a 96 well plate (3 × 104/well). After 24 h, cells were exposed to bile salts at pH 7.4 or pH 4, with two 10 min treatments per day for 3 days. The bile salts used (13 μM–1.3 mM) were glycocholate (GC), glycodeoxycholate (GCDC), glycochenodeoxycholate (GDC), taurocholate (TC), TCDC and taurodeoxycholate (TDC). Cells were then cultured for 24 h before determining cell metabolic activity by 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenyltetrazolium bromide (MTT-ESTA) assay.
MTT-ESTA assay
Cells were incubated with MTT-ESTA (0.5 mg/ml in PBS, 40 min, 37 °C). Acidified isopropanol (200 μl) was added and absorbance measured at 540 nm with a protein reference of 630 nm subtracted. Activity was calculated as a percentage of the control.
High content analysis of NF-κB translocation
Cells were exposed to bile salts (400 μM) at pH 7.4 or pH 4, with two 10-min treatments per day for 3 days. The bile salts used were as described above. Cells were cultured for a further 24 h, fixed in 1% formalin (30 min), washed ×2 in PBS and stored in PBS (4 °C) prior to imaging. Positive controls were treated with IL1-α (10 ng/ml). Immunofluorescent staining of NF-κB p65 subunit and high content analysis was performed by Imagen Biotech (Cheshire, UK). Six out of a possible 50 fields of view were analysed at random per well. Total fluorescence and nuclear–cytoplasmic ratios were calculated.
Primary human oesophageal squamous epithelial cell and human oesophageal fibroblast isolation and culture
Oesophageal tissue samples were taken from disease-free, background tissue proximal to any macroscopic pathology. Cells were isolated and cultured as described previously (Green et al. 2010). Human oesophageal squamous cells were used between passage 1 and 4, while HOFs were used between passages 4 and 9.
Ethical approval
Oesophageal tissue samples were obtained with informed consent and appropriate ethical approval (Human Tissue Bank licence number 12179 and SSREC 165/03) from patients undergoing gastric or oesophageal surgery.
Production of 3D oesophageal mucosal model
The production of a 3D oesophageal mucosal model has been described previously (Green et al. 2010). Briefly, porcine oesophagus was dissected along the lamina propria and the muscularis propria discarded. The epithelium was removed by incubating tissue pieces (5 cm2) in 1 M NaCl, 200 IU/ml penicillin, 200 μg/ml streptomycin, 1.25 μg/ml amphotericin B (72 h, 37 °C) and the tissue sterilized in glycerol solution (80%, 90% for 24 h each, 100% for >4 months). Prior to use, the de-epithelialized oesophagus (DEO) was re-hydrated in PBS and sterility tested (DMEM, 24 h, 37 °C).
Human oesophageal fibroblast cells (1 × 105) in 100 μl fibroblast culture medium (Green et al. 2010) were seeded on the DEO submucosal surface and medium replaced every 3–4 days. After 1 week, HOS cells (1 × 106) were seeded on the mucosal side in 100 μl Composite Medium I (Green et al. 2010). After 2 days, medium was switched to Composite Medium II, and after a further 2 days, the composites lifted to an air–liquid interface in Composite Medium III. The medium was replaced every 3–4 days and the composites maintained at air–liquid interface for 16 days.
Practical limitations prevent the model incorporating fibroblasts and epithelial cells from the same patient; consequently fibroblasts from one patient were used throughout to limit variability arising from epithelial/stromal interactions (Green et al. 2011).
Acid and taurochenodeoxycholate treatment of 3D cultures
From 5th day at air–liquid interface, the composites were exposed to simulated reflux twice a day for 11 days. Cultures were submerged (10 min, 37 °C) in one of four solutions – Composite Media III at pH 7.4 +/− 400 μM TCDC or pH 4 +/− 400 μM TCDC. Cultures were rinsed in PBS and returned to air–liquid interface.
The epithelial cells used in this study were obtained from three different patients, (Patients 1, 2 and 3). Two separate experiments (A and B) were performed using the cells from each patient to produce technical replicates. In a single experiment, four oesophageal composites were grown using the cells from one patient, and each was exposed to a different insult (control, pH 4, TCDC or TCDC pH 4). This regime resulted in 24 separate composites, referred to by patient number, experiment and treatment (i.e. 1A control through to 3B TCDC pH 4).
After the final treatment, each culture was bisected, half was fixed (10% formalin) for histological/immuno-histochemical study, and the other was submerged in Medium III and the epithelium removed by gentle scraping. The epithelial cells were pelleted, washed in PBS and stored (−80 °C) prior to RNA isolation. Total RNA was isolated using the mirVana RNA isolation kit (Ambion, Austin, TX, USA) following the manufacturer's procedure.
Histology and immunohistochemistry
Formalin fixed samples were wax embedded, sectioned and stained with haematoxylin and eosin (H&E). All immunostaining was performed with the Dako Autostainerplus using the Flex+ visualization kit and DAB. The following primary antibodies were used: Ki67 (Clone MIB1, 1:100; Dako, Ely, UK); claudin-4 (Clone 3E2C1, 1:200; Invitrogen, Paisley, UK), p63 (Clone 7JUL, 1:25; Leica, Milton Keynes, UK) and p27 (Clone SX53G8, 1:100; Dako), P1-HNF4α (Clone K9218, 1:200; Abcam, Cambridge, UK), P2-HNF4α (Clone H6939, 1;100; R&D Systems, Abingdon, UK), broad spectrum HNF4 (NB100-92338, 1:100, Novus Biologicals, Cambridge, UK), HNF4γ (Clone B6502A, 1:100; Abcam). Heat-induced epitope retrieval was performed for all antibodies using the Dako PT module with high pH buffer for claudin-4, p63, P1-HNF4α and HNF4γ and low pH buffer for Ki67, p27, P2-HNF4α and broad spectrum HNF4. Mouse IgG and TBS buffer were used as controls. Samples were counterstained with haematoxylin.
Affymetrix analysis
Twenty-four RNA samples were isolated and named, as described for the composites. RNA quality (2100 Bioanalyzer, RNA 6000 Nano LabChip; Agilent, Santa Clara, CA, USA) and quantity (NanoDrop 1000 spectrophotometer) were assessed. Equal amounts of RNA were combined as follows: 1A control, 2A control and 3A control were pooled to produce A control while 1B control, 2B control and 3B control produced the duplicate B control. This was performed for all four treatments, resulting in eight samples (A and B duplicates) for Affymetrix analysis. Pooling was performed to reduce the impact of interpatient variability.
Single-stranded DNA was produced from 150 ng of pooled total RNA, according to the manufacturer's instructions (Ambion WT Expression kit; Life Technologies, Paisley, UK). The Affymetrix sense target labelling protocol was used to fragment 5.5 μg single-stranded DNA and terminal deoxynucleotide transferase to end-label fragments with biotin. End-labelling was verified by gel shift assay as described in the Affymetrix protocol. At each stage, Agilent Bioanalyser and NanoDrop checked product quality and quantity.
DNA was applied to Human Exon 1.0 ST GeneChips (Affymetrix, High Wycombe, UK), following manufacturer's protocols. Following stringency washes, chips were stained and scanned and Expression Console (Affymetrix) used for quality control metrics. The data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and is accessible through GEO Series Accession Number GSE45380. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45380).
Bioinformatics
Downstream analysis of Affymetrix data was performed using Propagating Uncertainty in Microarray Analysis (puma) software (Pearson et al. 2009; Liu et al. 2013).
A posterior probability of positive log ratio (PPLR) >0.7 or <0.3 indicated upregulation or downregulation respectively. Posterior probability of positive log ratio utilizes a Bayesian approach to calculate the probability of differential gene expression based on the observed intensity and within-chip variance of each probe set (Liu et al. 2006).
Enrichment analysis was performed using the software packages Metacore™ (GeneGo, St Joseph, MO, USA) and DAVID Bioinformatics Resources 6.7 (Huang et al. 2009a,b) for canonical pathway and Gene Ontology (GO) analysis, and Metacore™ and Broad GSEA (Mootha et al. 2003; Subramanian et al. 2005) for transcription factor analysis. For Broad GSEA analysis, Ensembl Gene Ids generated by puma were first converted to Associated Gene Name by Ensembl Biomart (Kinsella et al. 2011). The Partek Genomics Suite was used to analyse alternative splicing events. The probeset intensities for each exon were normalized to the mean intensities for the gene as a whole.
qRT-PCR
250 ng total RNA from the Universal RNA and the samples was converted to cDNA using the QuantiTect system (Qiagen, Manchester, UK) following the manufacturer's instructions. Primers were designed and optimized for HNF4α exon1 (Forward: GCCATGGTCAGCGTGAAC, Reverse: CGTTGAGGTTGGTGCCTTCT) and human actin (Forward: TCCCCCAACTTGAGATGTATGAAG, Reverse: AACTGGTCTCAAGTCAGTGTACAGG). Reaction conditions were as follows, in duplicate: cDNA 12.5 ng (1 μl), Agilent Brilliant III SyBr green (5 μl), 1 μl primers (300 nM) and nuclease-free water to 10 μl; in PCR conditions, after initial denaturation (95 °C, 10 min), products were amplified (40 cycles – 95 °C, 15 s and 60 °C, 1 min). The relative amounts of exon 1D were compared to actin and the control results set to 1.
Statistical analysis
Cell metabolic activity, NF-κB activation and histology results were assessed by two-way anova with Bonferroni post-test. The overlap for the differentially regulated gene lists was compared using chi-squared test. Alternative splicing results were analysed by ancova.
Results
Cell metabolic activity and NF-κB activation in HET-1A cells following brief, pulsatile exposure to bile salts in the presence and absence of acid
Ten-minute exposure, twice a day for 3 days, to bile salts at pH 7.4 caused no significant decrease in HET-1A metabolic activity at the range of concentrations tested. However, at pH 4, 400 μM GDC and GCDC produced a significant decrease, suggestive of significant toxicity (Figure1). The taurine conjugates were without effect, at concentrations up to 1 mM, at pH 7.4 and pH 4.
Figure 1.
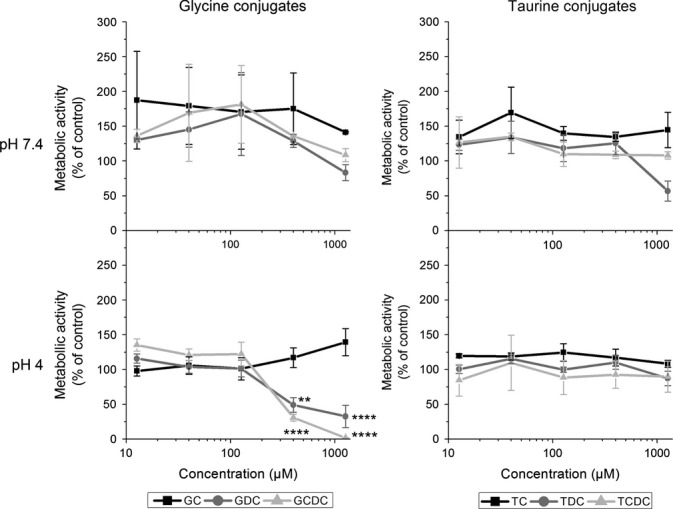
Effect of bile salts on metabolic activity of HET-1A cells. Cells were subjected to pulsatile exposure to individual bile salts at pH 7.4 and pH 4. Metabolic activity was measured as a percentage of control cells treated with pH 7.4 media. Differences were calculated between treatments and control by one-way anova and Bonferroni post-test. Significant decreases in metabolic activity are indicated by **(P < 0.01) and ****(P < 0.0001).
Subsequent experiments used 400 μM bile salts as a biologically relevant concentration. HET-1A cells exposed to pulsatile insults were analysed for NF-κB nuclear translocation. No significant change in the total amount of NF-κB within the cells was detected following any treatment at pH 7.4 (Figure2a). There was a significant decrease in NF-κB for all samples at pH 4; however, when compared to the pH 4 acid treatment alone, no significant differences were detected, indicating reduced levels were due to acid exposure alone.
Figure 2.
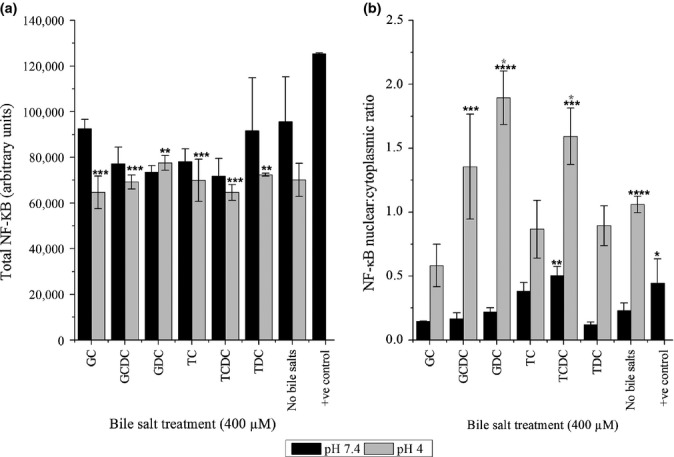
The effect upon (a) total levels of NF-κB and (b) NF-κB localization from pulsatile exposure of HET-1A cells to bile salts in the absence and presence of acid. Results from all treatments were compared to the pH 7.4 control using anova with Bonferroni post-test. Statistical significance is indicated by **(P < 0.01), ***(P < 0.001) and ****(P < 0.0001). Results from bile treatment at pH 4 were also compared to the pH 4 only control using anova with Bonferroni post-test (shown in grey *P < 0.05).
In contrast, significant increases were observed in the nuclear–cytoplasmic ratio (Figure2b) indicative of NF-κB translocation and activation. At pH 7.4, the ratio was low (<0.25), but pulsatile exposure to TCDC induced a doubling in nuclear translocation (0.5, P < 0.01). At pH 4, the ratio increased to ∼1.0 without bile salts (P < 0.0001) and was further increased by GDC and TCDC (1.9 and 1.6, respectively, P < 0.05).
Taurochenodeoxycholate was the only bile salt that induced NF-κB activation at pH 7.4 and pH 4 without a detrimental effect upon metabolic activity and so became the focus for the remaining experiments.
Gene expression profiling
Gene expression data for the oesophageal epithelium were obtained following pulsatile exposure to acid and/or TCDC. As we sought changes in gene expression patterns rather than individual, highly dysregulated genes, relatively liberal inclusion thresholds were applied at this stage, and all genes with a PPLR value of <0.3 or >0.7 were catalogued. Microarray data were obtained from the Human Exon 1.0 ST GeneChip, which provides data down to the exon level, but analysis primarily focussed upon gene level changes.
Propagating Uncertainty in Microarray Analysis identified ∼300 differentially regulated genes for each treatment, with 673 genes identified as differentially expressed with one or more of the treatments and a large and significant (P < 0.0001) overlap of genes affected between treatments (Figure3). Enrichment analysis examined the relationships between these genes. This was performed on the combined list of 673 differentially regulated genes and the individual gene lists for each treatment.
Figure 3.
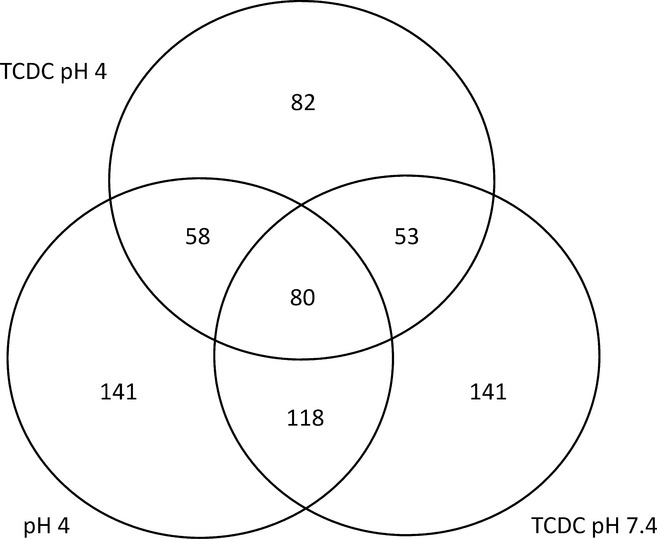
Venn diagram with values indicating the number and extent of overlap for the differentially expressed genes in the epithelium of the oesophageal model following pulsatile exposure to acid and/or taurochenodeoxycholate compared to the control. Results are given for posterior probability of positive log ratio <0.3 or >0.7.
Metacore™ and DAVID analysis of the dysregulated genes following acid, bile or acidified bile treatment highlighted enriched canonical pathways. Table1, ranked by lowest P-value (P < 0.05), indicates those pathways identified in more than one data set. A list of GO processes (Table2) over-represented in the differentially expressed lists and identified in more than one data set was also produced and ranked by lowest P-value (P < 0.005), together with analysis of the broad GO categories of the genes affected by the treatments (Figure4). Pulsatile exposure to acid or TCDC alone resulted in dysregulation of genes involved in cellular processes (GO:0009987), while TCDC exposure also resulted in changes in metabolic processes within the cell (GO:0008152). There was also a lesser impact upon genes involved in cellular organization (GO:0016043) and establishment and maintenance of cellular localization (GO:0051179). Following pulsatile exposure to acidified TCDC, 50% of the identified genes showing changes in expression were in the GO development process class (GO:0032502), while cell component organization (GO:0016043) and biological adhesion (GO:0022610) were also represented to a lesser extent. Significantly over-represented targets and transcription factors with over-represented interacting genes or proteins, identified in more than one data set, are listed (Table3, ranked by lowest P-value – P < 0.05).
Table 1.
Canonical pathways showing most significant over-representation in the differentially expressed gene list, ranked by lowest P-value (P < 0.05)
| Gene Set Name | P-value | ||||
|---|---|---|---|---|---|
| DAVID | Metacore™ | ||||
| All | pH 4 | TCDC | TCDC pH 4 | All | |
| Cadherin-mediated cell adhesion (Metacore) REACTOME adherens junctions interactions (DAVID) |
0.0533 | 0.0595 | 0.00007 | ||
| KEGG steroid biosynthesis | 0.0006 | 0.0253 | 0.00014 | ||
| WNT signalling pathway | 0.0561 | 0.03619 | 0.02416 | 0.0007 | |
| REACTOME transformation of lanosterol to cholesterol | 0.0112 | 0.0366 | 0.0030 | ||
| REACTOME metabolism of lipids and lipoproteins | 0.0086 | 0.03261 | 0.05383 | ||
| BIOCARTA IFN γ signalling pathway | 0.0091 | 0.08079 | |||
| BIOCARTA internal ribosome entry pathway | 0.0125 | 0.09364 | 0.04781 | ||
| BIOCARTA mTOR signalling pathway | 0.0300 | 0.05247 | 0.01395 | ||
| ST interleukin 13 pathway | 0.0438 | 0.0181 | |||
| BIOCARTA proteasome pathway | 0.0653 | 0.0197 | |||
| REACTOME cholesterol biosynthesis | 0.0833 | 0.0259 | |||
| ST Jak-STAT pathway | 0.0703 | 0.0298 | |||
| REACTOME gene expression | 0.0348 | 0.05401 | |||
| BIOCARTA role of ran in mitotic spindle regulation | 0.0362 | 0.0366 | |||
| REACTOME metabolism of proteins | 0.0708 | 0.04353 | |||
TCDC, taurochenodeoxycholate.
Table 2.
Gene Ontology terms showing most significant over-representation in the differentially expressed gene list, ranked by lowest P-value (P < 0.005)
| Gene Ontology Term | P-value | ||||
|---|---|---|---|---|---|
| DAVID | Metacore™ | ||||
| All | pH 4 | TCDC | pH 4 TCDC | All | |
| Regulation of protein kinase cascade | 0.0020 | 0.00014 | |||
| Regulation of cell adhesion (DAVID) Integrin-mediated cell matrix adhesion (Metacore) |
0.00054 | 0.00252 | 0.00349 | 0.04659 | 0.0019 |
| Peptidyl-tyrosine phosphorylation | 0.0025 | 0.00037 | |||
| Epithelial cell differentiation | 0.00054 | 0.01052 | 0.013748 | ||
| Epithelium development | 0.00087 | 0.03398 | 0.01628 | 0.09239 | |
| Proteolysis (DAVID) Proteolysis in cell cycle and apoptosis (Metacore) |
0.01442 | 0.04585 | 0.0009 | ||
| ATP metabolic process | 0.0015 | 0.00390 | |||
| Positive regulation of I-kappaB kinase/NF-kappaB cascade | 0.0039 | 0.00196 | |||
| Positive regulation of signal transduction | 0.0033 | 0.00218 | |||
| Purine ribonucleotide metabolic process | 0.0022 | 0.00363 | |||
Figure 4.
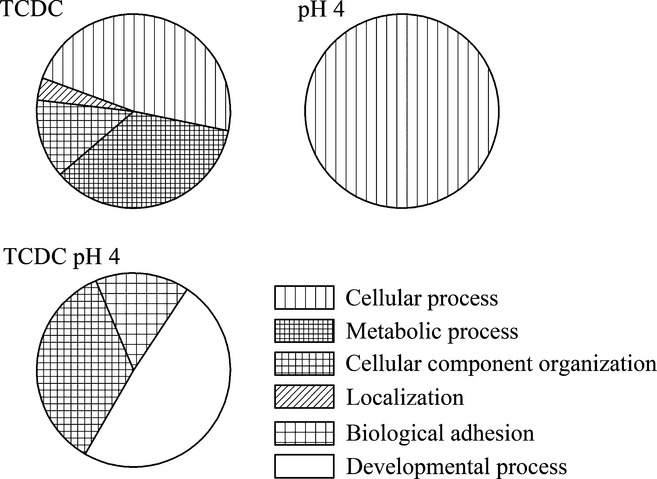
The broad Gene Ontology categories significantly represented by the dysregulated gene lists following pulsatile exposure of the model oesophagus to acid and/or taurochenodeoxycholate.
Table 3.
The transcription factors with over-represented targets or interacting genes and/or proteins in the list of dysregulated genes, ranked by lowest P-value (P < 0.05)
| Transcription factor | P-value | |||||
|---|---|---|---|---|---|---|
| Broad GSEA | Broad GSEA | Metacore™ | ||||
| All | pH 4 | TCDC | pH 4 TCDC | All – over-represented targets | All – over-represented interacting genes/proteins | |
| HNF4α | 0.00218 | 0.000732 | 0.00000382 | |||
| p65 NF-kB subunit | 0.000912 | |||||
| SF1 | 0.0040 | 0.0115 | 0.0295 | |||
| PTF1A | 0.0324 | 0.0231 | 0.0560 | 0.0245 | ||
| STAT1 | 0.1020 | 0.0255 | ||||
| TCF11 < br > MAFG | 0.0697 | 0.0306 | ||||
TCDC, taurochenodeoxycholate.
Similarities were observed in the processes, and transcription factors enriched following the three treatments. All treatments resulted in enrichment in cell adhesion, epithelium development and target enrichment for PTF1A. There were further similarities between the responses to TCDC or acid when applied separately, including epithelial differentiation and Wnt signalling. The responses to acid alone and bile alone were more similar than the response to acidified bile, where a greater number of transcription factor targets were enriched.
qRT-PCR quantification
Alternative splicing analysis revealed probeset ID 3886468 was upregulated in pH 4 and/or TCDC treatments. This corresponds to exon 1D in 5′ HNF4α UTR and is used when the P1 promoter is activated. qRT-PCR analysis was performed to confirm these results. Under these conditions, carrying out a ddCt calculation (Livak & Schmittgen 2001), the concentration of exon 1D increased following exposure to acid alone or in conjunction with TCDC (Figure5), although it did not reach statistical significance (with further analysis limited by material availability).
Figure 5.
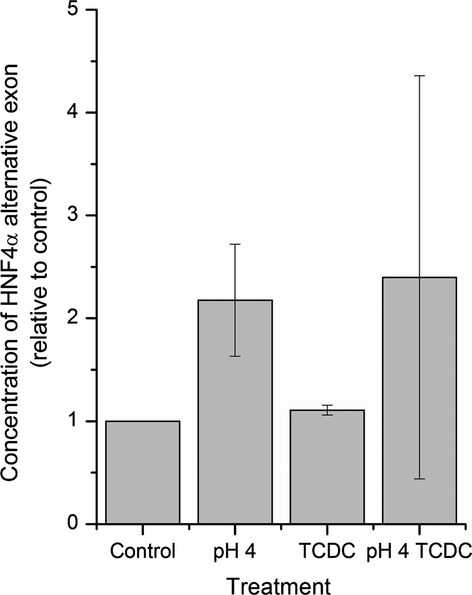
The concentrations of the alternative part of the 5′UTR of HNF4α, exon 1, which is driven by P1 promoter relative to the control, determined by qRT-PCR. Results are normalized against the pH 7.4 treated control.
Histology and immunohistochemistry of 3D composites following pulsatile exposure to taurochenodeoxycholate in the presence and absence of acid
Following 11-day pulsatile exposure to TCDC and/or acid, the epithelial morphology was compared to the control. In all cases, there was a mature, multilayered epithelium, with no significant differences in morphology (Figure6). Furthermore, no significant differences were observed in the extent of Ki67, claudin-4, p63 and p27 staining between the various treatments (results not shown).
Figure 6.
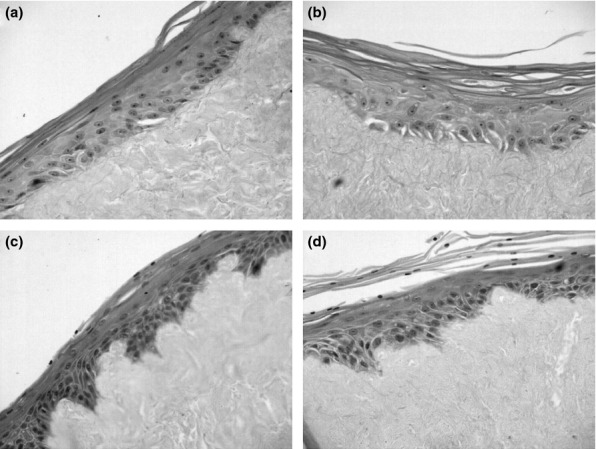
H + E staining of organotypic models following 2 weeks pulsatile (2 × 10 min per day) exposure to taurochenodeoxycholate (TCDC) +/− acid. (a) was exposed to pH 7.4, (b) was treated with TCDC, (c) received pH 4 treatment and (d) pH 4 and TCDC.
HNF4 expression was assessed in samples of clinically confirmed BM, adjacent normal oesophagus and composites (Figure7). Positive nuclear staining was observed for the metaplastic glandular elements of BM, the adjacent normal squamous epithelia and all composite epithelia with a broad spectrum anti-HNF4 antibody. The HNF4γ-specific and isoform 1–4 HNF4α-specific antibodies both produced positive nuclear staining in BM tissue but no staining in normal tissue or composites. In contrast, the isoform 7–9 specific HNF4α antibody produced positive nuclear staining in the metaplastic glandular elements of BM and discernable positive nuclear staining in the majority of adjacent normal squamous epithelia (six of seven), although there was variation in staining intensity and distribution between cases. In many cases, the composites also showed clear nuclear staining, generally within the basal layers. There was, however, no consistent relationship between intensity and treatment, with positive staining observed in both reflux-exposed and control samples.
Figure 7.
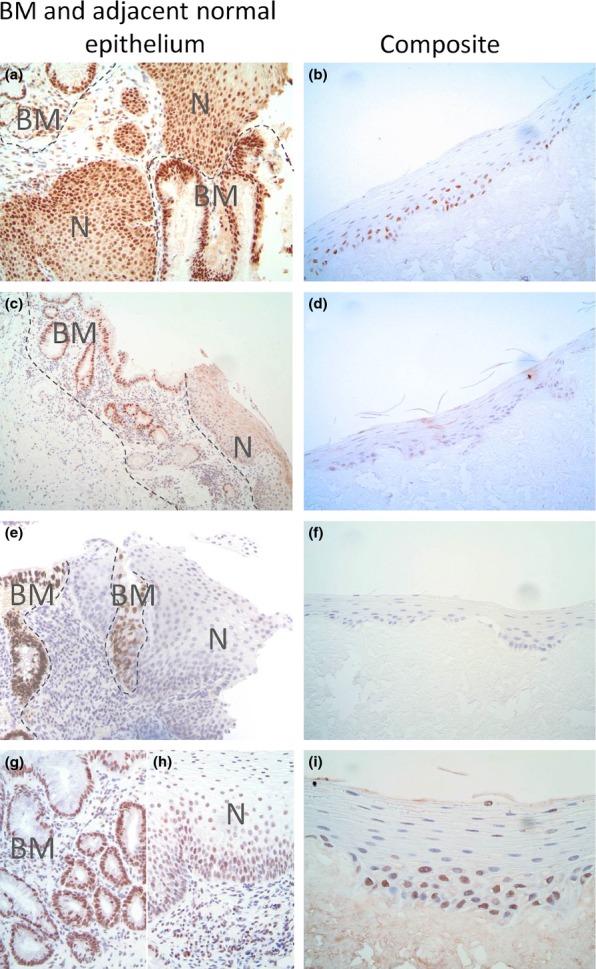
HNF4 immunohistochemical staining of clinically defined Barrett's metaplasia (BM) and adjacent normal oesophageal squamous epithelia (left) and composites (right). The antibodies used were anti-HNF4α/γ (a, b), anti-HNF4γ (c, d), anti-P1-HNF4α, specific for isoforms 1–4 (e, f) and anti- P2-HNF4α, specific for isoforms 7–9 (g, h and i). Regions of metaplasia (BM) and normal squamous epithelium (N) within the tissue are labelled. Where both BM and normal tissue are present in the same image, the BM boundary is indicated with a grey dotted line.
Discussion
The aim of this study was to investigate how medium-term pulsatile exposure to specific reflux components affected gene expression in a 3D model of the normal human oesophageal mucosa. Gastro-oesophageal refluxate is a cocktail of bile salts, acid, proteolytic enzymes and lysolecithin, all of which could potentially influence BM development, although studies usually focus upon bile and acid. As the 3D model does not facilitate the parallel testing of multiple compounds, preliminary work determined which bile salt should be applied to the 3D model.
Preliminary 2D experiments
We first used cells grown in 2D culture to devise a simulated reflux treatment likely to lead to changes in cell physiology. Taurochenodeoxycholate had the greatest impact upon NF-κB translocation in both neutral and acid conditions without a significant reduction in cell metabolic activity and became the focus for the remainder of the study. We did not investigate gene expression changes in the immortalized HET-1A cells; they do not recapitulate normal differentiation (Green et al. 2010), and changes in gene expression would bear little relevance to BM development.
Our identification of TCDC is in keeping with nutritional, epidemiological and experimental data pointing to a specific role for taurine conjugates in BM pathogenesis, with a link between the ‘western’ high fat diet and BM/OAC. Dietary sources of taurine are almost exclusively animal-derived; Wistar rats fed a high animal-fat diet, following surgery to induce reflux, have more taurine-conjugated bile salts and a higher incidence of BM and dysplasia (Chen et al. 2007); BM and severe oesophagitis patients have significantly greater taurine-conjugated concentrations in their gastric reflux (Nehra et al. 1999).
Gene expression changes in a 3D model of oesophageal epithelium following pulsatile exposure to taurochenodeoxycholate in the presence and absence of acid
Although mixed reflux is most common in GORD patients, it can also occur as acid only or bile only reflux (Hak et al. 2008), and consequently 3D experiments were carried out using acid and TCDC separately and in combination.
There is little consensus on the significance of reflux frequency and duration upon BM development, but total exposure time rather than frequency may be the most critical factor (Johnsson & Joelsson 1988). We therefore employed two 10 min exposures daily to provide 1.4% total exposure time, which is the same order of magnitude described in some studies (Richter et al. 1992).
Following 11-day reflux exposure, there were no observable morphological changes in the epithelium, such as columnar metaplasia, associated with BM. This was of little surprise following this relatively short period of reflux. However, by analysing changes in gene expression in the reflux-exposed normal epithelium, we sought to identify the very earliest events that occur in BM pathogenesis prior to phenotypic changes.
Enrichment analysis of the changes in genes expressed following reflux identified multiple gene sets related to cell signalling, inflammation, proliferation, differentiation and cell adhesion. Similar results were seen with oesophageal cell lines exposed to low pH, where changes in genes involved in inflammation, stress response, proliferation and differentiation were observed (Duggan et al. 2006).
The dysregulated gene lists for each treatment show a large amount of overlap, illustrating the systematic nature of the gene lists generated. However, GO analysis also revealed clear differences in the broad categories of the genes affected. Pulsatile exposure to TCDC alone induced changes in metabolic processes, which is unsurprising as bile acids play an important role in the regulation of metabolism (Staels & Fonseca 2009). With acidified TCDC, significant changes were observed in the developmental process category. Progression to BM is a metaplastic event characterized by the aberrant differentiation of cells and may involve genes normally active during development. Our data suggest mixed reflux has a greater impact upon developmental processes and may explain why patients with mixed reflux are more likely to develop BM than those with other reflux forms (Oh et al. 2006).
Analysis of the separate treatments and the combined list of 673 dysregulated genes highlighted a number of important areas, discussed below.
Simulated reflux produces changes in the development of the oesophageal epithelium and epithelial cell adhesion
All treatments impacted upon the expression of genes involved in epithelial development. Cell adhesion was also enriched in all cases, but only became a significant GO category following acidified TCDC treatment, suggesting an increased effect with mixed reflux. Tselepis et al. observed changes in intracellular adhesion with progression along the metaplasia–dysplasia sequence (Tselepis et al. 2000), and it is proposed that changes in cell adhesion may permit movement of clonally derived cells along the epithelium to create a cellular field change (Jenkins et al. 2002).
NF-κB activation is induced by pulsatile acid exposure
Positive regulation of the NF-κB cascade (DAVID) and interacting genes and proteins for the p65 subunit of NF-κB (Metacore™) underscore NF-κB activation in the model epithelium. Various reflux components have been suggested as candidates for NF-κB activation in BM. Our data indicate that acid reflux is sufficient to induce activation in the oesophageal model, but exposure to TCDC, at either pH, is not. Taurochenodeoxycholate was selected because it activated NF-κB in 2D cultures; however, as previously demonstrated in our laboratory (Sun et al. 2006), cells are significantly more resistant to insult in 3D co-culture compared to 2D monoculture. This is evidence of the necessity to evaluate results obtained in 2D using more biologically relevant 3D experimental models.
Wnt signalling is activated by exposure to acid or taurochenodeoxycholate alone
The Wnt signalling pathway is significantly overrepresented in the dysregulated genes following acid or TCDC exposure. Activated Wnt signalling is critical for normal intestinal development while oesophagus and stomach formation may be due, in part, to reduced Wnt signalling (Gregorieff et al. 2004). A recent publication suggests Wnt may play a role in the transformation from squamous to glandular mucosa (Moyes et al. 2012), and an increase in intestinal epithelial proteins was observed in an oesophageal model following increased Wnt activity (Kong et al. 2011). Increased Wnt signalling may therefore recapitulate the events that normally occur during development, promoting a switch from oesophageal to intestinal epithelium.
Developmental transcription factor targets are enriched by simulated reflux
Targets for the pancreas-specific transcription factor, PTF1A, are enriched in all treatments, although PTF1A expression levels do not change. PTF1A has a role in the development switch between intestinal and pancreatic fates (Kawaguchi et al. 2002) and its potential involvement in the switch from the normal oesophageal epithelium to the columnar; intestinal phenotype is worthy of further investigation.
HNF4α targets are also enriched following exposure to acidified bile, suggesting an increase in HNF4α transcriptional activity with mixed reflux. HNF4α plays a role in establishing the intestinal epithelial phenotype (Babeu et al. 2009); its induction results in intestinal marker expression in a mouse oesophageal model (Colleypriest et al. 2011), and it is enriched in BM (Wang et al. 2009). Furthermore, there is a putative HNF4α binding site upstream of CDX2, with HNF4α activating CDX2 in HeLa cells (Benahmed et al. 2008). CDX1/2 transcription factor targets were not enriched in our study, suggesting a very early role for HNF4α, potentially upstream of CDX2, in BM pathogenesis.
HNF4α isoform expression may change during Barrett's metaplasia development
HNF4α exists in at least nine isoforms, generated by both alternative promoter usage and C-terminal splicing (Hansen et al. 2002; Huang et al. 2009c). Isoforms α1 to α6 are initiated by the P1promoter, while promoter P2 initiates isoforms α7 to α9.
Our data indicate that while HNF4α targets are enriched following acidified bile exposure, HNF4α expression levels do not change. However, an alternative splicing event occurs following exposure to acid and/or bile, with upregulation of the exon encoding the P1 promoter region of HNF4α. This is in agreement with results obtained by qRT-PCR data (albeit not reaching significance).
HNF4α has been identified in BM (Wang et al. 2009), human oesophageal metaplastic tissue (Piessen et al. 2007) and gastric intestinal metaplasia (Takano et al. 2009) but has not previously been reported in normal squamous oesophageal epithelium. Our immunohistochemistry demonstrates P2-HNF4α isoforms are, in many instances, detected in both normal and BM tissue, while BM development is associated with activation of the P1 isoforms.
Conclusion
This study provides the first report on the impact of exposure to particular bile salts upon gene expression in tissue engineered human oesophageal epithelium, indicating possible changes in gene expression in the very earliest stages of BM pathogenesis. We have identified a number of potential targets for further research to validate these results and identify the mechanisms by which these changes result in BM development.
Significant changes in gene expression were observed in a number of pathways including
cell adhesion
epithelial development and differentiation
Wnt signalling
NF-κB activation
A high degree of overlap was observed between the dysregulated gene lists following acid, bile and acidified bile treatment, but only acidified bile produced significant changes in expression of genes within the GO developmental process category.
Two transcription factors with roles in organ development, PTF1A and HNF4α, also had enriched targets following reflux exposure and are worthy of further investigation in the switch to intestinal phenotype. P2-HNF4α isoforms are detectable in normal oesophageal epithelium, but the more transcriptionally active P1 isoforms (Eeckhoute et al. 2003) are observed in BM. We propose that the expression and activation of P1 HNF4 isoforms, driven by exposure to reflux, including taurine-conjugated bile salts, could be an early causal event in the development of columnar metaplasia in Barrett's oesophagus, upstream of CDX2 activation.
This preliminary study highlights new areas for further investigation and provides a biologically relevant experimental model of the normal human oesophagus for further in-depth analysis of other factors, including reflux composition and frequency, on the pathogenesis of BM.
Acknowledgments
We are grateful to Mr. Roger Ackroyd, Mr. Andrew Wyman and Mr. Chris Stoddard, Consultant Surgeons at Sheffield Teaching Hospitals NHS Foundation Trust, for their help in acquiring oesophageal tissue samples and their support for our work. We also thank Professor Magnus Rattray in Faculty of Life Sciences, University of Manchester, for his help and support with the puma analysis of the microarray data. We are grateful to Jennifer Stonard and Danielle McLuskey from the Department of Histopathology, Sheffield Teaching Hospitals NHS Foundation Trust for their efforts in the immunohistochemistry work. This study was supported by grants from the Bardhan Research and Education Trust (BRET) and Yorkshire Cancer Research (YCR).
Conflict of interests
The authors declare that they have no conflicting interests.
References
- Babeu JP, Darsigny M, Lussier CR, Boudreau F. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G124–G134. doi: 10.1152/ajpgi.90690.2008. [DOI] [PubMed] [Google Scholar]
- Benahmed F, Gross I, Gaunt SJ, et al. Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology. 2008;135:1238–1247. doi: 10.1053/j.gastro.2008.06.045. 1247. [DOI] [PubMed] [Google Scholar]
- Chen KH, Mukaisho K, Sugihara H, Araki Y, Yamamoto G, Hattori T. High animal-fat intake changes the bile-acid composition of bile juice and enhances the development of Barrett's esophagus and esophageal adenocarcinoma in a rat duodenal-contents reflux model. Cancer Sci. 2007;98:1683–1688. doi: 10.1111/j.1349-7006.2007.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleypriest BJ, Farrant JM, Slack JM, Tosh D. Hepatocyte Nuclear Factor 4a (HNF4a) provokes intestinal genes in squamous oesophageal cells. Gut. 2011;60:A171–A172. [Google Scholar]
- Debruyne PR, Witek M, Gong L, et al. Bile acids induce ectopic expression of intestinal guanylyl cyclase C Through nuclear factor-kappaB and Cdx2 in human esophageal cells. Gastroenterology. 2006;130:1191–1206. doi: 10.1053/j.gastro.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Duggan SP, Gallagher WM, Fox EJ, Abdel-latif MM, Reynolds JV, Kelleher D. Low pH induces co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis. 2006;27:319–327. doi: 10.1093/carcin/bgi211. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Moerman E, Bouckenooghe T, et al. Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology. 2003;144:1686–1694. doi: 10.1210/en.2002-0024. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sapinoso LM, Zhang H, et al. Altered expression of TFF-1 and CES-2 in Barrett's Esophagus and associated adenocarcinomas. Neoplasia. 2005;7:407–416. doi: 10.1593/neo.04715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LI, Esteves GH, Carvalho AF, et al. Expression profile of malignant and nonmalignant lesions of esophagus and stomach: differential activity of functional modules related to inflammation and lipid metabolism. Cancer Res. 2005;65:7127–7136. doi: 10.1158/0008-5472.CAN-05-1035. [DOI] [PubMed] [Google Scholar]
- Green N, Huang Q, Khan L, et al. The development and characterization of an organotypic tissue-engineered human esophageal mucosal model. Tissue Eng. Part A. 2010;16:1053–1064. doi: 10.1089/ten.TEA.2009.0217. [DOI] [PubMed] [Google Scholar]
- Green NH, Huang Q, Corfe BM, Bury JP, MacNeil S. NF-kappaB is activated in oesophageal fibroblasts in response to a paracrine signal generated by acid-exposed primary oesophageal squamous cells. Int. J. Exp. Pathol. 2011;92:345–356. doi: 10.1111/j.1365-2613.2011.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Grosschedl R, Clevers H. Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(-/-)/Tcf1(-/-) embryos. EMBO J. 2004;23:1825–1833. doi: 10.1038/sj.emboj.7600191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hak NG, Mostafa M, Salah T, et al. Acid and bile reflux in erosive reflux disease, non-erosive reflux disease and Barrett's esophagus. Hepatogastroenterology. 2008;55:442–447. [PubMed] [Google Scholar]
- Hansen SK, Parrizas M, Jensen ML, et al. Genetic evidence that HNF-1alpha-dependent transcriptional control of HNF-4alpha is essential for human pancreatic beta cell function. J. Clin. Invest. 2002;110:827–833. doi: 10.1172/JCI15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J, Levitsky LL, Rhoads DB. Novel P2 promoter-derived HNF4alpha isoforms with different N-terminus generated by alternate exon insertion. Exp. Cell Res. 2009c;315:1200–1211. doi: 10.1016/j.yexcr.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Jenkins GJ, Doak SH, Parry JM, D'Souza FR, Griffiths AP, Baxter JN. Genetic pathways involved in the progression of Barrett's metaplasia to adenocarcinoma. Br. J. Surg. 2002;89:824–837. doi: 10.1046/j.1365-2168.2002.02107.x. [DOI] [PubMed] [Google Scholar]
- Johnsson F. Joelsson B. Reproducibility of ambulatory oesophageal pH monitoring. Gut. 1988;29:886–889. doi: 10.1136/gut.29.7.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer WK, Peters JH, DeMeester TR, et al. Composition and concentration of bile acid reflux into the esophagus of patients with gastroesophageal reflux disease. Surgery. 1997;122:874–881. doi: 10.1016/s0039-6060(97)90327-5. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011 doi: 10.1093/database/bar030. 2011, bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Crissey MA, Stairs DB, Sepulveda AR, Lynch JP. Cox2 and beta-catenin/T-cell factor signaling intestinalize human esophageal keratinocytes when cultured under organotypic conditions. Neoplasia. 2011;13:792–805. doi: 10.1593/neo.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek PC, Nikiforuk A, Kania J, Raithel M, Hahn EG, Muhldorfer S. Activation of NFkappaB represents the central event in the neoplastic progression associated with Barrett's esophagus: a possible link to the inflammation and overexpression of COX-2, PPARgamma and growth factors. Dig. Dis. Sci. 2004;49:1075–1083. doi: 10.1023/b:ddas.0000037790.11724.70. [DOI] [PubMed] [Google Scholar]
- Liu X, Milo M, Lawrence ND, Rattray M. Probe-level measurement error improves accuracy in detecting differential gene expression. Bioinformatics. 2006;22:2107–2113. doi: 10.1093/bioinformatics/btl361. [DOI] [PubMed] [Google Scholar]
- Liu X, Gao Z, Zhang L, Rattray M. puma 3.0: improved uncertainty propagation methods for gene and transcript expression analysis. BMC Bioinformatics. 2013;14:39. doi: 10.1186/1471-2105-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Moyes LH, McEwan H, Radulescu S, et al. Activation of Wnt signalling promotes development of dysplasia in Barrett's oesophagus. J. Pathol. 2012;228:99–112. doi: 10.1002/path.4058. [DOI] [PubMed] [Google Scholar]
- Nancarrow DJ, Clouston AD, Smithers BM, et al. Whole genome expression array profiling highlights differences in mucosal defense genes in Barrett's esophagus and esophageal adenocarcinoma. PLoS One. 2011;6:e22513. doi: 10.1371/journal.pone.0022513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehra D, Howell P, Williams CP, Pye JK, Beynon J. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut. 1999;44:598–602. doi: 10.1136/gut.44.5.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DS, Hagen JA, Fein M, et al. The impact of reflux composition on mucosal injury and esophageal function. J. Gastrointest. Surg. 2006;10:787–796. doi: 10.1016/j.gassur.2006.02.005. [DOI] [PubMed] [Google Scholar]
- O'Riordan JM, Abdel-latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am. J. Gastroenterol. 2005;100:1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- Pearson RD, Liu X, Sanguinetti G, Milo M, Lawrence ND, Rattray M. puma: a Bioconductor package for propagating uncertainty in microarray analysis. BMC Bioinformatics. 2009;10:211. doi: 10.1186/1471-2105-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piessen G, Jonckheere N, Vincent A, et al. Regulation of the human mucin MUC4 by taurodeoxycholic and taurochenodeoxycholic bile acids in oesophageal cancer cells is mediated by hepatocyte nuclear factor 1alpha. Biochem. J. 2007;402:81–91. doi: 10.1042/BJ20061461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JE, Bradley LA, DeMeester TR, Wu WC. Normal 24-hr ambulatory esophageal pH values. Influence of study center, pH electrode, age, and gender. Dig. Dis. Sci. 1992;37:849–856. doi: 10.1007/BF01300382. [DOI] [PubMed] [Google Scholar]
- Staels B. Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32(Suppl 2):S237–S245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Jackson S, Haycock JW, MacNeil S. Culture of skin cells in 3D rather than 2D improves their ability to survive exposure to cytotoxic agents. J. Biotechnol. 2006;122:372–381. doi: 10.1016/j.jbiotec.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Takano K, Hasegawa G, Jiang S, et al. Immunohistochemical staining for P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha may complement mucin phenotype of differen-tiated-type early gastric carcinoma. Pathol. Int. 2009;59:462–470. doi: 10.1111/j.1440-1827.2009.02394.x. [DOI] [PubMed] [Google Scholar]
- Tselepis C, Perry I, Jankowski J. Barrett's esophagus: disregulation of cell cycling and intercellular adhesion in the metaplasia-dysplasia-carcinoma sequence. Digestion. 2000;61:1–5. doi: 10.1159/000007729. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhan M, Yin J, et al. Transcriptional profiling suggests that Barrett's metaplasia is an early intermediate stage in esophageal adenocarcinogenesis. Oncogene. 2006;25:3346–3356. doi: 10.1038/sj.onc.1209357. [DOI] [PubMed] [Google Scholar]
- Wang J, Qin R, Ma Y, et al. Differential gene expression in normal esophagus and Barrett's esophagus. J. Gastroenterol. 2009;44:897–911. doi: 10.1007/s00535-009-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong NA, Wilding J, Bartlett S, et al. CDX1 is an important molecular mediator of Barrett's metaplasia. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7565–7570. doi: 10.1073/pnas.0502031102. [DOI] [PMC free article] [PubMed] [Google Scholar]


