Abstract
Genetic risk scores (GRS), summing up the total effect of several single-nucleotide polymorphisms (SNPs) in genes associated with either coronary risk or cardiovascular risk factors, have been tested for association with ischemic stroke with conflicting results. Recently an association was found between a GRS based on 29 SNPs discovered by genome-wide association studies and hypertension. The aim of our study was to investigate the possible association of the same GRS with ischemic stroke on top of other ‘traditional risk factors', also testing its potential improvement in indices of discrimination and reclassification, in a Swedish case–control study. Twenty-nine SNPs were genotyped in 3677 stroke cases and 2415 controls included in the Lund Stroke Register (LSR), the Malmö Diet and Cancer (MDC) study and the Sahlgrenska Academy Study on Ischemic Stroke (SAHLSIS). The analysis was conducted in the combined sample, and separately for the three studies. After adjustment for hypertension, diabetes mellitus and smoking habits, the GRS was associated with ischemic stroke in the combined sample (OR (95% CI) 1.086 (1.029–1.147) per SD increase in the GRS P=0.003) with similar trends in all three samples: LSR (1.050 (0.967–1.140); P=0.25), MDC (1.168 (1.060–1.288); P=0.002) and SAHLSIS (1.124 (0.997–1.267); P=0.055). Measures of risk discrimination and reclassification improved marginally using the GRS. A blood pressure GRS is independently associated with ischemic stroke risk in three Swedish case–control studies, however, the effect size is low and adds marginally to prediction of stroke on top of traditional risk factors including hypertension.
Introduction
Stroke is one of the leading causes of mortality, morbidity and disability worldwide.1 Thanks to genome-wide association studies (GWAS), genetics of complex diseases has made dramatic steps forward in the last few years2 but very few single-nucleotide polymorphisms (SNPs) have been invariantly associated with ischemic stroke.3, 4, 5, 6, 7 Moreover, the relative risk conferred by individual genetic variants is usually low. Thus, in an attempt to estimate the aggregate effect of several gene variants, different genetic risk scores (GRS) have been constructed including a few studies on stroke with inconsistent results.8, 9, 10, 11, 12, 13, 14, 15 Interestingly, the selection of the SNPs to be included in GRS calculation have been based on different criteria: SNPs detected by GWAS either associated or not associated with known cardiovascular risk factors, as well as SNPs in candidate genes or pathways.8, 9, 10, 11, 12, 13, 14, 15
High blood pressure (BP) is the major risk factors for stroke and among the most important ones for other cardiovascular events.16,17 Despite BP and hypertension being heritable traits,18 the search for genetic variants associated with these traits has been more challenging compared with other cardiovascular risk factors. Only in recent years, GWAS have discovered several genetic variants which associate with BP-related traits.19, 20, 21 Indeed a GRS based on 29 SNPs has been associated with the prevalence of hypertension and the incidence of coronary events and stroke in a meta-analsyis,21 and more recently, also with the incidence of hypertension.22 The aim of the present study was to test the association of a GRS, consisting in the weighted allele sum of 29 SNPs previously associated with high BP, with ischemic stroke in a case–control collaboration consisting of three Swedish case–control studies, including more than 6000 individuals. Moreover, the possible improvement in the prediction of stroke over some ‘traditional risk factors', such as hypertension, diabetes and smoking habit, was assessed using indices of calibration, discrimination and reclassification applicable to a case–control design.
Materials and methods
All study participants or, when applicable, their next-of-kin provided informed consent. The procedures were in accordance with the institutional guidelines. The Ethics Committee of the Medical Faculty of Lund University and the University of Gothenburg approved the study.
Subjects
Lund stroke register (LSR)
LSR is an ongoing prospective study, recruiting consecutive patients with first-ever stroke from 2001 and later from the primary uptake area of Skåne University Hospital, Lund. The patients were 18 years or older at stroke onset and all patients diagnosed as having ischemic stroke were examined with neuroimaging or autopsy of the brain. The large majority of these patients are treated at the stroke unit of Skåne University Hospital, Lund.23 Control subjects were individuals without stroke, randomly selected from the same geographical uptake area and age and gender matched to patients included during the first year (2001–2002) of the LSR project. All included subjects (or if they were unable to answer, their next-of-kin) provided informed consent to participate. In the current study, patients with ischemic stroke were included if they also had provided a blood sample for DNA analysis.
Malmö diet and cancer (MDC)
Eight hundred and seventy-three ischemic stroke cases and 867 age- and sex-matched controls were selected from subjects recruited and followed over time in the MDC, a longitudinal study, ongoing in the urban-zone of Malmö, Sweden. Between 1991 and 1996, women aged 45–73 years and men aged 46–73 years, with residency in Malmö (~250 000 habitants), were invited by mail and by newspaper advertisement to participate in the MDC.24 In all, 28 449 participated out of an eligible population of 74 000. The participants were asked to complete a self-administered questionnaire at home, which included items on lifestyle factors, medication, previous and current diseases.
Sahlgrenska Academy Study on Ischemic Stroke (SAHLSIS)
The SAHLSIS is a case–control study on ischemic stroke before 70 years of age, the design of which has been described in detail before.25 Briefly, white patients who presented with first-ever or recurrent acute ischemic stroke before reaching the age of 70 years (n=844) were consecutively recruited between 1998 and 2008 at four stroke units in Western Sweden. White community-control subjects (n=668) from the same geographic area as the cases were randomly selected to match cases for age and sex.
Stroke definition
In all three samples, ischemic stroke was defined as rapidly developed clinical signs of local or global loss of cerebral function that lasted for 24 h or led to death within 24 h following the World Health Organization's definition. By definition, patients with transient ischemic attacks are excluded. CT, MRI or autopsy verified the infarction or excluded hemorrhage and nonvascular disease.23,25,26
Data about etiologic subtypes of ischemic stroke according to the modified ‘Trial of Org 10172 in Acute Stroke Treatment' (TOAST) criteria were available only for the LSR and SAHLSIS samples.27
Hypertension definition in different studies
In the three samples, hypertension was defined as being on antihypertensive treatment or having a systolic BP/diastolic BP ≥160/90 mm Hg.
Diabetes mellitus definition in different studies
In the LSR study, diabetes mellitus diagnosis was made based on fasting capillary or whole-blood glucose ≥6.1 mMol/l or plasma glucose ≥7.0 mMol/l at two time points usually during the hospitalization of the patient in the acute phase within 0–2 days interval; or non-fasting glucose above 11 mMol/l in addition with clinical symptoms; or previous treatment for diabetes mellitus (diet, oral medication or insulin). In the MDC study, diabetes mellitus was defined as use of antidiabetic medication or self-reported history of a physician's diagnosis of diabetes. In SAHLSIS, diabetes mellitus was defined by diet or pharmacological treatment, fasting plasma glucose ≥7.0 mmol/l, or fasting blood glucose ≥6.1 mmol/l.
Anamnestic data in different studies
In the LSR study, information about medical history was obtained from subjects (or when applicable from next-of-kin or previous medical records) at baseline investigation (for cases – in connection to stroke onset; for control subjects – in connection to clinical examination for inclusion in the LSR study). In the MDC and SAHLSIS studies, information about medical history and smoking habits was derived from a structured self-administered questionnaire. Smoking habits were coded as ‘current' vs ‘never' or ‘former'.
Genotype analysis
Information about the different SNPs included in the GRS is reported in the Online Supplementary Methods. The SNPs were genotyped using IPLEX on a MassARRAY platform (Sequenom, San Diego, CA, USA) and at KBioscience in the United Kingdom according to the manufacturer's standard protocols. Nearly 25% of the samples were run in duplicate. All genotypes were called by two different investigators. We pre-specified a threshold call rate of 90% per individual SNP and a threshold of P<10−03 for excluding SNPs according to Hardy–Weinberg equilibrium calculation in controls. Genetic data have been deposited at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001000936.
Genetic risk score
To create the multivariable GRS for each study participant we used the weighted method (weighted GRS) according to the beta value attributed to the 29 tested SNPs in previous association studies,19, 20, 21, 22 assuming each SNP to be independently associated with risk28 according to an additive genetic model. For each SNP included in the GRS calculation, weightings of 0, 1 and 2 were attributed according to the number of risk alleles (defined as coded) present. Successively, the number of corresponding coded alleles (0, 1 or 2) was multiplied for the absolute value of the beta-coefficient for hypertension, as detected in previous studies,19, 20, 21 and then these products were summed up. Thus, in our approach, different SNPs contribute with different weights to the GRS value, as opposed to an alternative approach in which no weighting of effects is used, and each SNP allele counts equally in the score. Successively, the risk score was divided by the number of effectively genotyped SNPs to produce an average measure, which takes into account the amount of effectively genotyped SNPs, and the ratio was standardized. The GRS was modeled as a continuous variable (increase in one unit means an increase in one SD of the GRS) and as tertiles (see the Online Supplementary Methods for the specific threshold, which were used to divide the different tertiles of GRS and for more details about the equation used to calculate the GRS). See also Supplementary Figure S1 for the distribution of GRS in the population. Only subjects with at least 27 valid genotypes were included in the final analysis (see also Supplementary Table S3 for the description of included/excluded subjects). This threshold was arbitrary chosen for allowing that not too many participants were excluded and, at the same time, that not too much noise was introduced to the GRS because of the fact that it is not based on all the genotypes.
Statistical analysis
Throughout the manuscript, continuous variables are reported as the mean±SD. The χ2-test (Pearson) was used to compare group frequencies and to test for deviations from Hardy–Weinberg equilibrium. Logistic regression (stepwise methods: backward Wald) analysis was used in the multivariate models with ischemic stroke as the dependent variables and either age and sex (model A), or age, sex, diabetes mellitus and smoking habits (model B) or age, sex, diabetes mellitus, smoking habits and hypertension (model C) as independent variables. The analyses were performed using the GRS modeled as a continuous variable and as tertiles (see above for more details).
An unbiased estimate of the variance explained by the GRS was obtained by evaluating the increase in explained variance of the trait when adding the GRS to all the covariates included in model C, when tested in logistic regression (Nagelkerke r2). Model calibration was assessed with the Hosmer–Lemeshow goodness-of-fit test.29 All these analyses were performed using SPSS statistical software (version 20.0; SPSS Inc. Chicago, IL, USA).
We assessed the improvement in risk discrimination by comparing the area under the receiver operator characteristic (ROC) curves (AUC) in models with all the nongenetic covariates significantly associated with stoke and in the same model plus the GRS. ROC curves were developed using a probability-weighted Cox model. The category-less net reclassification improvement (NRI) index for case–control studies and the integrated discrimination improvement (IDI) index were estimated according to Pencina et al.30,31 using the Hmisc library by Frank E Harrell Jr. implemented in the R statistical software (version 2.15.2).32
All tests were two-sided and P-values <0.05 were considered statistically significant. Bonferroni adjustments were performed when appropriate.
Results
The clinical characteristics of the participants in the three case–control studies and in the combined sample are summarized in Table 1. Results concerning Hardy–Weinberg equilibrium and details about individual markers are presented in Supplementary Table S1.
Table 1. Baseline characteristics for the three case–control samples (subjects with at least 27 valid genotypes).
| Combined samples | LSR | MDCa | SAHLSIS | |||||
|---|---|---|---|---|---|---|---|---|
| Control, n=2415 | IS, n=3677 | Control, n=910 | IS, n=2036 | Control, n=867 | IS n=873 | Control, n=638 | IS, n=768 | |
| Age, y (SD) | 65.3±12.3 | 67.8±13.3a | 74.0±12.0 | 74.3±12.4 | 62.9±6.6 | 62.9±6.6 | 56.1±10.4 | 56.1±10.7 |
| Male sex, n (%) | 1364 (56.5) | 2044 (55.5) | 518 (56.9) | 1060 (51.9)b | 471 (54.3) | 481 (55.1) | 375 (58.8) | 503 (65.5)b |
| Hypertension, n (%) | 1169 (48.5) | 2408 (66.5)a | 432 (47.6) | 1314 (66.0)a | 516 (59.6) | 646 (74.1)a | 221 (34.7) | 448 (59.2)a |
| Diabetes mellitus, n (%) | 122 (5.2) | 705 (20.1)a | 66 (7.3) | 481 (24.5)a | 25 (2.9) | 83 (9.5)a | 31 (4.9) | 141 (18.4)a |
| Smoking, n (%) | 404 (16.8) | 971 (26.8)a | 90 (9.9) | 394 (19.7)a | 188 (22.0) | 282 (33.1)a | 126 (19.7) | 295 (38.6)a |
Abbreviations: IS, ischemic stroke; LSR, Lund Stroke Register; MDC, Malmö Diet and Cancer study; SAHLSIS, Sahlgrenska Academy Study on Ischemic Stroke.
*P<0.001.
In the MDC the data, apart from age, refer to the baseline evaluation.
P<0.05 between cases and controls.
As expected, the GRS was significantly associated with hypertension, after adjustment for age and sex, in the combined sample (OR (95% CI): 1.175 (1.115–1.238) P=2.0E−09) and in the three samples separately (LSR: 1.194 (1.107–1.288) P=4.0E−06; MDC: 1.160 (1.050–1.282) P=0.004; SAHLSIS: 1.129 (1.005–1.268) P=0.041), whereas no association was evident between the GRS and either diabetes mellitus or smoking habits.
Association analysis between the GRS and ischemic stroke
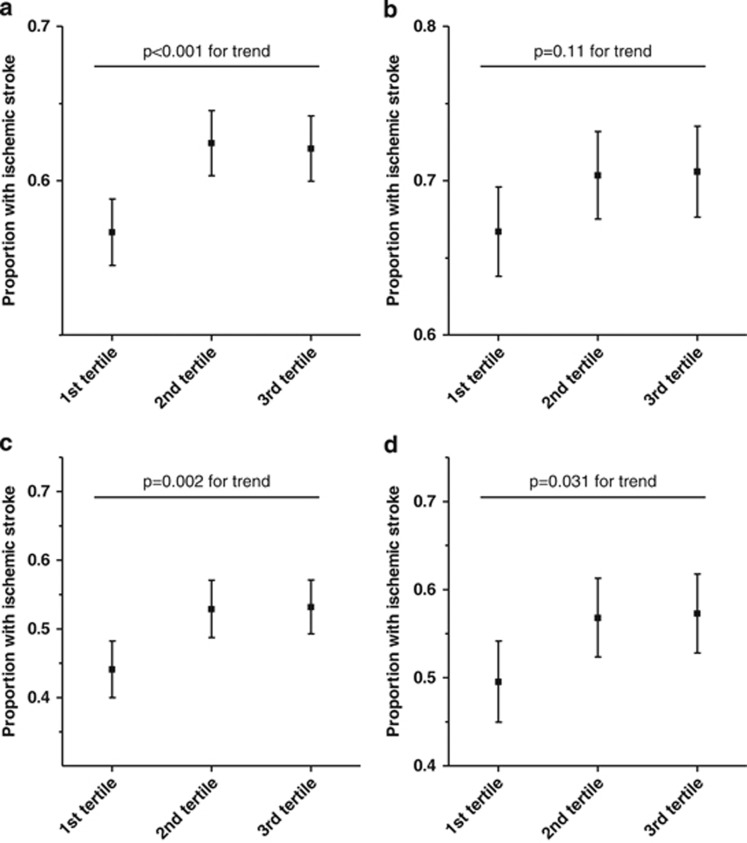
Figure 1 shows the association between the GRS in tertiles and ischemic stroke in the combined sample and in the three studies separately. In the combined sample, using regression model A, adjusting for age and sex, the GRS was associated with ischemic stroke (Table 2). When diabetes mellitus, smoking habits and hypertension also were included in the model, the association was somewhat attenuated but remained significant in the combined sample (model B and C). A similar trend was evident in the MDC, SAHLSIS and LSR, although this association did not reach statistical significance in the latter sample. The OR for ischemic stroke was nearly 25% higher in individuals classified in the 3rd tertile according to the GRS compared with those classified in the 1st tertile (Table 2). However, the magnitude of this OR was lower than that of the traditional risk factors, including hypertension (Table 3). The part of variance explained by the logistic regression model with the addition of the GRS was 0.170 as compared with 0.167 when tested after full adjustment but without the GRS (Nagelkerke r2) and model calibration was good for both models (P=0.671 for the model without the GRS and 0.544 for the model with the GRS). Only subjects with at least 27 valid genotypes were included in the analyses. However, the inclusion of subjects with 26 or 25 valid SNPs did not alter the results substantially (data not shown).
Figure 1.
Association between the weighted GRS (in tertiles) and ischemic stroke in the combined sample (a) and in the three studies separately (b=LSR, c=MDC, d=SAHLSIS).
Table 2. Association of the GRS with ischemic stroke in MDC, LSR, SAHLSIS and in the combined sample.
| Regression model | ||||||
|---|---|---|---|---|---|---|
| Model A | Model B | Model C | ||||
| OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | |
| Combined samples | ||||||
| GRS | 1.101 (1.046–1.160) | <0.001 | 1.111 (1.053–1.173) | <0.001 | 1.086 (1.029–1.147) | 0.003 |
| Tertiles | For trend | <0.001 | For trend | <0.001 | For trend | 0.001 |
| 1st vs 2nd | 1.278 (1.127–1.450) | <0.001 | 1.296 (1.137–1.479) | <0.001 | 1.261 (1.103–1.441) | 0.001 |
| 1st vs 3rd | 1.263 (1.114–1.433) | <0.001 | 1.304 (1.143–1.486) | <0.001 | 1.242 (1.087–1.418) | 0.001 |
| LSR | ||||||
| GRS | 1.093 (1.011–1.181) | 0.026 | 1.081 (0.997–1.172) | 0.058 | 1.050 (0.967–1.140) | 0.248 |
| Tertiles | For trend | 0.081 | For trend | 0.175 | For trend | 0.431 |
| 1st vs 2nd | 1.203 (0.996–1.452) | 0.055 | 1.181 (0.971–1.437) | 0.097 | 1.136 (0.930–1.386) | 0.211 |
| 1st vs 3rd | 1.208 (0.997–1.465) | 0.054 | 1.170 (0.958–1.430) | 0.125 | 1.098 (0.895–1.346) | 0.369 |
| MDC | ||||||
| GRS | 1.153 (1.051–1.265) | 0.003 | 1.187 (1.078–1.307) | <0.001 | 1.168 (1.060–1.288) | 0.002 |
| Tertiles | For trend | 0.002 | For trend | <0.001 | For trend | 0.002 |
| 1st vs 2nd | 1.422 (1.123–1.802) | 0.003 | 1.483 (1.161–1.894) | 0.002 | 1.445 (1.128–1.852) | 0.004 |
| 1st vs 3rd | 1.443 (1.147–1.815) | 0.002 | 1.556 (1.228–1.971) | <0.001 | 1.499 (1.180–1.905) | 0.001 |
| SAHLSIS | ||||||
| GRS | 1.118 (1.001–1.249) | 0.048 | 1.150 (1.023–1.291) | 0.019 | 1.124 (0.997–1.267) | 0.055 |
| Tertiles | For trend | 0.033 | For trend | 0.006 | For trend | 0.014 |
| 1st vs 2nd | 1.331 (1.027–1.723) | 0.030 | 1.407 (1.072–1.847) | 0.014 | 1.407 (1.062–1.865) | 0.017 |
| 1st vs 3rd | 1.367 (1.055–1.773) | 0.018 | 1.521 (1.158–2.000) | 0.003 | 1.472 (1.111–1.950) | 0.007 |
Abbreviation: GRS, genetic risk score.
Model A (age, sex adjusted); model B (age, sex, diabetes mellitus, smoking habits adjusted); and model C (model B+hypertension). Units are the unit of phenotypic measurement per SD of genetic risk score.
Table 3. Odds ratios (95% confidence intervals) as found in logistic regression (multivariate model C) for stroke incidence in the combined sample.
| OR (95%CI) | P-value | |
|---|---|---|
| Age, years | 1.012 (1.008–1.016) | <0.001 |
| Diabetes mellitus | 3.981 (3.246–4.883) | <0.001 |
| Smoking habits | 2.179 (1.897–2.503) | <0.001 |
| Hypertension | 1.867 (1.670–2.088) | <0.001 |
| GRS for trend | 0.001 | |
| GRS, 2nd tertile | 1.261 (1.103–1.441) | 0.001 |
| GRS, 3rd tertile | 1.242 (1.087–1.418) | 0.001 |
Abbreviation: GRS, genetic risk score.
Sex was discarded from the model; sex was coded as one male and two female.
In an exploratory association analysis of stroke subtypes, including only in the SAHLSIS and LSR samples, the stroke subtypes that were associated with the GRS in multivariate analysis were cryptogenic stroke and the combined group ‘other or undetermined causes' (see Supplementary Table S5) in the combined sample and in SAHLSIS. In addition, small vessel disease was associated with the GRS in SAHLSIS.
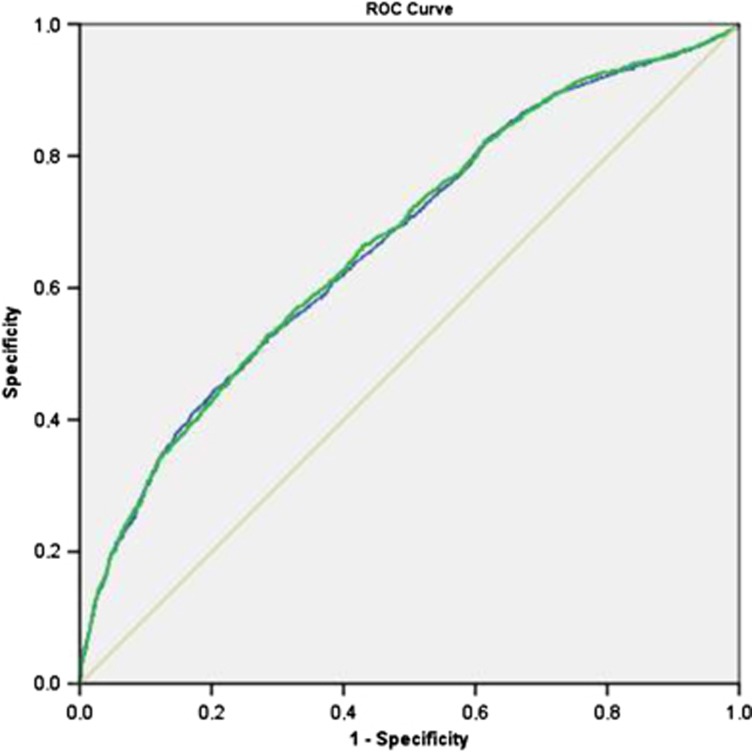
The AUC using the model with the GRS was not significantly improved as compared with the model without the GRS (0.672±0.007 vs 0.669±0.007; P>0.05, see Figure 2). The category free NRI (>0), which is applicable to case–control studies, was statistically significant (0.0659±0.0265; P=0.013; see also Supplementary Table S4), but not when evaluated in cases (0.0328±0.0169) separately from controls (0.0331±0.0205; P=0.106 and ). The IDI was significant (0.001452±0.000498; P=0.003)
Figure 2.
Receiver operator characteristics (ROC) curves for stroke discrimination using nongenetic risk factors (age, sex, hypertension, diabetes, smoking) as compared with nongenetic risk factors plus the GRS.
Association analysis between individual SNPs and ischemic stroke
Results from the association analysis between individual SNPs and ischemic stroke, after full adjustment (model C), are presented in Supplementary Table S2. A few SNPs showed a marginally significant P-value either in the combined sample or in the different studies separately. However, their significance did not withstand adjustment for multiple testing using the Bonferroni correction.
Discussion
The main finding of the present study is that a GRS, previously associated with hypertension incidence and prevalence, is associated with ischemic stroke in a large collaborative study pooling together three Swedish case–control studies. This result suggests that life-long exposure to small genetically mediated differences in BP may be causally related to ischemic stroke. However, the effect size of the tested GRS is relatively low and the improvement in the AUC, NRI and IDI indexes is too modest to add clinically meaningful predictive value when it is added on top of known risk factors. Our results are in line with recent studies using similar GRS,21,33 which showed an overall modest improvement in relative risk estimates (either odds ratio or hazard ratio): in the International Consortium for Blood Pressure GWAS, the increase in the risk of stroke passing from the 1st to the 5th quintile of the GRS spanned between 1.23 and 1.34, and in a study involving several Finnish cohorts between 1.23 and 1.35.21,33
There are a number of possible reasons why the effect size of the GRS was low in our study and even not significant in all studies separately. First, none of the individual SNPs included in the GRS, when analyzed separately, had a nominal association with ischemic stroke and someone had a paradoxical inverse effect (that is the ‘risk allele' confers protection in one or more subsamples of the present study). Second, it is possible that a different selection of beta-coefficients based on larger GWAS or on GWAS that analyze more appropriate population cohorts or even beta-coefficients, which take into account interaction between genes, could result in an improved GRS compared with the one that we used in this study. Furthermore, this is a GRS of hypertension and in the present samples the effect (OR) of hypertension on stroke is unexpectedly modest compared with other risk factors such as smoking and diabetes mellitus. Other reasons could refer to ischemic stroke per se. Ischemic stroke is a very heterogeneous disease with different etiological subtypes, whose causes, including genetic determinants, could be very different. In our exploratory analysis on stroke subtypes, the GRS showed association with cryptogenic stroke and with small vessels occlusion. However, the sample size for subtypes is small compared with genetic study standards, and thus further investigations of this GRS based on larger, well-characterized samples of patients with ischemic stroke are clearly warranted to see if there are subtype-specific associations. In addition, the heritability of ischemic stroke, as estimated in previous studies, is quite low making the discovery of SNPs constantly associated with it quite challenging.34, 35, 36, 37, 38 On the contrary, heritability of hypertension and BP-related traits seems higher18 and GWAS have reported different SNPs highly significantly associated with BP even if with quite low effect size in terms of mmHg.19, 20, 21,39 This evidence could support the hypothesis that the genetic component of stroke susceptibility that is mediated by hypertension is quite diluted. Another hypothesis is that rarer variants in genes implicated in Mendelian forms of stroke may have larger effects. It has already been shown that carriers of genetic variants in genes implicated in Mendelian forms of hyper- or hypotension, are associated with huge variation in BP40,41 as compared with common SNPs.21 In the same manner, rare mutations implicated in Mendelian forms of stroke, as well as maternally inherited mitrochondrial mutations, could contribute more than common SNPs to the occurrence of cerebrovascular events.42,43 Thus, we speculate that incorporating these rarer variants in GRS, as well as other SNPs directly related to stroke or stroke subtypes, could substantially augment the prediction of a genetic score.
Strengths of our study include the large sample size, with a number of stroke cases similar to that of studies on candidate genes in a recent meta-analysis,44 the genetic homogeneity of the populations, and the rigorous classification of stroke cases and controls. Even if longitudinal studies are better suited to assess predictivity with respect to case–control ones with a cross-sectional design, we tried to contrast this limitation by applying indices that have been adapted to this type of studies. Other major limitations of the GRS are that no interactions between single SNPs with other genetic variants and/or with other demographic or environmental factors were taken into account. The analysis of stroke subtypes is underpowered and was only possible to perform it on two samples. Furthermore, there is an overlap between nearly 300 stroke cases and controls in MDC (11% of the total sample) between the present study and the first study of the GRS,21 potentially overinflating the results. Finally, the effect of the GRS seems nonlinear, suggesting that the GRS we used, and especially the beta-coefficients derived from previous studies, could be non-optimal for a finer discrimination of stroke risk, at least in our samples.
Perspectives
In conclusion, we confirmed an independent association of a GRS with ischemic stroke in a case–control study. However, the low magnitude of the effect and the modest improvement in discrimination and reclassification indicate that the GRS, in its present form, is insufficient to add valuable information in the clinical setting. Larger genetic studies focusing on ischemic stroke subtypes could help clarify the pathophysiology of this worldwide burden disease. Further studies are needed to detect newer GRS, including well-validated SNPs along with rarer genetic variants, associated with stroke.
Acknowledgments
The authors acknowledge the Knut and Alice Wallenberg Foundation for its economic support of the SWEGENE DNA-extraction facility. DNA extraction and preparation for LSR was performed by Biobank Skåne, Labmedicin Skåne. This study was supported by grants from the European Research Council (StG-282255), the Swedish Research Council (K2011-65X-14605-09-6 and K2010-61X-20378-04-3), the Swedish state (ALFGBG-148861), the Swedish Heart and Lung Foundation, the Swedish Stroke Association, the Medical Faculty of Lund University, Malmö University Hospital, the Freemasons Lodge of Instruction EOS in Lund, the Region Skane, the Lennart Hansson Memorial Fund, the Rune and Ulla Amlöv, John and Brit Wennerström, Lars Hierta, Edit Jacobson, Tore Nilsson, Albert Påhlsson, Crafoord, Ernhold Lundströms, Hulda and Conrad Mossfelt, and the King Gustaf V and Queen Victoria research foundations.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Feigin VL, Lawes CM, Bennett DA, Barker-Collo SL, Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- Fava C, Montagnana M, Guidi GC, Melander O. From circulating biomarkers to genomics and imaging in the prediction of cardiovascular events in the general population. Ann Med. 2011;44:433–447. doi: 10.3109/07853890.2011.582511. [DOI] [PubMed] [Google Scholar]
- Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenguez C, Bevan S, Gschwendtner A, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram MA, Seshadri S, Bis JC, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday EG, Maguire JM, Evans TJ, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex A, Gaetani E, Papaleo P, et al. Proinflammatory genetic profiles in subjects with history of ischemic stroke. Stroke. 2004;35:2270–2275. doi: 10.1161/01.STR.0000140740.19421.fe. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- Paynter NP, Chasman DI, Pare G, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiannakouris N, Katsoulis M, Dilis V, et al. Genetic predisposition to coronary heart disease and stroke using an additive genetic risk score: a population-based study in Greece. Atherosclerosis. 2012;222:175–179. doi: 10.1016/j.atherosclerosis.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis-Ganella C, Subirana I, Lucas G, et al. Assessment of the value of a genetic risk score in improving the estimation of coronary risk. Atherosclerosis. 2012;222:456–463. doi: 10.1016/j.atherosclerosis.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim-Verbaas CA, Fornage M, Bis JC, et al. Predicting stroke through genetic risk functions: the CHARGE Risk Score Project. Stroke. 2014;45:403–412. doi: 10.1161/STROKEAHA.113.003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Bevan S, Nalls MA, et al. Multilocus genetic risk score associates with ischemic stroke in case-control and prospective cohort studies. Stroke. 2014;45:394–402. doi: 10.1161/STROKEAHA.113.002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- Vasan RS, Larson MG, Leip EP, et al. Impact of high-normal blood pressure on the risk of cardiovascular disease. N Engl J Med. 2001;345:1291–1297. doi: 10.1056/NEJMoa003417. [DOI] [PubMed] [Google Scholar]
- Fava C, Burri P, Almgren P, Groop L, Hulthen UL, Melander O. Heritability of ambulatory and office blood pressure phenotypes in Swedish families. J Hypertens. 2004;22:1717–1721. doi: 10.1097/00004872-200409000-00015. [DOI] [PubMed] [Google Scholar]
- Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava C, Sjogren M, Montagnana M, et al. Prediction of blood pressure changes over time and incidence of hypertension by a genetic risk score in Swedes. Hypertension. 2013;61:319–326. doi: 10.1161/HYPERTENSIONAHA.112.202655. [DOI] [PubMed] [Google Scholar]
- Hallstrom B, Jonsson AC, Nerbrand C, Petersen B, Norrving B, Lindgren A. Lund Stroke Register: hospitalization pattern and yield of different screening methods for first-ever stroke. Acta Neurol Scand. 2007;115:49–54. doi: 10.1111/j.1600-0404.2006.00738.x. [DOI] [PubMed] [Google Scholar]
- Berglund G, Elmstahl S, Janzon L, Larsson SA. The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med. 1993;233:45–51. doi: 10.1111/j.1365-2796.1993.tb00647.x. [DOI] [PubMed] [Google Scholar]
- Jood K, Ladenvall C, Rosengren A, Blomstrand C, Jern C. Family history in ischemic stroke before 70 years of age: the Sahlgrenska Academy Study on Ischemic Stroke. Stroke. 2005;36:1383–1387. doi: 10.1161/01.STR.0000169944.46025.09. [DOI] [PubMed] [Google Scholar]
- Zia E, Hedblad B, Pessah-Rasmussen H, Berglund G, Janzon L, Engström G. Blood pressure in relation to the incidence of cerebral infarction and intracerebral hemorrhage. Hypertensive hemorrhage: debated nomenclature is still relevant. Stroke. 2007;38:2681–2685. doi: 10.1161/STROKEAHA.106.479725. [DOI] [PubMed] [Google Scholar]
- Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- Horne BD, Anderson JL, Carlquist JF, et al. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Ann Hum Genet. 2005;69:176–186. doi: 10.1046/j.1529-8817.2005.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmer DW, Hosmer T, Le CS, Lemeshow S. A comparison of goodness-of-fit tests for the logistic regression model. Stat Med. 1997;16:965–980. doi: 10.1002/(sici)1097-0258(19970515)16:9<965::aid-sim509>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB, Sr, D'Agostino Jr, RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino Sr, RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core TeamR: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/. R Foundation for Statistical Computing, Vienna, Austria. 2013.
- Havulinna AS, Kettunen J, Ukkola O, et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32 669 Individuals. Hypertension. 2013;61 (5:987–994. doi: 10.1161/HYPERTENSIONAHA.111.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–161. doi: 10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- Touze E, Rothwell PM. Sex differences in heritability of ischemic stroke: a systematic review and meta-analysis. Stroke. 2008;39:16–23. doi: 10.1161/STROKEAHA.107.484618. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Silver LE, Heneghan C, et al. Relative familial clustering of cerebral versus coronary ischemic events. Circ Cardiovasc Genet. 2011;4:390–396. doi: 10.1161/CIRCGENETICS.110.959114. [DOI] [PubMed] [Google Scholar]
- Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Melander O, Johnson T, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava C, Montagnana M, Rosberg L, et al. Subjects heterozygous for genetic loss of function of the thiazide-sensitive cotransporter have reduced blood pressure. Hum Mol Genet. 2008;17:413–418. doi: 10.1093/hmg/ddm318. [DOI] [PubMed] [Google Scholar]
- Ji W, Foo JN, O'Roak BJ, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjot T, Yadav S, Hasan N, Bentley P, Sharma P. Genes associated with adult cerebral venous thrombosis. Stroke. 2011;42:913–918. doi: 10.1161/STROKEAHA.110.602672. [DOI] [PubMed] [Google Scholar]
- Silliman S. Mendelian and mitochondrial disorders associated with stroke. J Stroke Cerebrovasc Dis. 2002;11:252–264. doi: 10.1053/jscd.2002.129614. [DOI] [PubMed] [Google Scholar]
- Bentley P, Peck G, Smeeth L, Whittaker J, Sharma P. Causal relationship of susceptibility genes to ischemic stroke: comparison to ischemic heart disease and biochemical determinants. PLoS One. 2010;5:e9136. doi: 10.1371/journal.pone.0009136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.