Abstract
Germline variants affecting the exonuclease domains of POLE and POLD1 predispose to multiple colorectal adenomas and/or colorectal cancer (CRC). The aim of this study was to estimate the prevalence of previously described heterozygous germline variants POLE c.1270C>G, p.(Leu424Val) and POLD1 c.1433G>A, p.(Ser478Asn) in a Dutch series of unexplained familial, early onset CRC and polyposis index cases. We examined 1188 familial CRC and polyposis index patients for POLE p.(Leu424Val) and POLD1 p.(Ser478Asn) variants using competitive allele-specific PCR. In addition, protein expression of the POLE and DNA mismatch repair genes was studied by immunohistochemistry in tumours from POLE carriers. Somatic mutations were screened using semiconductor sequencing. We detected three index patients (0.25%) with a POLE p.(Leu424Val) variant. In one patient, the variant was found to be de-novo. Tumours from three patients from two families were microsatellite instable, and immunohistochemistry showed MSH6/MSH2 deficiency suggestive of Lynch syndrome. Somatic mutations but no germline MSH6 and MSH2 variants were subsequently found, and one tumour displayed a hypermutator phenotype. None of the 1188 patients carried the POLD1 p.(Ser478Asn) variant. POLE germline variant carriers are also associated with a microsatellite instable CRC. POLE DNA analysis now seems warranted in microsatellite instable CRC, especially in the absence of a causative DNA mismatch repair gene germline variant.
Introduction
Faithful DNA replication and the repair of errors are both essential for the maintenance of genomic stability and suppression of carcinogenesis.1 Duplication of genomes with high accuracy is achieved through three mechanisms: the high selectivity of DNA polymerases; exonucleolytic proofreading; and post replication mismatch repair.2 The DNA polymerases ɛ (POLɛ) and δ (POLδ) are required for the efficient genome replication in the eukaryotic replication fork.3 Their major component proteins, encoded by POLE and POLD1, respectively, possess an intrinsic 3′–5′ proofreading domain that removes incorrectly inserted nucleotides during DNA synthesis.4, 5, 6, 7, 8, 9 Studies in the yeast have shown that mutations in the proofreading domains of POLɛ or POLδ increase spontaneous mutation rates.8, 9 In addition, somatic mutations in the proofreading domains of POLD1 and POLE have been identified in microsatellite instable (MSI) and hypermutated subgroups of colorectal cancers (CRCs).10, 11, 12
Recently, Palles et al reported that heterozygous germline variants in the proofreading domain of the DNA polymerases POLE and POLD1 predispose, with a high penetrance, to multiple colorectal adenomas, early onset CRC (OMIM #114500) and endometrial cancer (OMIM #608089). These variants were found by whole-genome sequencing and linkage analysis in three large families with a dominant pattern of CRC and multiple adenomas.13 Subsequent screening of 3805 CRC patients revealed that these variants are relatively rare: POLE p.(Leu424Val) was found 12 times, and POLD1 p.(Ser478Asn) only once, in patients with a positive family history of adenomas or CRC. The tumours seen in POLE and POLD1 carriers were microsatellite stable and showed a hypermutator phenotype.13 Valle et al14 detected a single POLE p.(Leu424Val) variant in a screen of 858 familial/early onset CRC and polyposis patients.
The goal of our study was to estimate the prevalence of germline variants in POLE and POLD1 in a Dutch series of unexplained familial, early onset CRC and polyposis index cases. In addition, we analysed phenotypes and tumour characteristics in this patient series.
Materials and methods
Samples
DNA from index patients with colorectal polyposis15 and familial CRC16 was analysed for POLE NM_006231.2:c.1270C>G, p.(Leu424Val) and POLD1 NM_002691.1:c.1433G>A, p.(Ser478Asn). Leucocyte DNA from 485 polyposis cases was included. These patients had developed ≥10 colorectal adenomas and had been previously tested negative for APC and MUTYH germline mutations at the Laboratory for Diagnostic Genome Analysis in Leiden. Clinical data were collected from the Netherlands Foundation for the Detection of Hereditary Tumours and from clinical genetics departments in the Netherlands.17 The familial CRC cohort comprised 703 patients, mainly from the south-western region of the Netherlands, with most cases submitted by clinical genetics departments. These patients met clinical criteria for MSI testing, which are based on early onset of disease and/or familial clustering of CRC and associated cancers, corresponding to the revised Bethesda criteria. Samples were collected between 1997 and 2013, and DNA for this cohort was available from peripheral blood (340 cases) or from formalin-fixed paraffin embedded normal mucosa (363 cases). These samples were described before, only DNA that passes quality check was included in the study.16 The study was approved by the local medical ethical committee of the Leiden University Medical Center (P01-019).
Genotyping
POLE p.(Leu424Val) and POLD1 p.(Ser478Asn) were tested using the competitive allele-specific PCR (KASPar) assay, following the manufacturer's protocol (LGC Genomics, Berlin, Germany). The primers were designed using Primerpicker (KBioscience, Hoddesdon, UK). The following primers were used to analyse POLE c.1270C>G: POLE_L424V_C1; 5′- GGA TCA TAG CCT AGC TTG GCC TT–3′, POLE_L424V_A2; 5′–GAA GGT CGG AGT CAA CGG ATT CCT TCC TGT GGG CAG TCA TAA TG–3′ and POLE_L424V_A1; 5′–GAA GGT GAC CAA GTT CAT GCT CCT TCC TGT GGG CAG TCA TAA TC–3′. For POLD1 c.1433G>A, we used: POLD1_S478N_C2; 5′–TCT GCT CGC CCA GGA AGT GGA A–3′, POLD1_S478N_A2; 5′–GAA GGT CGG AGT CAA CGG ATT CCT ACA CGC TCA ATG CCG TGA A–3′ and POLD1_S478N_A1; 5′–GAA GGT GAC CAA GTT CAT GCT ACA CGC TCA ATG CCG TGA G–3′.
Variants were identified using the CFX manager software v3.0 (Bio-Rad, Veenendaal, the Netherlands). Formalin-fixed paraffin embedded and leucocyte DNA samples were genotyped in separate experiments for accurate genotyping results. Samples positive for POLE c.1270C>G, p.(Leu424Val) were subsequently validated by Sanger sequencing of leucocyte DNA and of DNA extracted from formalin-fixed paraffin embedded tissues, using both normal and tumour DNA where available. Sanger sequencing was performed by Macrogen (Amsterdam, the Netherlands). The following primers, with universal M13 tails (upper case), were used for POLE c.1270C>G; forward: 5′–TGT AAA ACG ACG GCC AGT cca tct gga tgc gtg cac a–3′ and reverse: 5′–CAG GAA ACA GCT ATG ACC gaa tca tcc tgg ctt ctg ttc tca–3′. For POLD1 validation we used the oligonucleotides, forward: 5′–TGT AAA ACG ACG GCC AGT ctg tcc ttg gaa ggc cact–3′ and reverse: 5′–CAG GAA ACA GCT ATG ACC gag gtc agg gag gca gca–3′. Sequencing primers were designed using Primer3 (http://primer3.wi.mit.edu/) and all oligonucleotides were manufactured by IDT (Leuven, Belgium).
The POLE p.(Leu424Val) carriers were submitted to the LOVD database http://databases.lovd.nl/shared/genes/POLE, IDs 00019773 (PT1), 00019821 (PT2) 00019822 (PT3) and 00019824 (PT4).
Immunohistochemistry
Immunohistochemistry (IHC) for the DNA MMR proteins was performed as previously described.18 POLE IHC was performed using 4 -μm thick, formalin-fixed paraffin embedded tissue sections on StarFrost adhesive slides (Waldemar Knittel, Braunschweig, Germany), dried overnight at 37 °C. Tissue sections were deparaffinised three times in xylene for a total of 15 min and subsequently rehydrated with 100% ethanol three times for a total of 9 min. Endogenous peroxidase activity was blocked by immersing the tissue sections in a 0.3% solution of hydrogen peroxide (H2O2) and methanol at room temperature for 20 min. This treatment was followed by hydration in a graded ethanol series to distilled water. Antigen retrieval was then performed by boiling the tissue sections in 0.01 M citrate buffer (pH 6.0) for 10 min using a microwave oven, after which the tissue sections were allowed to cool in the same buffer to room temperature. The tissue sections were then washed twice with distilled water, for 5 min each time, followed by two 5 min washes in 1 × phosphate buffered saline. This was followed by the addition of the primary antibody (ab110876, ABCAM, Cambridge UK, dilution 1:1600) and the incubation of the covered slides overnight in a humid chamber at room temperature.
After three 5 min washes in 1 × phosphate buffered saline the next morning, the tissue sections were incubated for 30 min with horseradish peroxidase-conjugated multimer secondary antibody (PowerVision Poly/HRP, DAKO, Heverlee, Belgium). The slides then received three 5 min rinses in 1 × phosphate buffered saline and the antigen–antibody reaction was visualised using 3,3′-diaminobenzidine in chromogenic solution, applied for 10 min. The tissue sections were subsequently rinsed in distilled water and then stained with haematoxylin for 20 s. The tissue sections were rinsed in running tap water for 5 min and washed twice with distilled water, followed by dehydration in a graded ethanol series (50, 70 and 100%) and cleared in xylene. Finally, the tissue sections were dried, mounted and covered for microscopic analysis. Tissue stroma and normal epithelium or lymph follicles served as positive internal controls while analysing POLE, MLH1, PMS2, MSH2 and MSH6 expression.
Somatic mutation screening
DNA from normal and tumour tissues of the POLE p.(Leu424Val) carriers PT1 and PT2 was screened for somatic mutations using a custom AmpliSeq panel (Life Technologies Europe, Bleiswijk, the Netherlands) of the DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6, PMS2, supplemented with APC and TP53. The panel was sequenced using the Ion PGM system at Leiden Genome Technology Center, the Netherlands (www.lgtc.nl) and analysed with the Nextgene Software package (Softgenetics, State College, PA, USA). MMR DNA sequencing was performed as previously described in the case of PT3.19 Somatic KRAS and BRAF hotspot mutations (KRAS exons 2 and 3 and BRAF p.V600E were tested as described previously.20 Somatic mutations are submitted to the COSMIC database (ID # 616).
Methylation
MSH2 methylation was studied as described earlier using the MSP1 and MSP3 primer sets for both methylated and unmethylated DNA.21 DNA was bisulfite treated using the EZ DNA methylation KIT (ZYMO Research Corp, Irvine, CA, USA).
Results and discussion
We assessed the prevalence of POLE p.(Leu424Val) and POLD1 p.(Ser478Asn) in a cohort of 1188 Dutch index patients with multiple polyps or familial CRC. All patients were originally referred for possible FAP (OMIM #175100), MAP (OMIM #608456) or Lynch syndrome (OMIM #120435) and no variants were found in the relevant genes. Although we did not detect POLD1 p.(Ser478Asn), three index patients with the POLE p.(Leu424Val) variant were identified. These patients developed multiple colorectal adenomas, of whom two showed early onset CRC. We can therefore confirm that POLE variants play a minor but tangible role in familial CRC and polyposis. Despite an enrichment in our cohort for inherited CRC and polyposis, the frequency (3 in 1188; 0.25%) is also comparable to the currently reported frequency.13, 14
The first patient (PT1) was diagnosed with a MSI caecum tumour and two adenomas at age 40. She developed, cumulatively, around 30 polyps and presented with a microsatellite stable endometrial cancer at age 50. Her son, patient 2 (PT2), is also a carrier of POLE p.(Leu424Val). In addition, he was clinically diagnosed with neurofibromatosis type I, the latter originating in his father's family. He developed a MSI caecum adenocarcinoma at age 30 and was diagnosed with an astrocytoma at age 15, presumably as a consequence of neurofibromatosis. Interestingly, a patient with multiple polyps, and two astrocytomas at age 26, has been described in a family with a POLD1 variant (family SM6).13
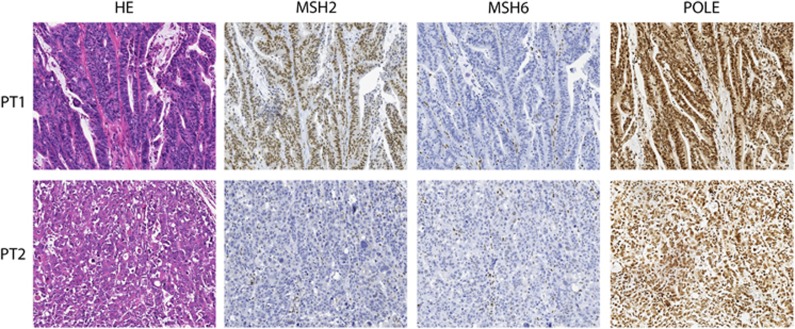
As tumour tissue was available for the POLE p.(Leu424Val) carriers PT1 and PT2 (mother and son, respectively), we performed IHC for POLE, MLH1, MSH2, MSH6 and PMS2. Considering that the POLE exonuclease domain is essential for the maintenance of replication fidelity, the lack of predictive value of POLE IHC is to be expected, in contrast to the predictive value for the MMR genes. However, the availability of tumour tissue from the POLE variant carriers allowed us to test this. Indeed, both MSI-H tumour tissues showed a positive nuclear POLE protein expression in tumour cells as compared with normal cells from the same tissue. POLE IHC therefore does not appear to be predictive for the effect of the variant.
The MSI-H caecum tumour from PT1 showed loss of only MSH6 protein expression in tumour cells (Figure 1), whereas the adenocarcinoma from PT2 showed loss of both MSH2 and MSH6 protein expression in tumour cells (Figure 1). The loss of nuclear expression of MSH6 and MSH6/MSH2 in the tumours from this family is indicative for Lynch syndrome because of an inherited MSH6 or MSH2 germline variant. However, no germline variants were found in MSH6 or MSH2, nor in any of the other MMR genes. We therefore screened PT1 and PT2 tumours for somatic mutations in MLH1, MSH2, MSH6, PMS2 and also for somatic mutations in APC and TP53 (Table 1). The mother's tumour (PT1) showed a hypermutator phenotype, with multiple somatic mutations in the MMR genes and in APC and TP53, whereas the adenocarcinoma from PT2 showed no APC or TP53 mutations. Interestingly, PT1 showed three somatic mutations in MSH6. The first is a pathogenic truncating mutation, c.2629G>T, p.(Glu877*), the second is a possibly pathogenic missense mutation, c.4000C>T, p.(Arg1334Trp) that might affect the splice donor site in exon 9, while the third mutation is probably non-pathogenic, c.3725G>A, p.(Arg1242His). A single truncating MSH2 mutation, c.643C>T, p.(Gln215*), was found in the son's tumour (PT2, Table 1), although a second hit in MSH2 was not detected. No somatic mutations were detected in KRAS and BRAF hotspots in the tumours from PT1 and PT2. These somatic mutations show an excess of G:C to T:A substitutions, similar to the reported EDM-associated mutator phenotype.13 No other somatic mutations or previously undetected germline mutations were found. In light of the hypermutator phenotype associated with POLE mutations, a second point mutation was to be expected. On the other hand, promoter methylation or loss of heterozygosity at the MMR locus as the second hit could explain the loss of MSH2 or MSH6. Recently it was demonstrated that the loss of heterozygosity accounts for the second hit in over 50% of MSI tumours, albeit mostly in MLH1 deficient tumours.19
Figure 1.
HE staining and MSH2, MSH6 and POLE immunohistochemistry (x20) of the MSI-H tumours from POLE variant carriers PT1 and PT2 (one family); both cases show MSH6 negative staining, with positive stromal cells (brown). PT2 also shows loss of MSH2 in tumour cells.
Table 1. Summary of somatic mutations in colorectal cancer genes in two tumours from POLE p.(Leu424Val) carriers (PT1 and PT2) from one family.
| Case | Gene.ref gene | DNA level (cDNA) | Protein | Chr | Start | End | Ref | Alt | Exonic func.ref gene | Pathogenic |
|---|---|---|---|---|---|---|---|---|---|---|
| PT1 | MSH6 | NM_000179.2:c.2629G>T | p.Glu877* | 2 | 48027751 | 48027751 | G | T | Stopgain SNV | Truncating |
| PT1 | MSH6 | NM_000179.2:c.2291C>T | p.Thr764Ile | 2 | 48027413 | 48027413 | C | T | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | MSH6 | NM_000179.2:c.3725G>A | p.Arg1242His | 2 | 48033421 | 48033421 | G | A | Non-synonymous SNV | Unknown pathogenicity |
| PT1 | MSH6 | NM_000179.2:c.4000C>T | p.Arg1334Trp | 2 | 48033789 | 48033789 | C | T | Non-synonymous SNV | Unknown pathogenicity |
| PT1 | MSH2 | NM_000251.2:c.49G>A | p.Val17Ile | 2 | 47630379 | 47630379 | G | A | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | MLH1 | NM_000249.2:c.31C>A | p.Leu11Met | 3 | 37035069 | 37035069 | C | A | Non-synonymous SNV | Unknown pathogenicity |
| PT1 | APC | NM_000038.5:c.680A>G | p.Asp227Gly | 5 | 112128177 | 112128177 | A | G | Non-synonymous SNV | Unknown pathogenicity |
| PT1 | APC | NM_000038.5:c.1778G>A | p.Trp593* | 5 | 112170682 | 112170682 | G | A | Stopgain SNV | Truncating |
| PT1 | APC | NM_000038.5:c.2662G>A | p.Ala888Thr | 5 | 112173953 | 112173953 | G | A | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | APC | NM_000038.5:c.4540C>T | p.Pro1514Ser | 5 | 112175831 | 112175831 | C | T | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | APC | NM_000038.5:c.5117C>T | p.Ser1706Leu | 5 | 112176408 | 112176408 | C | T | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | APC | NM_000038.5:c.8314T>C | p.Ser2772Pro | 5 | 112179605 | 112179605 | T | C | Non-synonymous SNV | Unlikely pathogenic |
| PT1 | TP53 | NM_000546.5:c.523C>T | p.Arg175Cys | 17 | 7578407 | 7578407 | G | A | Non-synonymous SNV | Likely pathogenic |
| PT1 | TP53 | NM_000546.5:c.742C>T | p.Arg248Trp | 17 | 7577539 | 7577539 | G | A | Non-synonymous SNV | Likely pathogenic |
| PT1 | TP53 | NM_000546.5:c.916C>T | p.Arg306* | 17 | 7577022 | 7577022 | G | A | Stopgain SNV | Truncating |
| PT2 | MSH2 | NM_000251.2:c.643C>T | p.Gln215* | 2 | 47637509 | 47637509 | C | T | Stopgain SNV | Truncating |
| PT2 | APC | NM_000038.5:c.2045G>A | p.Gly682Glu | 5 | 112173336 | 112173336 | G | A | Non-synonymous SNV | Likely pathogenic |
No mutations were found in KRAS, BRAF and PMS2.
Patient 3 (PT3) was diagnosed with multiple polyps at age 34. Two of the patient's brothers were affected with CRC at ages 18 and 37, while a third brother developed liver cancer at age 27. The patient's mother died aged 31 because of unknown causes, probably cancer. Tumour tissue from an adenoma with early cancer and one adenoma with high-grade dysplasia was available for examination. The adenoma with cancer (estimated tumour percentage 50%) showed microsatellite instability and immunohistochemical loss of MSH2 and MSH6 protein staining in the absence of germline variants in MSH2 and MSH6. A probably non-pathogenic heterozygous missense mutation, c.1550 C>T, p.(Ala517Val), in the MSH2 gene was detected in the adenoma with early cancer but not in normal tissue. Testing for somatic mutations in other genes could not be performed because of the limited availability of tumour DNA. Hypermethylation of the MSH2 promoter was not detected in the tumour or normal tissue of PT3. Microsatellite instability analysis of an adenoma with high-grade dysplasia, with an estimated percentage of cells with high-grade dysplasia of 65%, showed no microsatellite instability and normal staining of the DNA mismatch repair proteins MSH2 and MSH6, as well as MLH1 and PMS2. Tumour tissue and germline DNA from relatives of PT3 were not available.
Patient 4 (PT4) was diagnosed with a microsatellite stable colon cancer and polyposis at the age of 33. No tumour tissue was available for further analysis. Although a history of CRC was reported in both paternal and maternal branches of the family, the parents were not affected by polyposis and did not carry the POLE variant, indicating that the POLE variant in PT4 was probably de-novo. There was also no evidence for gonadal mosaicism in the parents; the sibling is not known to have polyposis. In a recent study, the same POLE variant was also detected as a de-novo occurrence in a patient with early onset CRC and polyposis.14
Interestingly, the germline POLE variant in the currently studied families (PT1, PT2 and PT3) is associated with a Lynch syndrome phenotype with MSI tumours and MSH6 or MSH2/MSH6 protein loss. This contrasts with previously identified POLE and POLD1 germline variant carriers who developed microsatellite stable tumours.13 Somatic POLE mutations have been reported in both microsatellite stable and MSI tumours.10, 12 The Lynch syndrome phenotype reported here is not because of the germline variants in the MMR genes but more likely the result of somatic inactivation (PT1 and PT2). The hypermutator phenotype associated with inherited POLE mutations suggests a causative role for the somatic mutations although the second hit could not be identified. A similar co-occurrence of somatic MMR and POLE mutations was recently shown in a sequencing effort of 147 CRC genomes. Eight of the eleven POLE-mutated tumours showed additional MSH6 somatic mutations, and of these, five cases also showed MSH2 mutations.12 Moreover, MSI tumours with two somatic MSH2 mutations, lacking MSH2 and MSH6 protein expression,22 or with loss of MLH1 protein staining in the tumour,23, 24, 25 have also been reported for patients with bi-allelic variants in the base excision repair gene MUTYH. Similarly to POLE germline variants, MUTYH missense variants can also induce somatic mutations in MMR genes, although the mechanism behind the co-occurrence of mutations in the different DNA repair defects remains elusive.
Conclusions
The three new families with POLE variants reported here bring the total number of reported families to 17. We have demonstrated that POLE germline variants can give rise to a Lynch syndrome-like phenotype, with MSI-H tumours displaying negative IHC for one of the MMR genes. IHC for POLE is not helpful in identifying currently known variants. We now recommend that testing for POLE should be considered when screening unexplained MSI-H tumours, and while clinical surveillance of POLE carriers appears to be indicated from a relatively young age, further conclusions regarding clinical management should be based on a larger series than the currently identified patients.
Acknowledgments
This work was supported by the Dutch Cancer Society (UL2010-4656); the Association for International Cancer Research (10-0619); COST Action BM1206. We thank the Dutch Cancer Genetics Group, represented by: Dr A Wagner, Dr RH Sijmons, Dr CM Aalfs, Dr I Kluijt, Dr N Hoogerbrugge, Dr E Gomez Garcia, Dr FH Menko, Dr TGW Letteboer and Dr FJ Hes for the contribution of multiple adenoma and familial CRC cases.
Author Contributions
FAE and TVW drafted the manuscript; FAE, ARM and DR performed analysis and interpretation of data; CMK, MN, HFAV, JTW, DCGG, CMT, HM, FJH, ARM, MJL provided clinical input and obtained samples; FAE, MS and BVDA technical support; All authors read and corrected the draft of the manuscript; TVW, FJH, HFAV and HM obtained funding; TVW supervised all aspects of the study.
The authors declare no conflict of interest.
References
- Venkatesan RN, Treuting PM, Fuller ED et al: Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol 2007; 27: 7669–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM: Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem 1993; 268: 23762–23765 [PubMed] [Google Scholar]
- Garg P, Burgers PM: DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 2005; 40: 115–128 [DOI] [PubMed] [Google Scholar]
- Seshagiri S: The burden of faulty proofreading in colon cancer. Nat Genet 2013; 45: 121–122 [DOI] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ: DNA replication fidelity and cancer. Semin Cancer Biol 2010; 20: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA: The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 2008; 18: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A: Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci 1995; 20: 319–323 [DOI] [PubMed] [Google Scholar]
- Morrison A, Bell JB, Kunkel TA, Sugino A: Eukaryotic DNA polymerase amino acid sequence required for 3'–5' exonuclease activity. Proc Natl Acad Sci USA 1991; 88: 9473–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Giot L, Faye G: The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J 1991; 10: 2165–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Miyashita K, Inoue M et al: Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet 2011; 19: 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network: The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Laird PW, Park PJ: The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 2013; 155: 858–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM et al: Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013; 45: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle L, Hernandez-Illan E, Bellido F et al: New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet 2014; 23: 3506–3512. [DOI] [PubMed] [Google Scholar]
- Hes FJ, Nielsen M, Bik CE et al: Somatic APC mosaicism: an underestimated cause of polyposis coli. Gut 2008; 57: 71–76. [DOI] [PubMed] [Google Scholar]
- Middeldorp A, Jagmohan-Changur S, van Eijk R et al: Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev 2009; 18: 3062–3067. [DOI] [PubMed] [Google Scholar]
- Hes FJ, Ruano D, Nieuwenhuis M et al: Colorectal cancer risk variants on 11q23 and 15q13 are associated with unexplained adenomatous polyposis. J Med Genet 2013; 51: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong AE, van Puijenbroek M, Hendriks Y et al: Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 2004; 10: 972–980. [DOI] [PubMed] [Google Scholar]
- Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA et al: Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 2014; 146: 643–646. [DOI] [PubMed] [Google Scholar]
- van Eijk R, Licht J, Schrumpf M et al: Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One 2011; 6: e17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK et al: Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 2006; 38: 1178–1183. [DOI] [PubMed] [Google Scholar]
- Morak M, Heidenreich B, Keller G et al: Biallelic MUTYH mutations can mimic Lynch syndrome. Eur J Hum Genet 2014; 22: 1334–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SP, Cotterchio M, Jenkins MA et al: Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology 2009; 136: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch A, Hitchins M, Williams R, Meagher A, Hawkins NJ, Ward RL: The role of MYH and microsatellite instability in the development of sporadic colorectal cancer. Br J Cancer 2006; 95: 1239–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre JH, Colas C, Coulet F et al: MYH biallelic mutation can inactivate the two genetic pathways of colorectal cancer by APC or MLH1 transversions. Fam Cancer 2010; 9: 589–594. [DOI] [PubMed] [Google Scholar]