Abstract
IMPORTANCE
The families evaluated in this study represent the second report of cone-rod dystrophy (CRD) cases caused by mutations in RAB28, a recently discovered gene associated with CRD.
OBJECTIVE
To determine the disease-causing gene in 2 families of Spanish descent presenting with CRD who do not have ABCA4 mutations.
DESIGN, SETTING, AND PARTICIPANTS
Molecular genetics and observational case studies of 2 families, each with 1 affected proband with CRD and 3 or 5 unaffected family members. The affected individual from each family received a complete ophthalmic examination including assessment of refractive errors and best-corrected visual acuity, biomicroscopy, color fundus photography, electroretinography analysis, and visual-evoked potential analysis. After complete sequencing of the ABCA4 gene with negative results, the screening for disease-causing mutations was performed by whole-exome sequencing. Possible disease-associated variants were determined by filtering based on minor allele frequency, predicted pathogenicity, and segregation analysis in all family members.
MAIN OUTCOMES AND MEASURES
The appearance of the macula was evaluated by clinical examination, fundus photography, and fundus autofluorescence imaging, and visual function was assessed by electroretinography. Disease-causing mutations were assessed by sequence analyses.
RESULTS
Ophthalmologic findings included markedly reduced visual acuity, bull’s eye maculopathy, foveal hyperpigmentation, peripapillary atrophy, dyschromatopsia, extinguished photopic responses, and reduced scotopic responses observed on electroretinography consistent with the CRD phenotype often associated with ABCA4 mutations. Although no ABCA4 mutations were detected in either patient, whole-exome sequencing analysis identified 2 new homozygous mutations in the recently described RAB28 gene, the c.172 + 1G>C splice site variant in IVS2 and the missense c.T651G:p.C217W substitution. Both variants were determined as deleterious by predictive programs and were segregated with the disease in both families. Sequencing of 107 additional patients of Spanish descent with CRD did not reveal other cases with RAB28 mutations.
CONCLUSIONS AND RELEVANCE
Deleterious mutations in RAB28 result in a classic CRD phenotype and are an infrequent cause of CRD in the Spanish population.
Autosomal recessive cone-rod dystrophies (arCRDs) represent a group of diseases involving a primary loss of cone photoreceptors resulting in severely reduced visual acuity, defects in color vision, atrophy in the macular region, and reduced cone responses observed on electroretinography (ERG).1 Genetic causes of arCRDs vary, but most cases (30%–50%) with a known genetic basis carry homozygous or compound heterozygous mutations in the ABCA4 (MIM601691) gene.2–4
The goal of our study was to identify genes that cause phenotypes usually attributed to ABCA4 mutations in families lacking ABCA4 disease-associated alleles. The approach combines a complete sequencing of the ABCA4 gene and adjacent intronic sequences5 or the entire ABCA4 genomic locus followed by whole-exome sequencing (WES) in cases in which no ABCA4 alleles are identified in either coding regions or introns.
Recently, RAB28 (MIM 612994), a newly identified gene responsible for a small fraction of arCRDs, was described.6 The RAB28 protein, encoded in 3 isoforms with alternative C termini, is a member of the Rab subfamily of the RAS-related small guanosine triphosphatases.7,8 The RAB28 protein is localized to the basal body and the ciliary rootlet of the photoreceptors and may be involved in ciliary transport.6 We describe 2 new, likely deleterious homozygous RAB28 mutations in 2 families of Spanish descent resulting in the CRD phenotype, suggesting that disease-associated variation in RAB28 is rare in the Spanish population.
Methods
Recruitment of Participants
The study was reviewed and approved by the ethics committee of the University Hospital Fundacion Jimenez Diaz in 2008, and it was performed according to the tenets of the Declaration of Helsinki. The participants signed a written informed consent form after the nature of procedures had been fully explained. The participants did not receive financial compensation. The collection of samples belongs to the Biobank of the University Hospital Fundacion Jimenez Diaz.
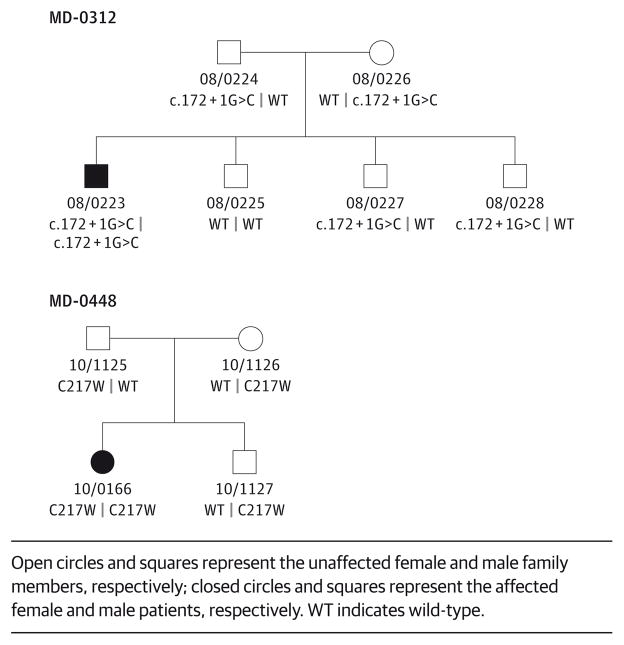
Members of 2 families, each harboring 1 affected person with CRD and compatible with a recessive mode of inheritance, were enrolled in the study in 2008 and followed up there-after. Both families were of Spanish descent with no reported consanguinity; however, parents of 1 of the 2 families (MD-0312) were from the same small town. Family members included the affected probands, both of their parents, and their unaffected siblings (Figure 1).
Figure 1. Pedigrees of the 2 Families and Segregation of the RAB28 Mutations With the Disease Phenotype.
Open circles and squares represent the unaffected female and male family members, respectively; closed circles and squares represent the affected female and male patients, respectively. WT indicates wild-type.
Clinical Evaluation
The diagnosis of arCRD was based on initial reports of poor visual acuity, blurred central vision, impairment of color vision, and intense photophobia since childhood without a history of night blindness. In addition, funduscopic evidence of atrophic macular degeneration and peripheral pigment clumping and earlier loss of cone than rod ERG amplitude were noticed.
Ophthalmologic examinations included assessment of refractive error, best-corrected visual acuity (BCVA [representative of the Snellen equivalent on a decimal scale]), biomicroscopic slitlamp examination, fundus examination, fundus autofluorescence, fluorescein angiography, chromatic sensitivity (Ishihara test), Goldmann perimetry, Ganzfeld ERG according to the International Society for Clinical Electrophysiology of Vision standards,9,10 and visual-evoked potentials.
Genetic Analyses
The probands were initially screened for variants in the ABCA4 gene by ABCA4 genotyping microarray (Asper Biotech Inc; http://www.asperbio.com) or by direct sequencing of all exons and adjacent intronic sequences, revealing no disease-associated variants. Since there are no other major CRD genes, the family underwent WES, which was performed on the affected proband, 1 unaffected parent, and 1 unaffected sibling in both families at the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC). Genomic DNA samples were constructed into paired-end precapture libraries according to the manufacturer’s protocol (Multiplexing_SamplePrep_Guide_1005361_D; Illumina, Inc) with modifications as described in the BCM-HGSC Illumina Barcoded Paired-End Capture Library Preparation11 protocol. Libraries were prepared using robotic workstations (Biomek NXp and FXp models; Beckman). The complete protocol and oligonucleotide sequences are accessible from the HGSC website (https://hgsc.bcm.edu/sites/default/files/documents/Illumina_Barcoded_Paired-End_Capture_Library_Preparation.pdf). Precaptured libraries were pooled and hybridized in solution to the HGSC CORE design (52 megabases; Roche NimbleGen), and exome capture was performed according to the manufacturer’s protocol12 with minor revisions. Library templates were prepared for sequencing using the cBot cluster generation system (TruSeq PE Cluster Generation Kits, part No. PE-401-3001; Illumina, Inc). Real-time analysis software was used to process the image analysis and base calling. Sequencing runs generated approximately 300 million to 400 million successful reads on each lane of a flow cell, yielding 9 to 10 gigabytes per sample. With these sequencing yields, samples achieved a mean of 90% of the targeted exome bases covered to a depth of 20 times or greater.
Illumina sequence analysis was performed using the HGSC Mercury analysis pipeline (https://github.com/dsexton2/Mercury-Pipeline), which addresses all aspects of data processing and analyses, moving data step by step through various analysis tools from the initial sequence generation on the instrument to annotated variant calls (single-nucleotide polymorphisms [SNPs] and intraread insertions and deletions). The pathogenicity of novel variants was assessed with predictive programs for splice sites and coding sequences (Alamut, version 2.3; http://www.interactive-biosoftware.com). All variants of interest were confirmed by Sanger sequencing, and segregation analyses were performed on all members of the 2 families.
Regions with absence of heterozygosity (AOH) were detected by analyzing the B-allele frequency data obtained from coding SNPs and WES. The B-allele frequency from WES was determined by computing the variant to total reads ratio for each SNP present in the variant call format file. The regions with AOH were identified using in-house scripts in R language (http://www.r-project.org) and the circular binary segmentation algorithm.13 A cohort of 107 unrelated patients of Spanish descent with arCRD were screened for variants on the RAB28 gene by direct Sanger sequencing of the 7 exons, including all 3 isoforms of RAB28.
Results
Clinical Examination
The clinical features of the affected individuals of both families are summarized in the Table. The affected member of the family MD-0448 was an 18-year-old woman with CRD diagnosed in her first decade of life (at 8 years). She presented with decreased visual acuity (BCVA, 0.5 on the Snellen-equivalent decimal scale [Snellen equivalent, 20/40]), high myopia, diplopia, and dyschromatopsia since childhood. Rapid deterioration of her vision was noticed at an early age; the BCVA progressed to 0.3 (Snellen equivalent, 20/65) at age 9 years and to 0.2 (Snellen equivalent, 20/100) at 12 years but has remained stable since her last examination at age 12. The funduscopic examination revealed a tigroid fundus, bull’s eye maculopathy, thin vessels, peripapillary atrophy, and remarkable optic pallor. Her peripheral retina had no signs of abnormality during initial examinations. Photopic responses were undetectable by ERG, and scotopic responses were residual. Visual-evoked potential recorded pathologic responses, including low amplitude and enlarged latency (implicit time).
Table.
Clinical Data From the 2 Probands With CRD and Mutations in the RAB28 Gene
| Family | Age at Diagnosis/Last Visit, y | Refractive Error at Last Visit | BCVAa | Observations |
|---|---|---|---|---|
| MD-0448; Spanish ancestry | 8/17 | RE: 0.3 with −8.75 (−1 to 50); LE: 0.2 with −10.25 (−1 to 160) | 0.2 (20/100) (stable at 17 y); 0.2 (20/100) (12 y); 0.3 (20/65) (9 y); 0.5 (20/40) (8 y) | Biomicroscopy: normal; funduscopy: bull’s eye maculopathy, peripapillary atrophy, optic pallor, thin vessels, tigroid fundus, no signs of peripheral involvement; color vision (Ishihara test): dyschromatopsia; electroretinogram, photopic response: absent; scotopic response: residual; visual-evoked potentials; pathologic: low amplitude of P100 wave, enlarged latency (implicit time); other: magna myopia, diplopia |
| MD-0312; Spanish ancestry | 12/22 | RE: −1 (−1.50 × 35°); LE: −0.50 (−1.25 × 140°) | RE: 0.1 (20/200); LE: 0.3 (20/65) (22 y); RE: 0.3 (20/65); LE: 0.4 (20/80) (21 y); RE: 0.5 (20/40); LE: 0.5 (20/40) (20 y) | Biomicroscopy: normal; funduscopy: loss of foveal reflex, atrophic areas in the macula, optic pallor, no flecks; fluorescein angiography: hyperfluorescent area at central and peripapillary areas, no choroidal silence; color vision (Ishihara test): dyschromatopsia (deuteranopia); visual field (Goldmann perimetry): central scotoma, development of peripheral field involvement; electroretinogram: photopic response absent, scotopic response diminished; visual-evoked potentials; diminished amplitude of P100 wave, enlarged latency (implicit time); other: intense photophobia |
Abbreviations: BCVA, best-corrected visual acuity; CRD, cone-rod dystrophy; LE, left eye; NA, not available; RE, right eye.
Best-corrected visual acuity is reported as the Snellen equivalent on a decimal scale (Snellen equivalent).
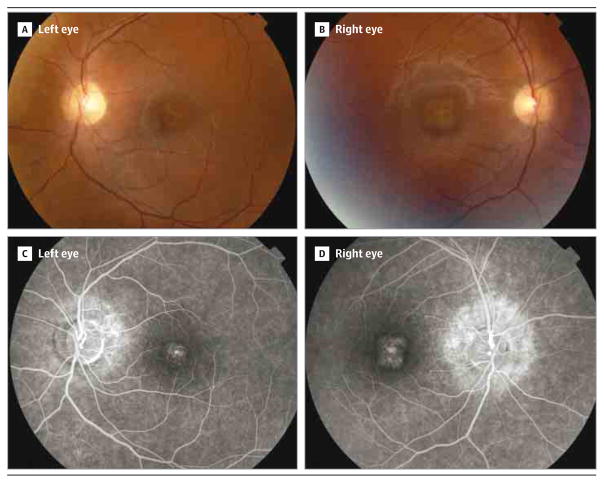
The affected member of family MD-0312 was a 30-year-old man presenting with low visual acuity, deuteranopia, and intense photophobia since diagnosis at age 12 years. At age 20, the patient’s BCVA was 0.5 (Snellen equivalent, 20/40), which decreased to 0.3 OS (Snellen equivalent, 20/65) and 0.1 OD (Snellen equivalent, 20/200) in 2 years. Funduscopic examination demonstrated a loss of the foveal reflex and optic pallor. Fluorescein angiography revealed hyperfluorescent central and peripapillary areas, with no silent choroid (Figure 2). Goldmann perimetry, performed when the patient was aged 20 years, showed central scotoma developing toward the peripheral field. At his last visit at age 22, full-field ERG testing revealed no photopic responses, and both scotopic and flicker responses were reduced. Visual-evoked potential examination recorded diminished amplitude and enlargement of the latency (implicit time).
Figure 2. Clinical Presentation of the Disease in the Patient From Family MD-0312.
A and B, Funduscopy revealed normal retinal papilla and parenchyma, with normal vessels. A, Left eye; note changes at the foveal and perifoveal retinal pigment epithelium, including yellowish deposits, together with both nasal and inferior peripapillary atrophy. B, Right eye; note similar changes with the exception of the peripapillary atrophy, which was observed only at the nasal region. C and D, Fluorescein angiography demonstrated foveal hyperfluorescent dots and peripapillary hyperflourescence in both eyes. D, A higher extension of the affected area was observed in the right eye.
In both patients, the onset of the disease started during childhood (ages 8 and 12 years), with markedly reduced visual acuity and dyschromatopsia as the first symptoms. The proband from the MD-0312 family reported intense photophobia as one of the worst initial clinical signs. However, this patient was not affected by high myopia, which was a common feature among previously described patients with mutations in the RAB28 gene.6 The proband from the MD-0448 family presented with both high myopia and diplopia.
No signs of peripheral involvement were initially observed in either patient by funduscopic examination. The patients did not report night blindness despite the reduced scotopic responses. However, the peripheral visual field involvement was observed in subsequent ophthalmologic examinations (Table). In summary, both patients presented with a remarkably similar CRD phenotype despite differences in the molecular genetic findings (splice-site mutation vs missense variant).
Genetic Analyses
Considering the apparent autosomal recessive inheritance of CRD, the observed WES variants were filtered by all of the following criteria: (1) variants were present in less than 0.5% in ESP6500 (http://evs.gs.washington.edu/EVS/), 1000 Genomes,14 and ARIC, an internal control database of 6250 exomes at Baylor Genome Center; (2) variants were in protein coding regions, in canonical splice sites, or both; (3) variants were missense, nonsense, frameshift, or splice site; and (4) variants were compound heterozygous or homozygous in the affected proband, heterozygous in a parent, and either heterozygous or absent in unaffected siblings. These analyses identified likely disease-associated homozygous variants in RAB28 in both families (Figure 1). In MD-0312, the homozygous c.172 + 1G>C variant is located in intron 2 in the splice donor site of exon 2. The change is predicted to eliminate the splice donor site, resulting in skipping of the second exon and, consequently, by conceptual translation, a nonfunctional protein. In family MD-0448, the c.T651G substitution results in a missense variant p.C217W in exon 8 of RAB28. There is a large physicochemical difference between cysteine and tryptophan (Grantham15 distance, 215), and the variant is predicted to be deleterious by all predictive programs, SIFT (score, 0.00),16 PolyPhen2 (score, 1.00),17 and MutationTaster (probability value, 1.00).18 The amino acid Cys217 is highly conserved up to Caenorhabditis elegans. Neither variant is present in 1000 Genomes, in the Exome Variant Server, or in the 6520 Baylor in-house database of control exomes.
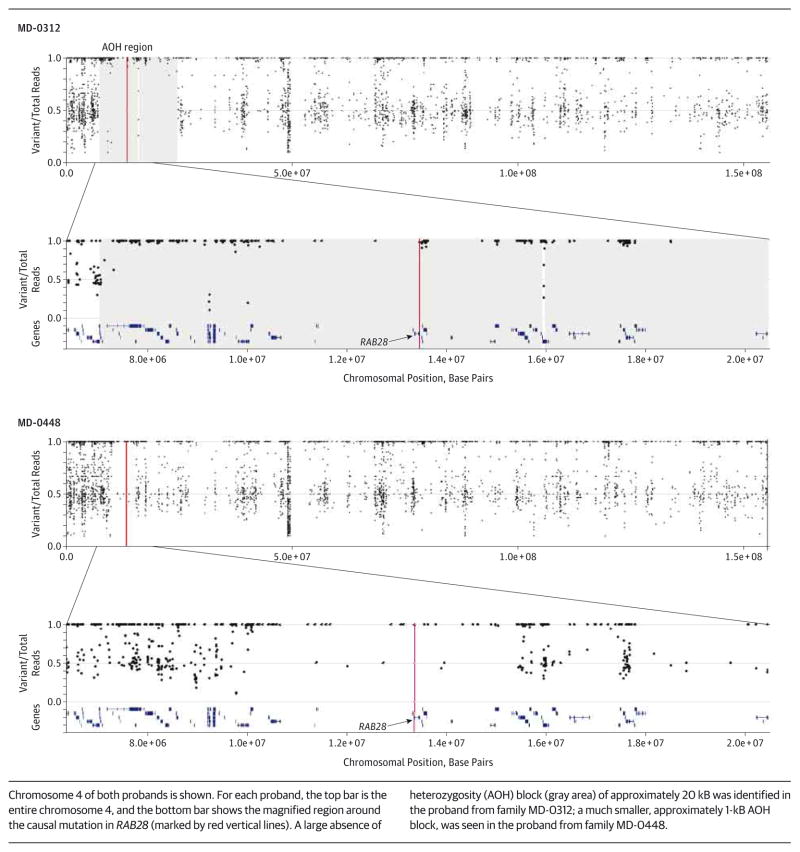
The WES and coding SNP data confirmed that the RAB28 mutation in family MD-0312 is located within a large, approximately 20-kB block of AOH (Figure 3), suggesting that the parents of the affected proband are distantly related. Considering that both parents are from the same small town and that the variant is very rare in the general population, the causal c.172 + 1G>C mutation probably arose relatively recently in the local population. The AOH was much smaller, approximately 1 kB, around the RAB28 locus in family MD-0448 (Figure 3).
Figure 3. B-Allele Frequencies Generated From Whole-Exome Sequencing Data for Both Affected Patients From Families MD-0312 and MD-0448.
Chromosome 4 of both probands is shown. For each proband, the top bar is the entire chromosome 4, and the bottom bar shows the magnified region around the causal mutation in RAB28 (marked by red vertical lines). A large absence of heterozygosity (AOH) block (gray area) of approximately 20 kB was identified in the proband from family MD-0312; a much smaller, approximately 1-kB AOH block, was seen in the proband from family MD-0448.
The RAB28 gene encodes for 3 isoforms with alternative C termini.7 The c.172 + 1G>C variant affects all 3 isoforms, and the p.C217W variant is in exon 8, which is present only in isoform 2 (RAB28L [RefSeq NM_004249.3]). Sanger sequencing of all coding exons of RAB28 in 107 unrelated Spanish patients with CRD did not identify additional mutations. No pathogenic variants were identified in the genes known to be associated with the arCRD phenotype (ABCA4, ADAM9, C8orf37, CERKL, EYS, RPGRIP1, and TULP1). After the multimodal analysis under the hypothesis of autosomal recessive inheritance, no other plausible candidate gene remained in either family.
Discussion
The advent of next-generation sequencing has allowed the identification of many rare disease genes, including genes for retinal dystrophies.6,19,20 Most often, the phenotype resulting from mutations in these genes is indistinguishable from that caused by known genes, therefore offering few clues for gene discovery. Cone-rod dystrophies are a good example of a group of diseases in which causal genes have many different functions but mutations in them cause a very similar phenotype.21 The gene most frequently mutated in arCRD is ABCA4, the gene for recessive Stargardt disease22; approximately one-third of all arCRDs is caused by the pathogenic ABCA4 variant.4,21 Cone-rod dystrophy is often considered a severe or advanced form of Stargardt disease and is usually associated with deleterious mutations in ABCA4.4,21
Screening for genetic causality is often performed in several steps, starting from the most obvious candidate gene based on the phenotype that is screened first followed by, in case of a negative result, screening of a group of genes implicated in similar phenotypes or all genes known to cause retinal phenotypes, such as the entire RetNet panel (https://sph.uth.edu/retnet/). However, because WES is now extremely robust and affordable, it is becoming more labor-and cost-efficient to prefer WES to other means of screening, especially in large families with recessive diseases in whom comprehensive segregation analysis can be performed.
The present study exemplifies this approach. We first screened ABCA4 in all patients presenting with phenotypes compatible with ABCA4 defects and, in case of negative results, performed WES on all or most available family members. The affected members of the 2 families of Spanish descent described here were identified as having classic early-onset, rapidly progressing CRD, suggesting no specific clue as to the genetic causality after the ABCA4 gene was ruled out. Evaluation using WES identified 2 new deleterious mutations in a very recently described6 CRD gene, RAB28. Screening of 107 unrelated individuals with CRD did not identify any more Spanish patients with RAB28 mutations. Roosing and colleagues6 screened more than 600 patients with CRD and cone dystrophy and assessed the sequence data on more than 400 additional individuals with CRD, Leber congenital amaurosis, and retinitis pigmentosa that are available at the European Retinal Disease Consortium; this screening resulted in identification of only 2 families segregating RAB28 mutations with a CRD phenotype. With our data and those of Roosing and colleagues, we can conclude that CRD-associated mutations in RAB28 are very rare and that they are exclusively or at least predominantly deleterious. The clinical phenotype of the 2 patients in the present study has extensive similarities with those described by Roosing et al,6 including high myopia in 1 of the 2 cases, foveal hyperpigmentation, bull’s eye maculopathy, defects in color vision, extinguished photopic responses on ERG, and peripapillary atrophy. The accumulated clinical and genetic data allow us to also conclude that RAB28 mutations cause only CRD phenotype and are not associated with related diseases, such as Stargardt disease or retinitis pigmentosa.
Expression analysis presented by Roosing and colleagues6 (as shown in Figure S1 in their article) suggests that all RAB28 isoforms, including isoform 2 (also called RAB28L), in which the mutation was detected in family MD-0448, are expressed in various human tissues including, most importantly for associated eye disease traits, retinal pigment epithelium and photoreceptors. The functional role of RAB28 and its isoforms in photoreceptors and retinal pigment epithelium is unknown, but it has been suggested6 that the localization of RAB28 to the basal body and ciliary rootlet plays a role in ciliary transport. Three other proteins (ie, C8orf37, RPGRIP1, and TULP1), with mutations causing CRD, are located in the connecting cilia of photoreceptor cells.
Conclusions
Whole-exome sequencing identified new rare, deleterious mutations in RAB28 in 2 families of Spanish descent. Whole-exome sequencing is a powerful tool for identification of rare genes in which disease-associated variants can convey a shared clinical phenotype and is especially effective in family-based analyses.
Acknowledgments
Funding/Support: This work was supported in part by National Institutes of Health grants EY021163, EY019861, HG006542, and EY019007 (Core Support for Vision Research); by research grants FIS PI13/00226, FIS PS09/00459, and RD09-0076 -00101 (Retics Biobank); CIBERER Intra/07/704.1 and Intra/09/702.1; ONCE; Fundaluce and Fundacion Conchita Rabago de Jimenez Diaz; and unrestricted funds from Research to Prevent Blindness to the Department of Ophthalmology, Columbia University.
Role of the Funder/Sponsor: The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
Author Contributions: Dr Riveiro-Álvarez and Ms Xie contributed equally to this study. Drs Ayuso and Allikmets had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Riveiro-Álvarez, López-Martínez, Ayuso, Allikmets.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Riveiro-Álvarez, Xie, Pérez-Carro, Ávila-Fernández, López-Molina, Zernant, Allikmets.
Critical revision of the manuscript for important intellectual content: Riveiro-Álvarez, Xie, López-Martínez, Gambin, Jhangiani, Muzny, Yuan, Boerwinkle, Gibbs, Lupski, Ayuso, Allikmets.
Statistical analysis: Xie, Gambin.
Obtained funding: Boerwinkle, Ayuso, Allikmets.
Administrative, technical, or material support: Riveiro-Álvarez, Ávila-Fernández, López-Molina, Zernant, Jhangiani, Muzny, Boerwinkle, Gibbs, Lupski, Allikmets.
Study supervision: Riveiro-Álvarez, Muzny, Gibbs, Ayuso, Allikmets.
References
- 1.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cremers FP, van de Pol DJ, van Driel M, et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene. ABCR Hum Mol Genet. 1998;7(3):355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 3.Maugeri A, Klevering BJ, Rohrschneider K, et al. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67(4):960–966. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riveiro-Álvarez R, López-Martínez MA, Zernant J, et al. Outcome of ABCA4 disease-associated alleles in autosomal recessive retinal dystrophies: retrospective analysis in 420 Spanish families. Ophthalmology. 2013;120(11):2332–2337. doi: 10.1016/j.ophtha.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zernant J, Schubert C, Im KM, et al. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci. 2011;52(11):8479–8487. doi: 10.1167/iovs.11-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roosing S, Rohrschneider K, Beryozkin A, et al. European Retinal Disease Consortium. Mutations in RAB28, encoding a farnesylated small GTPase, are associated with autosomal-recessive cone-rod dystrophy. Am J Hum Genet. 2013;93(1):110–117. doi: 10.1016/j.ajhg.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brauers A, Schürmann A, Massmann S, et al. Alternative mRNA splicing of the novel GTPase Rab28 generates isoforms with different C-termini. Eur J Biochem. 1996;237(3):833–840. doi: 10.1111/j.1432-1033.1996.0833p.x. [DOI] [PubMed] [Google Scholar]
- 8.Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: molecular cloning of YPT-related cDNAs from a rat brain library. Proc Natl Acad Sci U S A. 1987;84(23):8210–8214. doi: 10.1073/pnas.84.23.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marmor MF, Brigell MG, McCulloch DL, Westall CA, Bach M. International Society for Clinical Electrophysiology of Vision. ISCEV standard for clinical electro-oculography (2010 update) Doc Ophthalmol. 2011;122(1):1–7. doi: 10.1007/s10633-011-9259-0. [DOI] [PubMed] [Google Scholar]
- 10.Marmor MF, Holder GE, Seeliger MW, Yamamoto S. International Society for Clinical Electrophysiology of Vision. Standard for clinical electroretinography (2004 update) Doc Ophthalmol. 2004;108(2):107–114. doi: 10.1023/b:doop.0000036793.44912.45. [DOI] [PubMed] [Google Scholar]
- 11.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5(6):57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NimbleGen SeqCap EZ Library LR user’s guide. Roche; [Accessed September 19, 2014]. http://www.nimblegen.com/products/lit/06560881001_SeqCapEZLibraryLR_Guide_v2p0.pdf. [Google Scholar]
- 13.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 14.Abecasis GR, Altshuler D, Auton A, et al. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185 (4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 16.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 19.Ozgül RK, Siemiatkowska AM, Yücel D, et al. European Retinal Disease Consortium. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89(2):253–264. doi: 10.1016/j.ajhg.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang PW, Wang J, Chen Y, et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat Genet. 2012;44 (9):972–974. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 21.Thiadens AAHJ, Phan TML, Zekveld-Vroon RC, et al. Writing Committee for the Cone Disorders Study Group Consortium. Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology. 2012;119(4):819–826. doi: 10.1016/j.ophtha.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Allikmets R, Singh N, Sun H, et al. A photoreceptor cell–specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15(3):236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]