Abstract
KAT6B sequence variants have been identified previously in both patients with the Say-Barber-Biesecker type of blepharophimosis mental retardation syndromes (SBBS) and in the more severe genitopatellar syndrome (GPS). We report on the findings in a previously unreported group of 57 individuals with suggestive features of SBBS or GPS. Likely causative variants have been identified in 34/57 patients and were commonly located in the terminal exons of KAT6B. Of those where parental samples could be tested, all occurred de novo. Thirty out of thirty-four had truncating variants, one had a missense variant and the remaining three had the same synonymous change predicted to affect splicing. Variants in GPS tended to occur more proximally to those in SBBS patients, and genotype/phenotype analysis demonstrated significant clinical overlap between SBBS and GPS. The de novo synonymous change seen in three patients with features of SBBS occurred more proximally in exon 16. Statistical analysis of clinical features demonstrated that KAT6B variant-positive patients were more likely to display hypotonia, feeding difficulties, long thumbs/great toes and dental, thyroid and patella abnormalities than KAT6B variant-negative patients. The few reported patients with KAT6B haploinsufficiency had a much milder phenotype, though with some features overlapping those of SBBS. We report the findings in a previously unreported patient with a deletion of the KAT6B gene to further delineate the haploinsufficiency phenotype. The molecular mechanisms giving rise to the SBBS and GPS phenotypes are discussed.
Introduction
The Say-Barber-Biesecker syndrome (SBBS, MIM 603736), also referred to as the Say-Barber-Biesecker variant of Ohdo syndrome1 is a rare multiple congenital anomaly syndrome, which is usually diagnosed clinically on the basis of a striking facial phenotype. Typically, patients have a distinctive mask-like face with severe blepharophimosis and ptosis, a broad nasal bridge, bulbous nasal tip, small mouth, thin upper lip and small, low set ears. Long thumbs and long great toes may be present along with congenital heart defects, thyroid abnormalities, hypoplastic teeth and absent or hypoplastic patellae.2 Genital anomalies, usually cryptorchidism, are universally present in males, and cleft palate has been observed in a number of cases. All individuals with SBBS, reported so far, have had global developmental delay and severe intellectual disability. Patients with features overlapping those of SBBS were reported by Young and Simpson3 in 1987, and it is now generally agreed that Young–Simpson syndrome and SBBS are in fact the same entity. Young and Simpson were the first to draw attention to the presence of hypothyroidism in this condition. Clayton-Smith et al4 identified KAT6B, a histone acetyltransferase, as the causative gene for SBBS by a whole-exome sequencing approach and reported sequence variants in 13 individuals. Subsequently, Szakszon identified a further two SBBS individuals with KAT6B sequence variants5 and another individual was reported recently by Yu et al.6 In addition, KAT6B sequence variants have been reported in 11 patients with genitopatellar syndrome (GPS, MIM 606170), a further multiple anomaly phenotype previously considered to be quite distinct from SBBS.7, 8 In GPS, the overall clinical picture tends to be more severe than in SBBS.9 The main diagnostic features of GPS include the presence of large joint contractures, ambiguous genitalia and absent patellae. Agenesis of the corpus callosum, hydronephrosis and congenital heart disease are frequently present, and thyroid abnormalities have been reported in rare cases. The facial phenotype of GPS has not been well described but is not considered so distinctive as in SBBS, with the diagnosis usually being made based on the other clinical features.
To further delineate the clinical and molecular spectrum of KAT6B, we have studied a new cohort of 57 patients with a clinical diagnosis of SBBS (n=47), GPS (n=5) or features overlapping both of these entities (n=5). We have gathered detailed clinical information on this group and screened for sequence variants in the coding exons of KAT6B by Sanger sequencing. Our study aimed to define differences in clinical features between KAT6B variant-positive and KAT6B variant-negative patients, and to explore the breadth of the phenotypic spectrum, examining in particular, the clinical overlap between SBBS and GPS.
Patients and methods
Ascertainment of study patients
In 2011, we ascertained a cohort of 19 patients with SBBS for genetic studies and identified likely pathogenic KAT6B sequence variants in 13 of these.4 For this current study, we have gathered a cohort of a further 57 patients with features suggestive of SBBS or GPS who were recruited via clinical geneticists from within the UK or abroad. Forty-five patients were tested for KAT6B sequence variants as part of a research study, exploring the genetic basis of blepharophimosis syndromes (REC reference 10/H1016/12). Variants in a further nine patients were identified in the diagnostic laboratory within the Manchester Centre for Genomic Medicine. Three further individuals (23, 24, 25) were referred to our study after KAT6B variants had been identified in laboratories in Ulm (2) and Freiburg (1). Individuals 23 and 24 had originally been ascertained as part of a study of the UBE3B gene,10 but were considered to have features more suggestive of a KAT6B phenotype. Referring clinicians provided detailed phenotypic information and photographs of participants for review. Consent to publish clinical details was obtained from all participating families and consent to publish photographs was obtained for those included in this report.
Sequencing of KAT6B
Samples from the 54 individuals studied in Manchester were screened for sequence variants in all 18 exons of KAT6B by Sanger sequencing. Details of primer design and synthesis are given in Supplementary Information Table 1. The coding exons and the intron–exon boundaries of KAT6B were amplified by polymerase chain reaction. Post-PCR purification was conducted to remove unwanted reaction components using the Agencourt AMpure XP (Beckman Coulter Genomics, Takeley, UK) system on a Biomek NX Liquid Handler (Beckman Coulter). The purified PCR products were then subjected to sequencing using BigDye Terminator v3.1 (Invitrogen, Paisley, UK). Reactions were purified using Agencourt CleanSEQ solution (Beckman Coulter) by Beckman Coulter NX robotics. Automated capillary electrophoresis was conducted on the ABI 3730XL Genetic Analyzer (Applied Biosystems, Paisley, UK) using POP-7 polymer (Life Technologies, Paisley, UK) according to local protocols. The reference sequence of KAT6B NG_032048.1, covering transcript NM_012330.3 was uploaded as a txt.file to Staden package Pregap4 1.4b1. The sequence data were aligned and analysed graphically using the DNA sequence analysis software, STADEN (http://staden.sourceforge.net/). Any variants identified were checked against the NCBI SNP database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/) and against the Exome Variant Server (http://evs.gs.washington.edu/EVS/) to distinguish common variants from likely pathogenic variants. The variant interpretation software Alamut, (http://www.interactive-biosoftware.com/software/alamut/features) was used to interpret and predict the consequences of identified variants. Sequence variants identified were reported according to the Human Genome Variation Society nomenclature. Where available, DNA from the parents of the patients with possible pathogenic variants was screened to investigate if the change was de novo. Three patients (numbers 23, 24 and 25) were sequenced in other genetic laboratories as reported above using similar methodology, but in cases 23 and 24, only exon 18 was sequenced. Taking into account the clinical context, that is, all patients had been selected on the basis of a clinical diagnosis of SBBS or GPS, sequence variants were classified as causative or not. Variants resulting in a truncated protein were classified as causative. Variants deemed to be likely causative were missense changes, which were proven to be de novo, or if in silico analysis predicted them to affect splicing or highly conserved amino acids. Sequence changes that were synonymous and did not affect splicing, missense changes reported in EVS or those seen in a parent were classified as being unlikely to cause disease. Details of causative variants were submitted to the LOVD public database at http://databases.lovd.nl/shared/genes/KAT6B, (patient IDS 00018483-00018515).
Table 1. Genotype–phenotype differences between KAT6B-positive and KAT6B-negative patients.
| KAT6B+(n1=47) | KAT6B−(n2=23) | ||||||
|---|---|---|---|---|---|---|---|
| Present | Not present | Not known | Present | Not present | Not known | P-value | |
| Hypotonia | 44 | 2 | 1 | 9 | 14 | 0 | <0.0001 |
| Feeding difficulties | 42 | 3 | 2 | 15 | 8 | 0 | 0.0051 |
| Contractures | 27 | 7 | 13 | 6 | 12 | 5 | 0.0020 |
| Dental anomalies | 28 | 14 | 5 | 7 | 16 | 0 | 0.0087 |
| Long thumbs | 33 | 14 | 0 | 3 | 20 | 0 | <0.0001 |
| Long great toes | 31 | 16 | 0 | 5 | 18 | 0 | 0.0008 |
| Thyroid abnormalities | 23 | 22 | 2 | 1 | 22 | 0 | 0.0001 |
| Congenital heart defects | 27 | 19 | 1 | 12 | 11 | 0 | 0.6177 |
| Genital anomalies | 25 | 22 | 0 | 7 | 15 | 1 | 0.1237 |
| Abnormal patella | 15 | 31 | 1 | 1 | 22 | 0 | 0.0134 |
| Microcephaly | 12 | 23 | 12 | 8 | 12 | 3 | 0.7734 |
| Cleft palate | 14 | 33 | 0 | 2 | 21 | 0 | 0.0690 |
Bold values indicate significance (P<0.05).
Genotype–phenotype analysis
Patients were classified as having either a SBBS phenotype (n=47), a GPS phenotype (n=5) or an overlapping phenotype (n=5) by dysmorphologists from the Manchester Centre For Genomic Medicine on the basis of clinical information and photographs. In accordance with previous literature, large joint contractures, patellar abnormalities and ambiguous genitalia/severe genital anomalies were considered as core features of GPS.9 For SBBS, the chracteristic facial features of blepharophimosis and mask-like face were the key diagnostic features. To analyse differences in phenotypic features between variant-positive and variant-negative patients, we studied 45 variant-positive patients for whom we had detailed clinical information (Supplementary Table 2a). This group included the 13 patients, which we had reported previously.4 We compared their clinical phenotypes with those of 23 variant-negative patients (Supplementary Table 2b), including five reported previously (patients 71–75).4 Differences between the presence of 11 commonly presenting features in SBBS and GPS were analysed using the Fisher's Exact test. We noted the type and position of the causative and likely causative variants within the SBBS group, the GPS group and the group with the overlapping phenotype. Comparison was also made with sequence data from 28 patients with KAT6B sequence variants previously reported in the literature (11 GPS and 17 SBBS).
Results
Sequence analysis
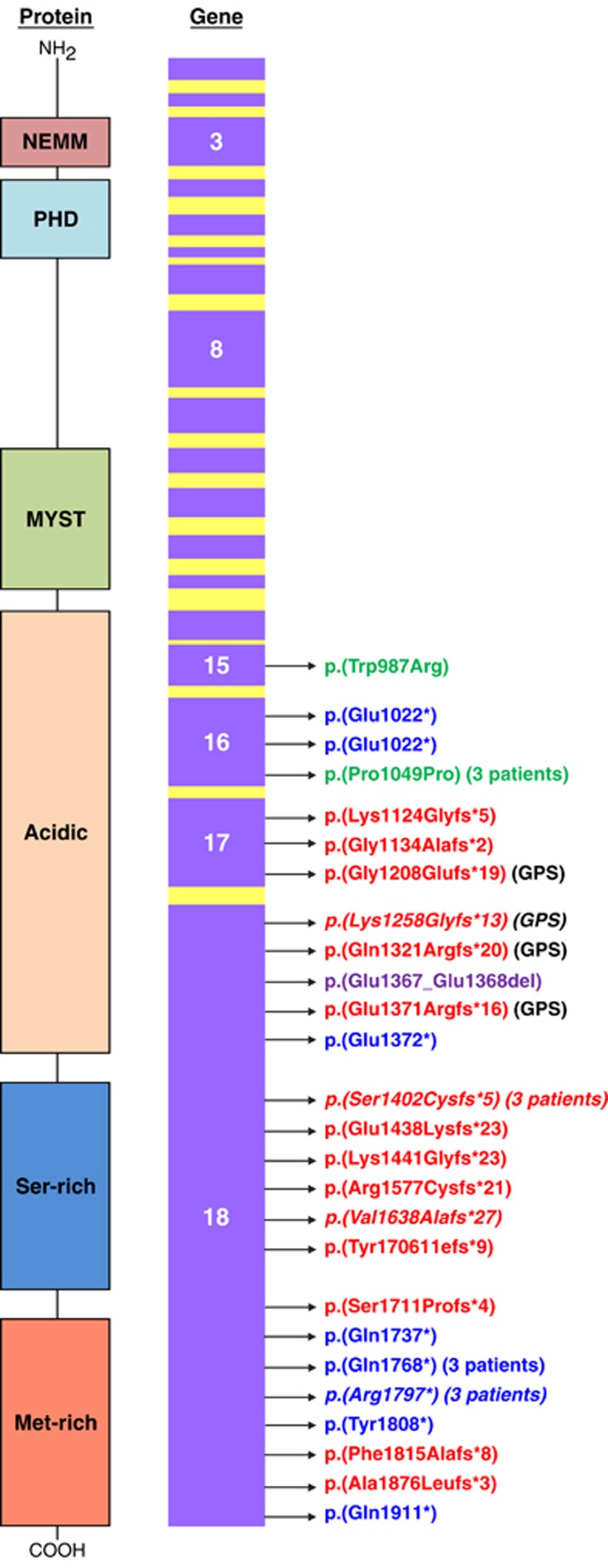
Sequencing of all 18 coding exons and intron–exon boundaries of KAT6B in our new cohort of 57 patients identified 34 causative or likely disease-causing KAT6B variants. Of these, 26/47 SBBS individuals had such variants, 4/5 GPS patients and 4/5 of those with an overlapping phenotype. The majority (29/34) were truncating variants, but one was a missense variant, one was a synonymous change shared by three unrelated SBBS patients and one was an in-frame deletion. The sequence variants are summarised in Supplementary Table 3, which also incorporates the findings from our original cohort3 and previous reports.4, 5, 6 In addition, we observed a recurrent sequence variant, which we considered not causative of disease, consisting of a 12 bp sequence in exon 16 c. 3252_3263 p.(Glu1086_1089), which was either deleted or duplicated. The majority of the causative sequence variants identified clustered at the C-terminal end of the gene in exon 18, as documented previously. This exon encodes a highly conserved serine- and methionine-rich transcriptional activation domain. However, we also identified variants that occurred more proximally in exon 18 and in exons 15, 16 and 17. Three individuals shared an identical de novo synonymous variant in exon 16, c.3147 G>A p.(Pro1049Pro). The Alamut 2.2 prediction programme suggested that this change was likely to create a cryptic splice site. All three individuals had the typical SBBS phenotype. One patient had a missense variant in exon 15, c.2959 T>C p.(Trp987Arg). The parents of this patient were not available for study, so the question as to whether this variant is causative remains, but it has not been reported as a SNP on dbSNP, and the amino acid 987 is well conserved. In silico analysis predicted that this variant may be causative. In 18/34 cases where parental samples could be tested, all of the sequence changes identified had occurred de novo. Several of the variants were recurrent, having been seen in more than one individual or reported previously in the literature (see supplementary Table 3).
Genotype/phenotype correlation
The position of the variants in the GPS patients relative to the SBBS patients is shown in Figure 1. In previously reported cases, the sequence variants in GPS patients have tended to cluster more proximally to those in SBBS.7, 8 This was the case overall in our four GPS patients who had variants in exon 17 and more proximally in exon 18. However, we observed that patient 54 who had patellar abnormalities, contractures, genital anomalies and agenesis of the corpus callosum (all features, which overlap between GPS and SBB) had the c.4205_4206del p.(Ser1402Cysfs*5) variant. This variant has been reported in patients with typical SBBS4 who lacked the features of large joint contractures, agenesis of the corpus callosum and renal anomalies, although one patient with this variant had patellar abnormalities. The variant occurs more distally to the other GPS variants. Patient 56 whose sequence change occurred more proximally in exon 18 had significant genital anomalies and was also considered to have an overlapping phenotype. The remaining two patients with overlapping phenotypes (53 and 55) had the variants c.4320_4321del p.(Lys1441Glyfs*23) and c.4074_4079del p.Glu1367_Glu1368. Patients 1 and 17 had truncating variants occurring more proximally to those seen in GPS, yet they had a working diagnosis of SBBS and patient 20 who had one of the more proximal variants in exon 17 did not have any abnormalities of the patellae, genitalia, kidneys or corpus callosum. The three individuals (12,13,14) who all shared the same exon 16 de novo synonymous variant fitted better with SBBS, as all had the typical facies, normal patellae, and none had significant contractures, agenesis of the corpus callosum or renal anomalies. All of the above observations suggest that the clinical distinction between GPS and SBBS is not always clear, and it might be more important for the clinician to recognise when a patient has clinical findings within the broader KAT6B spectrum rather than to only consider the possibility of a KAT6B phenotype if the clinical features fit clearly within either the GPS or SBBS entities. This broader presentation of a KAT6B spectrum disorder was the basis upon which testing KAT6B was suggested in case 51, a 24-week fetus with contractures, agenesis of the corpus callosum and hydronephrosis.
Figure 1.
Schematic representation of the spectrum of KAT6B variants identified in this study. All coding exons are shown in purple. Arrows point to the exonic location of the variants identified. Key: frameshift variants, in-frame deletion, missense variant, synonymous variant, nonsense variants. Variants identified in other studies are in italics. The protein structure shows the protein domains. NEMM: highly conserved N-terminal domain with a role in transcriptional repression. PHD: zinc finger domain, MYST: responsible for histone acetyltransferase activity.
Comparison of individuals with and without KAT6B sequence variants
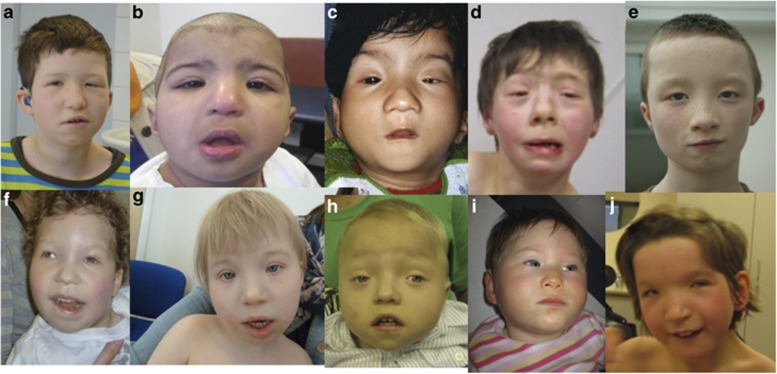
Comparison and analysis of clinical features from variant-positive and variant-negative patients indicated that dysmorphic facial features, in particular blepharophimosis, severe intellectual disability and developmental delay were common to both groups (Figure 2,Supplementary Figure 1). Genital anomalies, usually cryptorchidism, was universal in males with KAT6B sequence variants and tended to be more severe in GPS, where the genitalia may be ambiguous. Genital anomalies (hypoplastic labia) were noted in one affected female, but we suspect that genital anomalies in females are under-ascertained. Significant features, which predicted whether a pathogenic KAT6B variant would be found, were the presence of hypotonia, thyroid abnormalities, patellar abnormalities, long thumbs and great toes, feeding difficulties and dental anomalies. A pathogenic KAT6B sequence variant was much more likely to be identified if the latter features were present (Table 1). The commonest cardiac anomalies observed in the KAT6B-positive group were atrial and ventricular septal defects and patent ductus arteriosus, though one patient had a Tetralogy of Fallot, and right-sided aorta and pulmonary stenosis were seen in several patients. Dental anomalies, particularly hypoplastic teeth, emphasised as a salient feature of SBBS in the early literature, were reported as present in only 21/44 individuals with KAT6B variants though they were a significant distinguishing feature. Hearing loss, often remarked upon in the early literature on SBBS was present in around a third of individuals with likely pathogenic KAT6B variants and was usually sensori-neural in origin. Cleft palate was seen in 14/44 patients, drawing attention to the fact that SBBS and GPS deserve consideration as a differential diagnosis for syndromic clefting. Thyroid abnormalities were present in 22/42 KAT6B-positive patients where these had been searched for and only 1/22 KAT6B-negative patients, again making this one of the most important distinguishing features. The commonest abnormality was congenital hypothyroidism, with raised TSH. Thyroid agenesis was also documented in some patients. There was considerable clinical overlap between the clinical features of the SBBS and the GPS patients. Blepharophimosis was less striking in GPS (Figure 3) though this group did still have a distinctive, but less obvious facial phenotype. Several patients with SBBS had either agenesis of the corpus callosum, hydronephrosis and joint contractures, features more typical of GPS. In SBBS, the joint contractures were less severe and manifested more frequently as talipes and overlapping fingers and toes (Supplementary Figure 2) rather than large joint contractures. Renal anomalies and agenesis of the corpus callosum were probably under-ascertained in the SBBS patients where MRI brain and renal scans had not always been undertaken. Several GPS patients have had the long, straight thumbs and great toes seen in SBBS, and this was even a feature in the fetus observed at 24 weeks of gestation. An interesting observation was the presence of malrotation of the bowel in two patients with KAT6B sequence variants. A further patient had a laryngeal cleft. When analysing the relationship between the position of KAT6B variants and individual clinical features, no genotype/phenotype correlation could be established. For patients with thyroid and patellar abnormalities, for example, sequence changes were found across the region from exon 15 to the distal part of exon 18. The three patients who all shared the same exon 16 synonymous change had a similar facial gestalt (Figure 2f–h), which had been easily recognisable as that of SBBS by their referring geneticists, though ptosis was not as prominent in these three patients as others with SBBS. All three had small, low set ears, intellectual disability, global developmental delay, feeding abnormalities and genital anomalies but lacked some of the other structural malformations such as congenital heart defects, cleft palate and patellar abnormalities.
Figure 2.
2a, 2b, 2c, 2d show individuals 24, 20, 25 and 7 who all have sequence variants within KAT6B and show typical facial features of blepharophimosis, mask-like face, small mouth and bulbous nose with depressed nasal tip. 2e shows the facial features of the boy with a large chromosome deletion encompassing KAT6B, and who therefore has KAT6B haploinsufficiency. Individuals 2f, 2g, 2h are individuals 12, 14 and 13 who all have the same de novo synonymous variant, predicted to affect splicing in exon 16 of KAT6B. 2i and 2j (cases 44 and 35) have blepharophimosis and intellectual disability but neither have a KAT6B sequence variant and an alternative diagnosis is likely.
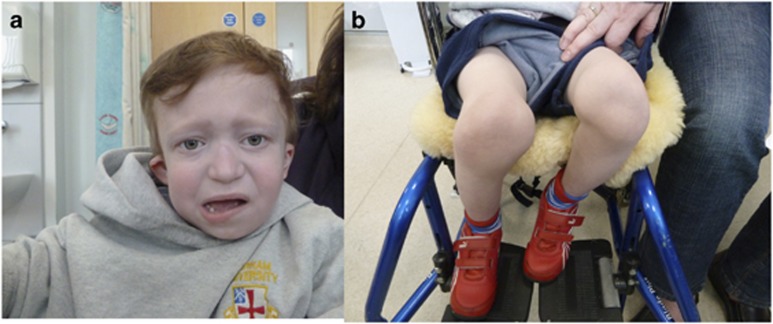
Figure 3.
Study patient 49 who shows typical features of genitopatellar syndrome. Note in 3a that blepharophimosis is less apparent, and in 3b the are knees are held in flexion and the patellae are displaced and hypoplastic.
Discussion
This study confirms previous findings that both SBBS and GPS are caused by de novo truncating variants in KAT6B. However, it is clear that not all variants are confined to the terminal exon of this gene, making it important to screen other exons, too, for diagnostic testing. The study identified several novel splice site variants, including a common synonymous variant in exon 16. Sequence changes causing the more severe phenotype of GPS did appear overall to occur more proximally as seen in the previous reports.5, 6 Campeau et al11 have hypothesised that features that occur commonly in both GPS and SBBS are due to either haploinsufficiency or loss of a function mediated by the C-terminal acidic domain, with the more severe features seen in GPS being due to alterations in binding capacity of the proteins arising as a result of specific GPS sequence variants. Our results would partially confirm these hypotheses, as they suggest that the sequence variants cluster into three domains; (a) those occurring more proximally, which are subject to nonsense-mediated decay (NMD). The phenotype is a milder SBBS phenotype caused by haploinsufficiency. (b) Variants that are not subject to NMD but cause a more severe GPS phenotype if they affect critical binding sites of KAT6B. (c) variants that occur more distally and again escape NMD but cause the SBBS phenotype rather than GPS, as they do not interfere with the critical binding sites associated with GPS. All of these three groups could share common features if, as Campeau suggest, haploinsufficiency for the very distal C-terminal region is crucial, but it is also possible that the more distal variants are also contributing to a more severe SBBS phenotype due to a gain-of-function effect. Further studies of the individual sequence variants are needed to clarify the mechanisms involved. KAT6B deletions have been documented very rarely in the literature; Tzschach et al12 described four patients with 10q22 deletions encompassing KAT6B. They had a mild phenotype with common features of hypotonia and developmental delay, especially in the area of speech. Facial features were nonspecific, though on scrutiny of the available photographs, blepharophimosis did appear to be a feature. Interestingly, one of the patients was noted to have unusually long and straight thumbs. The DDD study (DECIPHER@sanger.uk)13 identified a patient, DECIPHER number 258813, with a 2.5-Mb deletion encompassing KAT6B, HGVS description chr10.hg19:g.75, 971,593_78,526,861del. This deletion involved nine genes, of which KAT6B is the only one known to be associated with a developmental disorder. This child, a male, weighed 2.1 kg at 37 weeks and presented with hypotonia and developmental delay, particularly in the area of expressive speech. He had short palpebral fissures and prominent epicanthic folds (Figure 2e), but the facial features were less striking than in SBBS. He had normal patellae and a normal TSH level. He was macrocephalic and an MRI scan showed only mild frontal atrophy. Thiel et al14 have previously reported a patient with a translocation disrupting KAT6B and went on to demonstrate that this individual had lowered the expression of KAT6B. He had much milder features than patients with SBBS but did have blepharophimosis and was reported as being ‘Noonan-like.' Like the patients reported by Tzschach and the DDD patient, there were no major malformations present. The demonstration by Thiel et al that RAS-MAP pathway gene expression was affected in this patient is interesting in view of the fact that in two of our patients, a diagnosis of Cardio-facio-cutaneous syndrome had been suggested initially and that pulmonary stenosis and malrotation of the gut, both features seen in RAS-MAP disorders were documented as clinical features in our patient cohort. Only one convincing KAT6B sequence variant has been seen in the proximal part of the gene (exon 3). This was documented in the series by Clayton-Smith et al 4 in a patient with a much milder blepharophimosis-intellectual disability phenotype. Taken together, all of the above observations strengthen Campeau's case that a mild blepharophimosis-intellectual disability phenotype is caused by haploinsufficiency of KAT6B, whereas disease-causing variants toward the terminal end interfere with the transcriptional activity of the gene, and therefore cause a more severe phenotype through a gain-of-function effect. These hypotheses remain to be tested in further studies.
From a clinical point of view, clinicians should be aware that the KAT6B phenotypic spectrum is broad, encompassing presentations from mild blepharophimosis and intellectual disability to the severe, life threatening phenotype seen in GPS. Naming of the KAT6B-related disorders has been problematic. Though it is possible to classify some patients as having typical SBBS or GPS, this is not possible in all cases, and we propose that considering this whole group as ‘KAT6B spectrum disorders' may be more helpful. Good diagnostic handles, which might indicate a causative KAT6B sequence variant, include severe blepharophimosis, patellar abnormalities, thyroid abnormalities, hypothyroidism, long thumbs and toes and agenesis of the corpus callosum. There are some similarities with RAS-MAP disorders, and abnormalities of KAT6B may act in part by influencing this pathway. The relatively broad clinical spectrum of KAT6B variants means that clinical diagnosis is not always easy and suggests that affected individuals might be identified in large-scale exome and genome sequencing programmes rather than as a result of targeted testing. Apart from one family, reported so far, where two affected siblings were born to parents who did not carry the variant in blood, and where gonadal mosaicism was presumed, all KAT6B variants have occurred de novo and recurrence risks for most families are low, though prenatal testing should be offered as an option.
No alternative diagnoses have yet been confirmed in those patients who did not have causative variants in KAT6B. On careful clinical review, we considered that the majority of these had some atypical features and that, rather than KAT6B variants or deletions being missed, other diagnoses were more likely. A diagnosis of Cohen syndrome was suggested in patient 57 based on the facial features and additional finding of visual problems. Chromosomal abnormalities, one of the main differential diagnoses, have been excluded by microarray analysis. A further differential diagnosis, which was excluded in suggestive cases, was that of a UBE3B phenotype.10 Verloes et al reviewed the classification of the group of patients with blepharophimosis-intellectual disability syndromes. He classified this group into five main groups, including both SBBS and chromosomal abnormalities. The remaining groups comprised the original patients reported by Ohdo et al,15 The X-linked disorder originally reported by Maat-Kievit et al16 and now known to be due to sequence variants within the MED12 gene,17 and a recessive entity associated with microcephaly and multiple structural anomalies. None of our patients had features, which were consistent with any of these other groups identified by Verloes. It is likely, therefore, that there are other causes of blepharophimosis and intellectual disability, which remain to be elucidated.
Acknowledgments
Cranio-facial research within the University of Manchester is supported by the Healing Foundation UK Cleft Collective. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003) a parallel funding partnership between the Wellcome Trust and the Department of Health and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK research ethics approval (10/H0305/83 granted by the Cambridge South REC and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Verloes A, Bremond-Gignac D, Isidor B et al: Blepharophimosis-mental retardation (BMR) syndromes: A proposed clinical classification of the so-called Ohdo syndrome, and delineation of two new BMR syndromes, one X-linked and one autosomal recessive. Am J Med Genet 2006; 140A: 1285–1296. [DOI] [PubMed] [Google Scholar]
- Day R, Beckett B, Donnai D et al: A clinical and genetic study of the Say/Barber/Biesecker/Young-Simpson type of Ohdo syndrome. Clin Genet 2008; 74: 434–444. [DOI] [PubMed] [Google Scholar]
- Young ID, Simpson K: Unknown syndrome: abnormal facies, congenital heart defects, hypothyroidism, and severe retardation. J Med Genet 1987; 24: 715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, O'Sullivan J, Daly S et al: Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet 2011; 89: 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakszon K, Salpietro C, Kakar N et al: De novo mutations of the gene encoding the histone acetyltransferase KAT6B in two patients with Say-Barber/Biesecker/Young-Simpson syndrome. Am J Med Genet 2013; 161A: 884–888. [DOI] [PubMed] [Google Scholar]
- Yu HC, Geiger EA, Medne L et al: An individual with blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) and additional features expands the phenotype associated with mutations in KAT6B. Am J Med Genet 2014; 164A: 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Kim JC, Lu JT et al: Mutations in KAT6B, encoding a histone acetyltransferase, cause genitopatellar syndrome. Am J Hum Genet 2012; 90: 282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson MA, Deshpande C, Dafou D et al: De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause genitopatellar syndrome. Am J Hum Genet 2012; 90: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier-Daire V, Chauvet ML, Lyonnet S et al: Genito-patellar syndrome: a new condition comprising absent patellae, scrotal hypoplasia, renal anomalies, facial dysmorphism and mental retardation. J Med Genet 2000; 37: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Dallapiccola B, Ramirez-Solis R et al: Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome. Am J Hum Genet 2012; 91: 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Lu J, Dawson JT et al: The KAT6B-related disorders genitopatellar syndrome and Ohdo/SBBYS syndrome have distinct clinical features reflecting distinct molecular mechanisms. Human Mutation 2012; 33: 1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Bisgaard AM, Kirchhoff M et al: Chromosome aberrations involving 10q22; report of three overlapping interstitial deletions and a balanced translocation disrupting C10orf11. Eur J Hum Genet 2010; 18: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth HV, Wright CF: DDD Study. The Deciphering Developmental Disorders (DDD) study. Dev Med Child Neurol 2011; 53: 702–703. [DOI] [PubMed] [Google Scholar]
- Kraft M, Cirstea IC, Voss AK et al: Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Invest 2011; 121: 3479–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdo S, Madokoro H, Sonoda T et al: Mental retardation associated with congenital heart disease, blepharophimosis, blepharoptosis and hypoplastic teeth. J Med Genet 1986; 23: 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat-Kievit JA, Milla PJ, Collins JE et al: A case with blepharophimosis resembling Ohdo syndrome. Clin Dysmorphol 1994; 3: 125–127. [PubMed] [Google Scholar]
- Anneke T, van Silfhout V, Bert BA, de Vries, Bregje WM van Bon et al: Mutations in MED12 Cause X-Linked Ohdo Syndrome. Am J Hum Genet 2013; 92: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.