Abstract
Ultraviolet photodissociation (UVPD) mass spectrometry (MS) was used to characterize the sequences of proteins in native protein–ligand and protein–protein complexes and to provide auxiliary information about the binding sites of the ligands and protein–protein interfaces. UVPD outperformed collisional induced dissociation (CID), higher-energy collisional dissociation (HCD), and electron transfer dissociation (ETD) in terms of yielding the most comprehensive diagnostic primary sequence information about the proteins in the complexes. UVPD also generated noncovalent fragment ions containing a portion of the protein still bound to the ligand which revealed some insight into the nature of the binding sites of myoglobin/heme, eIF4E/m7GTP, and human peptidyl-prolyl cis–trans isomerase 1 (Pin1) in complex with the peptide derived from the C-terminal domain of RNA polymerase II (CTD). Noncovalently bound protein–protein fragment ions from oligomeric β-lactoglobulin dimers and hexameric insulin complexes were also produced upon UVPD, providing some illumination of tertiary and quaternary protein structural features.
INTRODUCTION
The macromolecular structures of protein and protein complexes determine their biological functions; therefore, understanding how proteins assemble and interact is pivotal to understand a myriad of cellular functions.1 X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy are time-tested protein structural analysis tools; however, it has become clear that a wider array of complementary technologies are needed to characterize native protein structures. The development of mass spectrometry (MS) for characterization of intact proteins has accelerated over the past two decades, and new advances have demonstrated the ability to lift proteins in native conformations and even noncovalent protein complexes into the gas phase for high accuracy mass assignment and structural analysis by a variety of MS/MS methods.2–5 Native mass spectrometry (native-MS) utilizes nanoelectrospray ionization (nanoESI) and native-like buffer solutions to facilitate the preservation of noncovalent interactions and facilitate the transfer of intact complexes into the gas phase.6,7 MS-based technologies have been used to characterize even very complex macromolecular assemblies, including the intact V-type ATP synthase8 and intact 18 MDa capsids of bacteriophage HK97.9
The use of tandem mass spectrometry (MS/MS) has afforded one of the most versatile means to characterize proteins and protein complexes. Several ion activation methods, including collisional activation, electron-based activation, and photoactivation, have been developed to disassemble protein complexes and characterize the constituents.8,10–28 A common outcome of activation of macromolecular assemblies is the disruption of noncovalent interactions, leading to separation of the protein constituents which provides information about the subunit composition of the complexes but not necessarily any sequence detail about the individual proteins. Collisional induced dissociation (CID) has been the standard activation method used for every type of MS/MS application. In the context of macromolecules, low energy CID typically causes disassembly of the complexes and removal of one or multiple subunits.10 This information is useful for confirming the stoichiometry of the complexes and revealing insight into quaternary structure.11 However, little sequence information is obtained upon CID of protein complexes, an outcome that is problematic for the examination of macromolecules containing large or unknown proteins or ones with post-translational modifications, especially as the frontier of interactomics accelerates.
Other higher energy and alternative MS/MS methods, such as higher kinetic energy beam type CID12 and higher energy collisional dissociation (HCD),13,14 surface induced dissociation (SID),15–17 electron capture dissociation (ECD),18–26 electron transfer dissociation (ETD),27 and photodissociation,28 have been explored to impart more sequence and structural information from intact proteins and protein complexes. Higher kinetic energy CID has been implemented on a modified quadrupole/time-of-flight instrument, offering the benefit of providing some sequence information about the individual proteins in protein complexes.12 A two-step activation/HCD method applied to protein complexes as large as 800 kDa13 allowed disassembly of the complexes and high accuracy mass detection of b/y sequence ions arising from backbone fragmentation of the constituent proteins. The sequence ions were more prevalent from the terminal ends of each protein with less extensive information from the midsection,13 as also noted for characterization of individual proteins by HCD.29 Surface induced dissociation (SID) is an activation technique based on the high energy collision of an ion with a surface, promoting unique fragmentation pathways of macromolecues.15–17 SID causes disassembly of macro-molecular complexes prior to unfolding of the individual proteins, an outcome that aligns well with the ability to map subunit contacts and reconstruction of quaternary structure. Auxiliary ion mobility studies of SID product ions have confirmed that SID yields structurally compact fragment ions that retain native-like conformations.17 However, SID does not provide many fragment ions that reveal primary sequence information. Electron capture dissociation (ECD) has been shown to be a powerful tool for characterization of intact proteins30–32 and macromolecular protein assemblies.18–26 ECD results in an array of informative sequence ions from intact proteins, and interestingly it has been found that noncovalent interactions can be retained during the electron capture activation process, resulting in direct information about protein–ligand contacts.20,21,23 The Loo and Gross groups in particular have capitalized on the benefits of ECD for the top-down characterization of noncovalent protein complexes.18–22,24 These ECD studies have yielded many diagnostic fragment ions to characterize the polypeptide backbone sequences and to support the determination of protein tertiary structures. Aspects of tertiary structure can be elucidated based on the propensity of ECD for fragmenting more flexible regions of proteins or complexes.18–26 Very recently, Sobott et al. successfully implemented ETD on a hybrid ion-mobility time-of-flight mass spectrometer and elucidated the more flexible regions of alcohol dehydrogenase through the observation of diagnostic c and z ions.27 The effectiveness of electron-based methods is partially impeded by the low charge states (and thus more compact structures) of proteins and protein complexes generated from native ESI conditions. ECD and ETD have shown to be more efficient for dissociation of more unfolded ions with high charge densities,33,34 thus tempering the scope of ECD for determining primary sequence information on the proteins in native complexes (which typically have much lower charge states than denatured proteins). More recently vacuum UVPD was demonstrated for analysis of a small protein (IB5)/tannin complex using 16 eV synchrotron radiation.28 Product ions included those comprised of part of the IB5 protein with retention of the tannin ligand, thus allowing localization of the tannin binding site.
Ultraviolet dissociation (UVPD) has been shown to be an effective activation method for the characterization of a wide array of biological molecules,35–47 including intact proteins.48–50 The amide backbone serves as an effective chromophore for absorption of UV photons at 193 nm, leading to high energy activation and dissociation into primarily a, b, c, x, y, Y, and z ions.44–46,49,50 The implementation of UVPD on a hybrid linear ion trap/Orbitrap mass spectrometer generated near-complete sequence coverage of proteins up to 30 kDa49 and has been successfully adapted for the high-throughput top-down analysis of mixtures of ribosomal proteins.50 UVPD has also very recently revealed conformerspecific fragmentation for ion mobility-separated ubiquitin ions in which there was evidence for cis/trans isomerization of a proline peptide bond based on variation of the UVPD fragmentation yields.51 Here we report the application of 193 nm UVPD for characterization of native protein–ligand and oligomeric complexes. Native-ESI paired with top-down 193 nm UVPD yields unique and comprehensive fragment ions to characterize proteins in noncovalent complexes.
EXPERIMENTAL SECTION
Materials and Proteins
Myoglobin from equine skeletal muscle, bovine β-lactoglobulin variant A, insulin from bovine pancreas, ammonium acetate, and all other reagents were purchased from Sigma-Aldrich and were utilized without further purification. A truncated version of the cap binding protein eukaryotic initiation factor 4E (eIF4E) was expressed and purified as previously described.52,53 Pin1 was expressed and purified as previously described.49 The synthetic 10-mer CTD phosphopeptide was purchased from AnaSpec Inc (Fremont, CA).
Mass Spectrometry
All mass spectrometry experiments were performed on a Thermo Scientific Orbitrap Elite mass spectrometer (Bremen, Germany) modified to perform UVPD in the HCD collision cell as described previously.49 UVPD experiments were performed using a Coherent Excistar ArF (193 nm) excimer laser (Santa Clara, CA) which produced 5 ns laser pulses and a repetition rate of 500 Hz. Native-ESI experiments were performed using a Proxeon nano ESI source with Pd/Au borosilicate nanoESI emitters (Thermo Scientific). The HCD collision cell pressure was increased from 10 to 20 mTorr to enhance stabilization of the noncovalent complexes. Protein solutions (5–10 μM) for native ESI were prepared in 200 mM ammonium acetate with the exception of eIF4E (in 350 mM ammonium acetate) and insulin (2.5 μM zinc chloride, 1.67 μM ammonium chloride, and 5 mM phenol in 300 mM aqueous ammonium acetate). ESI and MS/ MS mass spectra were collected using the extended m/z range to record up to m/z 15 000 using a resolution of 120 000–240 000 (at m/z 400). MS and MS/MS spectra were collected using between 100 and 400 spectral averages (of five microscans each). CID and ETD experiments were performed in the linear ion trap and typically performed using 5–40 ms activation times. Ions selected for MS/MS activation were typically isolated using a window of 25–30 m/z. Normalized collisional energies (NCE) of 7–40% were used for all CID and HCD experiments. Using higher NCE values did not necessarily result in the production of greater abundances or types of sequence ions, so the NCE values were set to maximize dissociation efficiencies and spectral quality and minimize precursor ejection. UVPD experiments were performed in the HCD cell using one or two laser pulses at 2–4 mJ per pulse. However, we estimate that only a fraction of the beam (<1%) enters the HCD cell due to the high divergence of the laser and lack of light collimation.
Data Processing
MS/MS spectra were interpreted using a customized version of ProSight PC 3.0 (Thermo Scientific) to include a, b, c, x, y, z fragment ion types. Mass spectra were deconvoluted using the Xtract data processing algorithm (Thermo Fisher Scientific) with an S/N threshold of 3. All searches were performed using the Δm mode and with a mass tolerance set to 15 ppm. Manual interpretation was required for identification of most of the noncovalent fragment ions; this was performed by creating monoisotopic charge deconvoluted mass spectra using the Xtract algorithm (Thermo Scientific). The deconvoluted mass spectra were then searched using Protein Prospector (v 5.10, http://prospector.ucsf.edu/prospector/mshome.htm) for potential a/b/c/x/y/z and internal fragment ions within a 10 ppm mass accuracy threshold. In these searches, fragments containing a part of the protein plus the bound ligand or one protein bound to part of another protein were identified via exact mass losses from the original intact protein complexes. Normalized cleavage frequencies were calculated by summing the abundances of all a, b, c, z, y, and z fragment ions associated with a particular backbone position and dividing this by the summed abundance obtained for the backbone position having the largest summed abundance. The MS/MS sequence coverages were calculated as percentages based on the total number of observed nonredundant backbone cleavages divided by the total number of possible backbone cleavages in the protein. B-factor values were obtained from the RCSB Protein Data Bank (PDB, http://www.rcsb.org/pdb/home/home.do), and images of protein crystal structures were generated using Pymol (version 1.3). All spectra are available at IU Scholar Works site.
RESULTS AND DISCUSSION
Myoglobin–Heme
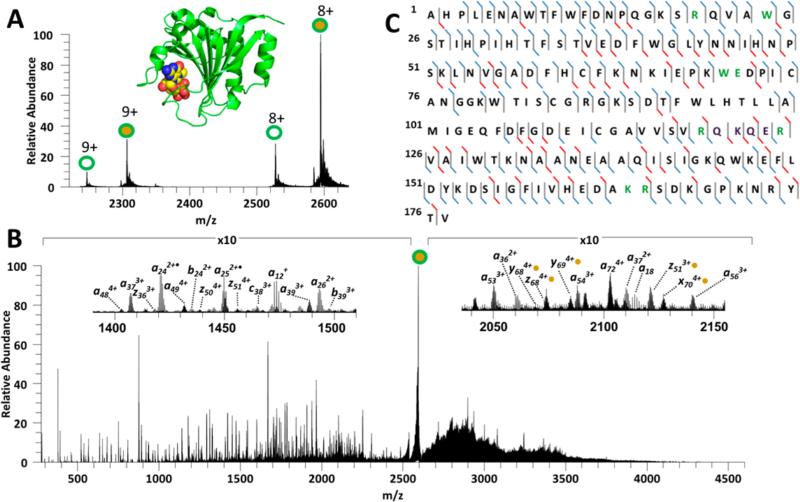
Myoglobin (16.9 kDa, 153 residues) is an oxygen carrying-protein found in muscle tissues and was the first protein structure solved by X-ray crystallography.54 Myoglobin forms a noncovalent complex with heme (net complex is 17.6 kDa; X-ray structure is shown in Figure 1A) and can be readily observed using native ESI conditions. The ESI mass spectrum for the intact myoglobin–heme complex is shown in Figure S1A, displaying complexes in the 7+, 8+, and 9+ charge states. The myoglobin–heme complexes were isolated in the linear ion trap region and transferred to the HCD cell where they were subjected to a single 5 ns laser pulse (at 2 mJ/pulse). A representative UVPD mass spectrum is shown in Figure 1B for the 9+ complex. UVPD generated a wide range of fragment ions arising from cleavages of the polypeptide backbone, leading to sequence-type ions both with and without retention of the heme group, as well as intact apomyoglobin (without heme) (unfilled blue circle in Figure 1B) and the heme moiety alone, all within 15 ppm mass error and typically a 10:1 or greater signal-to-noise. The insets in Figure 1B show two expanded m/z regions which highlight the wide range of assignable product ions observed using UVPD which were used to characterize the primary sequence of myoglobin in the noncovalent complex. The 7+, 8+, and 9+ myoglobin/heme complexes were subjected to CID, HCD, and UVPD, and the resulting MS/MS spectra analyzed to assess the ability to probe protein primary sequence information. Examples of CID, HCD, and ETD mass spectra for the 9+ complexes are shown in Figure S2. CID of the myoglobin/heme complexes (7+, 8+, and 9+) mainly resulted in release of the heme group and yielded little primary sequence information (Figure S2A). HCD also led to release of the heme group as well as production of some diagnostic b and y fragment ions from large stretches of the protein (aside from one poorly characterized 25 residue stretch close to the N-terminus) (Figure S2B). ETD yielded primarily charge reduction of the intact protein complexes, and only a few c and z ions of low abundance were identified (Figure S2C). UVPD of the myoglobin/heme protein complexes generated the widest array of fragment ions and yielded between 85% and 95% sequence coverage depending on the charge state of the selected precursor complex (Figures 1C and S2D).
Figure 1.
(A) X-ray crystal structure of myoglobin in complex with heme (PDB 1AZI). (B) UVPD mass spectrum of the native heme/myoglobin complex (9+). The filled blue circle indicates the [myoglobin + Heme]9+ precursor ion and the unfilled blue circle indicates intact apo-myoglobin (no heme) (8+). (C) Resulting UVPD cleavage map with known heme binding sites shaded in pink. The UVPD cleavage map displays the backbone cleavages that lead to sequence ions. Cleavages that result in a, b, c, x, y, z fragment ions without heme are shown as blue slashes, and cleavages that lead to fragments identified both with and without heme are red slashes. Cleavage marks slant to the left or right to denote N- and C-terminal product ions, respectively.
Despite the high energy deposition of 193 nm UVPD (6.4 eV per photon), many products that retained the heme group were identified after photodissociation (Figure 1C). The presence of noncovalent heme-containing fragment ions in the native UVPD spectra is consistent with previous studies that suggest that 193 nm UV photodissociation may occur from excited electronic states prior to extensive vibrational redistribution of energy;49 this could rationalize the retention of noncovalent ligands even upon high energy deposition. This result suggests there is a strong potential for UVPD characterization of noncovalent complexes akin to other activation methods (ECD, ETD) that might exhibit nonergodic behavior. One representative sequence coverage map for the myoglobin complexes (9+) is shown in Figure 1C, where blue tick marks show all the N-terminal and C-terminal products that lack the heme group and the red tick marks indicate the various N-terminal and C-terminal products that retain the heme group. There are cases in which product ions that originate from specific backbone cleavage sites are observed both with and without retention of the heme group; in these cases red tick marks are used. This type of behavior (i.e., analogous product ions with and without the bound ligand or protein partner) was observed for all examined native protein complexes. A total of 78 a, b, c x, y, and z ions that retained the heme group were identified using ProSight PC (Table S1), and a total of 325 product ions did not retain the heme group. Many of the diagnostic cleavages that led to products retaining the heme group occurred sequentially near the H64 and H93 heme binding residues (shaded in pink in Figure 1C). A histogram showing the normalized backbone cleavage frequencies for myoglobin (9+) (calculated as described in the Experimental Section) along with the B-factors associated with each of the amino acid residues are shown in Figure S3A. Additionally the crystal structure of the myoglobin/heme complex is shown as B-factor putty in Figure S3B. Examination of the normalized backbone cleavage frequencies (blue bars in Figure S3A) suggests that there are some correlations between the regions of greatest protein disorder (B-factors, shown via the red line in Figure S3A) and the UVPD spectra. In particular, the N-terminus and C-terminus exhibited both high degrees of protein structural disorder and high cleavage frequencies. However, it is likely that a number of other factors also contribute to the production and abundances of UVPD fragment ions, including the extent of hydrogen bonding and salt bridging in the gas-phase structures. The favored cleavage sites upon UVPD differed from the dominant ones observed upon HCD (Figure S3), with the latter favoring cleavages adjacent to acidic or proline residues (ones often associated with preferential cleavages upon collisional activated dissociation of peptides) and exhibiting a less substantial correlation with higher order structure.
The apo and holo a/x product ions were plotted with respect to the product ion charge state to reveal a correlation between the expected binding interaction region of the myoglobin/heme complex and the association (or release) of the heme group (as reflected in the product ions) (Figure S4). The distribution of holo a product ions demonstrates that the heme group is retained only by those product ions that contain residue 63 and higher, and similarly for the holo x ions, the heme group is retained only by those product ions that contain 68 residues or more (starting from the C-terminus). The absence or presence of holo a and x ions brackets the presumed binding region of the heme group and mirrored the results presented by Enyenihi et al. for intact supercharged myoglobin/heme complexes (12+, 14+, and 16+) obtained via ETD.55 This suggests that UVPD can be used to elucidate primary sequence information, along with production of fragment ions that retain the binding ligand in a manner that is consistent with the presumed location of the binding ligand.
Eukaryotic Initiation Factor 4E
Eukaryotic initiation factor 4E (eIF4E) plays a pivotal role in eukaryotic protein translation, and its functional role entails the interaction with the 7-methylguanosine (m7G) cap group at the 5′ end of mRNAs.52 This protein in complex with the triphosphate form of m7G (7-methylguanosinetriphosphate ((m7GTP), Mr = 537.0) was transferred to the gas phase using native ESI conditions (Figure 2A), resulting in a mixture of apo-eIF4E (shown as unfilled circles) and holo-eIF4E protein complexes (1:1 eIF4E:m7GTP complexes shown as filled circles). Holo-and apo-eIF4E were characterized using CID, HCD, and UVPD in a similar manner to that described above. Examples of the HCD, CID, and UVPD mass spectra are shown in Figures S5, S6, and 2B, respectively. The sequence coverages obtained for several charge states of the protein and its m7GTP complex are summarized in the histograms in Figure S7. CID of apo-eIF4E (8+) resulted in a mixture of b and y fragment ions, whereas CID of the eIF4E/m7GTP complex (8+) primarily resulted in ejection of m7GTP and release of intact apo-eIF4E (7+). Free m7GTP (1+) was not detected in the CID spectrum as its mass falls below the low mass cutoff (LMCO) of the linear ion trap. HCD yielded significantly more sequence ions for both the eIF4E protein alone and the eIF4E/m7GTP complex. CID of apo-eIF4E yielded less than 45% sequence coverage, whereas HCD of both the apo-eIF4E and holo-eIF4E varied from 41% to 62%. The HCD spectra of apo-eIF4E (8+ and 9+) and holo-eIF4E (8+ and 9+) were both dominated by a series of fragment ions arising from preferential cleavages C-terminal to Asp or Glu residues or N-terminal to Pro residues, also in conjunction with water or ammonia losses, thus providing limited information about the protein or its ligand interactions. A representative sequence coverage map for holo-eIF4E derived from the UVPD (8+) mass spectrum is shown in Figure 2C. UVPD of the apo-eIF4E (Figure S6A) and holo-eIF4E protein complexes (Figure 2B) resulted in extensive backbone cleavages which allowed more complete characterization of the primary sequence of eIF4E, translating into the greatest sequence coverage (~85%). Several of the m7GTP-containing fragment ions observed in the UVPD mass spectrum of the eIF4E/m7GTP complexes (Figure 2) could be easily identified through exact mass shifts corresponding to the m7GTP ligand (Figure S6B). Furthermore, the sequence coverage was similar for both the 8+ and 9+ charge states, suggesting that there was little dependence on the charge state of the precursor protein– ligand complex.
Figure 2.
(A) Native ESI mass spectrum of eIF4E complexed to m7GTP. Unfilled circles represent apo-eIF4E, filled circles represent holo-eIF4E. (B) UVPD mass spectrum of the eIF4E/m7GTP (8+) complex. The insets are expansions of the m/z regions 1350–1550 and 2040–2155. Fragments with retained m7GTP interactions are shown with gold circles. (C) Cleavage map for the eIF4/m7GTP (8+) complex generated using UVPD. The UVPD cleavage map displays the backbone cleavages that lead to sequence ions. Residues that engage in key electrostatic interactions with m7GTP are shown in green font. Cleavages that result in a, b, c, x, y, z fragment ions without m7GTP are shown as blue slash marks, and cleavages that lead to fragments identified both with and without retention of m7GTP are shown as red slash marks.
It is known that wheat eIF4E binds the m7G cap structure through several types of hydrophobic and electrostatic interactions,52 which are highlighted in the crystal structure of eIF4E illustrated in Figure S8. In particular, the side chains of Arg20, Arg120, and Arg125 all form strong hydrogen bonds with the m7G phosphate groups, whereas the guanosine moiety of m7G interacts via hydrogen bonds with Glu71 and the N–H backbone of Trp70 and by π–π interactions involving the aromatic side chains of Trp70 and Trp24. There is also experimental evidence that the looped region around the Lys165 and Arg166 residues interacts with the mRNA cap.53 UVPD of the holo-eIF4E (Figure 2) generated a wide variety of fragment ions that retained m7GTP (a subset of m7GTP-bound fragment ions is shown in Table S2). The UVPD fragmentation map of the eIF4E/m7GTP complex (8+) displays the backbone cleavage sites that lead to fragment ions that retain m7GTP (red cleavage marks in Figure 2C). The subset of apo and holo a/x fragment ions is shown graphically in Figure S9. Several interesting features are observed upon inspection of the fragments that retain the m7GTP moiety upon UVPD. In particular, a predominance of fragment ions retain m7GTP (holo product ions in Figure S9) for those that contain 120 or more residues for the N-terminal a ions and contain 107 or more residues for the C-terminal x ions, again appearing to “bracket” the presumed binding region of the eIF4E/m7GTP complex.
The relative cleavage frequencies were calculated for apo and holo eIF4e (Figure S10A) and compared to the B-factors obtained from the known protein crystal structure (Figure S8). Overall the apo and holo fragment ions showed similar characteristics and fragment ion abundances. However, one notable difference between apo-eIF4E and holo-eIF4E is apparent with the sharp decrease in the cleavage frequencies in the regions with the highest structural disorder (Lys165 to Pro171, red looped region in Figure S10B) and a few minor decreases from the N-terminus to Cys75. These differences in fragment ion abundances may reflect the increasing stabilization of the protein upon ligand complexation, a known outcome of eIF4E cap binding.55 This also suggests that UVPD could be utilized for the characterization of tertiary structural changes upon disulfide bond formation.
Peptidyl-prolyl Cis–Trans Isomerase 1
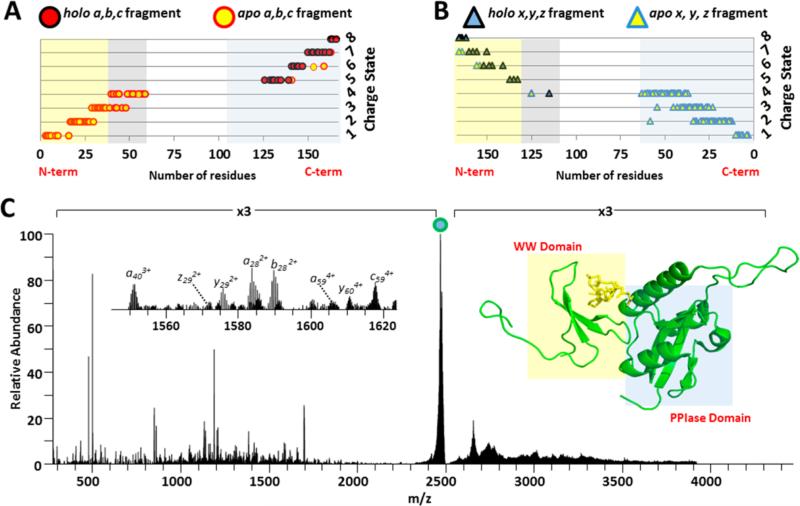
Pin1 (Figure 3) is an 18 kDa protein which expedites the conversion of cis–trans isomeric states of prolines when preceding phosphorylated Ser/Thr residues.57,58 This conformational change of the peptidyl backbone has been shown to affect the catalytic activity, protein–protein interactions, cellular location, and stability of proteins; thus Pin1 can rewire the signal transduction pathways and is believed to play a critical role in the pathological development of cancer and Alzheimer's disease.59 One of the major physiological substrates for Pin1 is phosphorylated RNA polymerase II, which is the primary form of RNA polymerase II during active transcription.60–62 Although the X-ray structure of Pin1 has been resolved, a substantial portion of the protein (i.e., the linker connecting the WW and PPIase domains) is totally disordered (residue 43–54, shaded gray in Figure 3A and 3B), rendering the interpretation of ligand interaction with the protein incomplete.62,63
Figure 3.
Plot of Pin1 (A) N-terminal and (B) C-terminal backbone cleavages with respect to number of amino acid residues in the products. (C) UVPD mass spectrum of the Pin1/CTD peptide (8+) complex. The precursor ion is labeled with a filled circle. The inset shows the m/z 1545–1625 expanded region of the UVPD mass spectrum. On the right side of C is shown the crystal structure of Pin1 (PDB:IF8A) with the CTD peptide structure. Red filled circles and blue filled triangles represent holo-product ions (retaining CTD peptide), whereas yellow filled circles and triangles represent apo-product ions (without CTD peptide). The N-terminal WW domain of the Pin1 protein is shaded in yellow, and the C-terminal PPIase domain is shaded in blue in the fragment ion plots (A and B) and the protein crystal structure (bottom right). The disordered region between residues 43–54 are highlighted in gray in (A) and (B).
Pin1 was incubated at room temperature with a 5 molar excess of the 1.2 kDa CTD phosphodecapeptide (YSPTpSPSYSP) and subsequently analyzed using native ESI-MS/MS. A 1:1 noncovalent complex between Pin1 and the CTD peptide was observed, and the 8+ charge state was subjected to HCD, ETD, and UVPD for structural characterization. Both ETD and HCD resulted in rather uninformative spectra (Figure S11). ETD resulted in charge reduction of the intact complex and virtually no sequence ions (nor disassembly of the complex). The HCD spectrum was dominated by the intact CTD peptide, some CTD peptide b/y sequence ions, and very few diagnostic Pin1 fragments (<20% sequence coverage). UVPD of the complex yielded ~70% sequence coverage of Pin1 (Figure 3C), and a number of the resulting N-terminal and C-terminal fragments were identified both with and without retention of the CTD peptide. Importantly, the linker region missing from the crystal structure is well covered in the UVPD mass spectrum (Figure 3C), yet shows no association with CTD peptide. Even though it is suspected that the disorder of the linker region in the crystal structure suggests that it plays little role in ligand binding, the present MS/MS data provide concrete evidence for this notion.
Examination of the array of Pin1 C-terminal and N-terminal UVPD fragments (Figure 3A and 3B) provided insight into the CTD peptide binding site. In particular, those C-terminal fragment ions that retained the CTD peptide were the ones containing the WW domain (shaded yellow in Figure 3). Only those N-terminal fragments that spanned the Pin1 PPIase domain (shaded blue in Figure 3A and 3C) retained the CTD peptide. Interestingly, it was necessary for the N- and C-terminal fragments to contain both the WW and PPIase domains in order to retain the CTD peptide; this reflects the requirement for the existence of both domains to stabilize the noncovalent interactions necessary for retention of the CTD peptide upon photodissociation of the intact protein complex. Indeed, the CTD peptide is bound at the cleft formed at the interface of the WW and PPIase domains in the crystal structure.62 Around 50 proteins have been reported as substrates for Pin1, including tumor suppressor p53 and oncoprotein Myc, yet how Pin1 recognizes these substrates remains largely unknown due to the difficulty in obtaining the crystals for complexes between Pin1 and substrate peptides. NMR studies have been reported using only the WW domain with various ligand peptides from substrates, but the NMR results show dramatic differences in the association mode between the enzyme and substrate compared to the only reported crystal structure (PDB code 1F8A).62,64 Our UVPD MS results provide an explanation for this discrepancy and emphasizes the requirement for both WW and PPIase domains for effective substrate binding.
Multimeric Protein Complexes
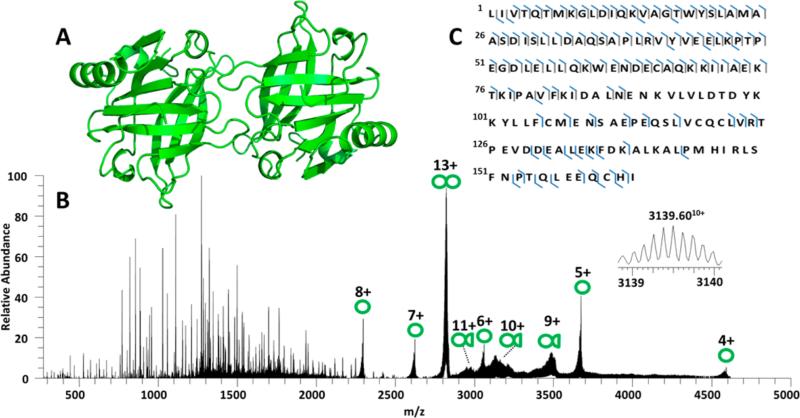
β-Lactoglobulin (18.3 kDa, 162 residues) is believed to be a lipid transporter protein which readily forms dimers in solution (Figure 4A).65–67 β-Lactoglobulin was transferred to the gas phase using native ESI conditions, and the resulting ions were examined using CID, HCD, and UVPD. The sequence coverage obtained for the monomer and dimer forms using CID, HCD, ETD, and UVPD are summarized in Figure S12, and the UVPD spectrum for the 13+ dimer is shown in Figure 4B. (The CID, HCD, and ETD spectra for the monomer and dimer are shown in Supplemental Figure S13, and the UVPD spectrum for the monomer is shown in Supplemental Figure S14.) CID, HCD, and ETD of the dimer yielded rather low sequence coverage (40–50% for HCD and <15% for CID and ETD), with CID causing primarily separation of the 13+ dimer into monomers (8+, 7+, 6+, and 5+). HCD yielded similar fragmentation patterns for both the monomeric or dimeric states, including formation of a mixture of b and y fragments. ETD mainly yielded charge reduction of both the monomer and the dimer. The dimer complexes also produced a few diagnostic c and z ions upon ETD as well as several unidentified products of large abundance (marked with a “@” in Figure S13F), the latter which, although not confidently unidentified, might be associated with cleavages in the more disordered regions with retention of noncovalent interactions. The low sequence coverage obtained by HCD, CID, and ETD translated into large uncharacterized stretches of the protein sequence for both the monomer and dimer. UVPD generated a broader array of informative sequence ions for both the monomers and dimers as demonstrated by the rich spectrum for the 13+ dimer in Figure 4B and 8+ monomer in Figure S14. Sequence coverage for the monomer ranged from 65% to 72% and averaged 67% for the dimer upon UVPD (Figure S12); however UVPD of the β-lactoglobulin monomers generated a greater number of C-terminal fragments (Figure S15) relative to the dimerized species (Figure 4C). The difference in the number of C-terminal fragments could be related to suppression of cleavages in the dimerized form due to a greater number of intermolecular interactions involving the C-terminus (as shown from prior crystallographic analysis)66 and in part due to greater congestion of the UVPD mass spectrum for the dimer due to unassigned internal ions. UVPD of the 13+ dimer led to release of intact monomeric proteins in several charge states (designated by single green circles in Figure 4B), as well as products that were consistent with one (presumably) intact protein and a portion of the second protein (shown by single circles attached to half circles). The charge deconvoluted UVPD mass spectrum of the 13+ dimer (Figure S16) revealed a large number of poorly resolved product ions primarily between 29.5 and 32.5 kDa which corresponded to various 9+, 10+, and 11+ fragment ions. An initial effort to interpret these fragments was undertaken by subtracting the mass of β-lactoglobulin from the identified fragments, thus yielding the mass of the remaining portion that could be matched to conventional a/b/c/x/y/z-type fragment ions using ProteinProspector with a mass cutoff of 10 ppm. This methodology was based on the assumption that the product ions contained one molecule of the intact protein plus an assignable portion of the second protein. Other possible products, such as those containing cleaved portions of each of the two proteins in the dimer, could not be uniquely differentiated. This level of mass accuracy did not allow particularly confident assignments but was a first step toward making assignments. Other complementary methods such as ion mobility, the development of new searching algorithms to facilitate assignment of product ions that contain portions of more than one protein, and higher resolution analysis are needed to elucidate confidently the structures of the dimer fragment ions generated using UVPD. In any case, UVPD provided the most extensive series of product ions that contain potential insight into the contact regions of the two proteins in the dimer, in addition to the greatest sequence coverage for each protein in the dimer.
Figure 4.
(A) X-ray crystal structure of β-lactoglobulin (PDB: 1BSY). (B) UVPD mass spectrum of β-lactoglobulin dimer (13+). Single circles indicate intact monomers; double circles indicate dimers and circles with half circles are dimer fragments. The inset is an expansion of an identified a116 + β-lactoglobulin (10+) product ion. (C) UVPD sequence coverage map obtained for dimeric β-lactoglobulin (13+).
As a final example, insulin (5.7 kDa, 51 residues) is a highly conserved protein responsible for regulation of carbohydrate metabolism, and insulin insufficiency is the primary source of diabetes. Each molecule of insulin is composed of two separate polypeptide chains (chain A and chain B) connected via three disulfide bonds. In the inactive form, insulin forms a hexamer (see inset of Figure 5A) bound through a network of noncovalent interactions involving both zinc coordination and hydrogen bonding between the C-terminal β-sheets of insulin chain B. CID and HCD spectra of the insulin hexamer (11+) acquired at several collision energies are shown in Figure S17, displaying formation of both monomers and pentamers upon CID and low energy HCD. HCD spectra acquired at a higher collision energy also exhibited some sequence ions from insulin (Figure S17E). The UVPD mass spectra of hexameric insulin (12+ and 11+) are shown in Figures 5A and S18, respectively. UVPD generated the greatest array of sequence ions from the insulin subunits, including a, b, c, x, y, z and cross-linked fragment ions (Figure 5B). There were several missed cleavages in chain A, likely due to the impact of intact disulfide bonds, but UVPD outperformed both HCD (~33% sequence coverage) and CID (no sequence coverage) in overall sequence coverage. Similarly, a complete array of subunit cleavages was observed, leading to formation of pentamers, tetramers, trimers, dimers, and monomers and release of intact insulin chain A and insulin chain B, revealing that some of the disulfide bonds were in fact cleaved by UVPD. Furthermore, UVPD resulted in cleavages of the intact oligomeric complexes corresponding to fragmentation of one subunit while still coordinated to the other insulin chains (Figure 5C), an outcome not observed in the CID or HCD spectra. Closer examination of the deconvoluted UVPD spectrum (Figure S19A) reveals that some of the products could be attributed to secondary fragmentation of tetramers and were consistent with losses from the C-terminus of insulin chain B (black fragments) and the N-terminus of insulin chain A (red fragments) from the intact tetrameric product. The X-ray structure of insulin is shown as B-factor putty in Figure S19B. A possible explanation is that the losses from the tetrameric species arise from the structurally disordered areas of the complexes. Overall UVPD yielded more structural information relative to HCD and CID via formation of a wide array of diagnostic polypeptide sequence ions, disassembly of the hexamer, and formation of product ions from smaller oligomeric species.
Figure 5.
(A) UVPD mass spectrum of hexameric insulin (12+). Each red circle represents one monomeric insulin unit. The inset shows the X-ray structure of hexameric insulin, where zinc is colored yellow and the coordinated His10 residues are colored green. (B) UVPD cleavage maps are shown for insulin chain A and insulin chain B for the hexameric insulin complex (12+). (C) Expansion of the m/z region between 3400 and 4000 m/z.
CONCLUSIONS
UVPD, as a high energy activation method, yields a diverse array of fragment ions that provide primary sequence information as well as reveal some insight into the interacting regions of the proteins or proteins and ligands. The mixture of structurally informative noncovalent fragment ions for the protein complexes showcase the ability to probe native structures, giving information about tertiary and quaternary arrangements. CID and HCD, on the other hand, lead to preferential disruption of the noncovalent interactions of the protein complexes, thus releasing the subcomponents. ETD mainly yielded charged reduced native complexes and little to no sequence information. Improvements to the database searching method for the product ions that contain one intact protein plus some portion of a second protein or a part of a protein still bound to a ligand are needed to facilitate data analysis.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the following funding sources: NSF (CHE-1012622 to J.S.B.), NIH (R01GM104896 to Y.Z), and the Welch Foundation (F-1155 to J.S.B. and F-1778 to Y.Z.).
Footnotes
Supporting Information
The contents of the Supporting Information include the native ESI mass spectrum of myoglobin, numerous MS/MS spectra (CID, HCD, ETD, UVPD) and sequence coverage maps, histograms showing sequence coverages for various proteins and various activation methods, histograms showing backbone cleavage frequencies, crystal structures of proteins, and tables of fragment ions for various proteins showing accurate m/z values, fragment ion assignments, and ppm errors. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Robinson CV, Sali A, Baumeister W. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 2.Loo JA. Mass Spectrom. Rev. 1998;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 3.Konermann L, Vahidi S, Sowole MA. Anal. Chem. 2014;86:213–232. doi: 10.1021/ac4039306. [DOI] [PubMed] [Google Scholar]
- 4.Benesch JL, Ruotolo BT. Curr. Opin. Struct. Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcoux J, Robinson CV. Structure. 2013;21:1541–1550. doi: 10.1016/j.str.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Light-Wahl KJ, Schwartz BL, Smith RD. J. Am. Chem. Soc. 1994;116:5271–5278. [Google Scholar]
- 7.Heck AJ. R. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 8.Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinković D, Murata T, Bernal RA, Stock D, Robinson CV. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Angew. Chem., Int. Ed. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benesch JLP. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Hernández H, Robinson CV. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 12.Benesch JLP, Ruotolo BT, Sobott F, Wildgoose J, Gilbert A, Bateman R, Robinson CV. Anal. Chem. 2009;81:1270–1274. doi: 10.1021/ac801950u. [DOI] [PubMed] [Google Scholar]
- 13.Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, Kelleher NL. Anal. Chem. 2013;85:11163–11173. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- 14.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. Nat. Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 15.Ma X, Zhou M, Wysocki VH. J. Am. Soc. Mass Spectrom. 2014;25:368–379. doi: 10.1007/s13361-013-0790-y. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Jones CM, Wysocki VH. Anal. Chem. 2013;85:8262–8267. doi: 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Dagan S, Wysocki VH. Angew. Chem., Int. Ed. 2012;51:4336–4339. doi: 10.1002/anie.201108700. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Wolff JJ, Van Orden SL, Loo JA. Anal. Chem. 2014;86:317–320. doi: 10.1021/ac4033214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. J. Am. Soc. Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin S, Loo JA. J. Am. Soc. Mass Spectrom. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Xie Y, Zhang J, Yin S, Loo JA. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Cui W, Gross ML, Blankenship RE. FEBS Lett. 2013;587:1012–1020. doi: 10.1016/j.febslet.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke DJ, Murray E, Hupp T, Mackay CL, Langridge-Smith PRR. J. Am. Soc. Mass Spectrom. 2011;22:1432–1440. doi: 10.1007/s13361-011-0155-3. [DOI] [PubMed] [Google Scholar]
- 24.Wen J, Zhang H, Gross ML, Blankenship RE. Biochemistry. 2011;50:3502–3511. doi: 10.1021/bi200239k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Anal. Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Cui W, Gross ML. Int. J. Mass Spectrom. 2013:354–355. 288–291. doi: 10.1016/j.ijms.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lermyte F, Konijnenberg A, Williams JP, Brown JM, Valkenborg D, Sobott F. J. Am. Soc. Mass Spectrom. 2014;25:343–350. doi: 10.1007/s13361-013-0798-3. [DOI] [PubMed] [Google Scholar]
- 28.Canon F, Milosavljević AR, van der Rest G, Réfrégiers M, Nahon L, Sarni-Manchado P, Cheynier V, Giuliani A. Angew. Chem., Int. Ed. 2013;52:8377–8381. doi: 10.1002/anie.201304046. [DOI] [PubMed] [Google Scholar]
- 29.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Müller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach J-J, Cox J, Horning S, Mann M, Makarov A. Mol. Cell. Proteomics. 2012;11:O111.013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 31.Han X, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 32.Zubarev RA. Curr. Opin. Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Ledvina AR, McAlister GC, Gardener MW, Smith SI, Madsen JA, Schwartz JC, Stafford GC, Syka JEP, Brodbelt JS, Coon JJ. Angew. Chem., Int. Ed. 2009;48:8526–8528. doi: 10.1002/anie.200903557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horn DM, Breuker K, Frank AJ, McLafferty FW. J. Am. Chem. Soc. 2001;123:9792–9799. doi: 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- 35.Ly T, Julian RR. Angew. Chem., Int. Ed. 2009;48:7130–7137. doi: 10.1002/anie.200900613. [DOI] [PubMed] [Google Scholar]
- 36.Diedrich JK, Julian RR. Anal. Chem. 2010;82:4006–4014. doi: 10.1021/ac902786q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T-Y, Thompson MS, Reilly JP. Rapid Commun. Mass Spectrom. 2005;19:1657–1665. doi: 10.1002/rcm.1969. [DOI] [PubMed] [Google Scholar]
- 38.Devakumar A, O'Dell DK, Walker JM, Reilly JP. J. Am. Soc. Mass Spectrom. 2008;19:14–26. doi: 10.1016/j.jasms.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Gabelica V, Rosu F, De Pauw E, Lemaire J, Gillet J-C, Poully J-C, Lecomte F, Grégoire G, Schermann J-P, Desfrançois C. J. Am. Chem. Soc. 2008;130:1810–1811. doi: 10.1021/ja077321w. [DOI] [PubMed] [Google Scholar]
- 40.Rosu F, Gabelica V, De Pauw E, Antoine R, Broyer M, Dugourd PJ. Phys. Chem. A. 2012;116:5383–5391. doi: 10.1021/jp302468x. [DOI] [PubMed] [Google Scholar]
- 41.Smith SI, Brodbelt JS. Anal. Chem. 2010;82:7218–7226. doi: 10.1021/ac100989q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madsen JA, Cullen TW, Trent MS, Brodbelt JS. Anal. Chem. 2011;83:5107–5113. doi: 10.1021/ac103271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodbelt JS. J. Am. Soc. Mass Spectrom. 2011;22:197–206. doi: 10.1007/s13361-010-0023-6. [DOI] [PubMed] [Google Scholar]
- 44.Madsen JA, Boutz DR, Brodbelt JS. J. Proteome Res. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodbelt JS. Chem. Soc. Rev. 2014;43:2757–2783. doi: 10.1039/c3cs60444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reilly JP. Mass Spectrom. Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Reilly JP. J. Proteome Res. 2009;8:734–742. doi: 10.1021/pr800766f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ly T, Julian RR. J. Am. Chem. Soc. 2007;130:351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 49.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. J. Am. Chem. Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cannon JR, Cammarata MB, Robotham SA, Cotham VC, Shaw JB, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Anal. Chem. 2014;86:2185–2192. doi: 10.1021/ac403859a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warnke S, Baldauf C, Bowers MT, Pagel K, von Helden G. J. Am. Chem. Soc. 2014;136:10308–10314. doi: 10.1021/ja502994b. [DOI] [PubMed] [Google Scholar]
- 52.Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS. Plant Physiol. 2007;143:1504–1518. doi: 10.1104/pp.106.093146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Brien JP, Mayberry LK, Murphy PA, Browning KS, Brodbelt JS. J. Proteome Res. 2013;12:5867–5877. doi: 10.1021/pr400869u. [DOI] [PubMed] [Google Scholar]
- 54.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 55.Enyenihi AA, Yang H, Ytterberg AJ, Lyutvinskiy Y, Zubarev RA. J. Am. Soc. Mass Spectrom. 2011;22:1763–1770. doi: 10.1007/s13361-011-0182-0. [DOI] [PubMed] [Google Scholar]
- 56.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, Miura K, Ishida T. J. Mol. Biol. 2003;328:365–383. doi: 10.1016/s0022-2836(03)00314-0. [DOI] [PubMed] [Google Scholar]
- 57.Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld J-U, Lu KP, Fischer G. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- 58.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 59.Lu KP, Zhou XZ. Nat. Rev. Mol. Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 60.Albert A, Lavoie S, Vincent M. J. Cell Sci. 1999;112:2493–2500. doi: 10.1242/jcs.112.15.2493. [DOI] [PubMed] [Google Scholar]
- 61.Morris DP, Phatnani HP, Greenleaf AL. J. Biol. Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 62.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Nat. Struct. Mol. Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 63.Ranganathan R, Lu KP, Hunter T, Noel JP. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 64.Wintjens R, Wieruszeski J-M, Drobecq H, Rousselot-Pailley P, Buée L, Lippens G, Landrieu I. J. Biol. Chem. 2001;276:25150–25156. doi: 10.1074/jbc.M010327200. [DOI] [PubMed] [Google Scholar]
- 65.Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB. Biochemistry. 1998;37:14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 66.Brownlow S, Cabral JHM, Cooper R, Flower DR, Yewdall SJ, Polikarpov I, North AC, Sawyer L. Structure. 1997;5:481–495. doi: 10.1016/s0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 67.Oliveira KM, Valente-Mesquita VL, Botelho MM, Sawyer L, Ferreira ST, Polikarpov I. Eur. J. Biochem. FEBS. 2001;268:477–483. doi: 10.1046/j.1432-1033.2001.01918.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.