Abstract
Transcription is the primary step in the retrieval of genetic information. A substantial proportion of the protein repertoire of each organism consists of transcriptional regulators (TRs). It is believed that the differential expression and combinatorial action of these TRs is essential for vertebrate development and body homeostasis. We mined the zebrafish genome exhaustively for genes encoding TRs and determined their expression in the zebrafish embryo by sequencing to saturation and in situ hybridisation. At the evolutionary conserved phylotypic stage, 75% of the 3302 TR genes encoded in the genome are already expressed. The number of expressed TR genes increases only marginally in subsequent stages and is maintained during adulthood suggesting important roles of the TR genes in body homeostasis. Fewer than half of the TR genes (45%, n=1711 genes) are expressed in a tissue-restricted manner in the embryo. Transcripts of 207 genes were detected in a single tissue in the 24 h embryo, potentially acting as regulators of specific processes. Other TR genes were expressed in multiple tissues. However, with the exception of certain territories in the nervous system, we did not find significant synexpression suggesting that most tissue-restricted TRs act in a freely combinatorial fashion. Our data indicate that elaboration of body pattern and function from the phylotypic stage onward relies mostly on redeployment of TRs and post-transcriptional processes.
Keywords: Transcription, Chromatin, Basal transcription, RNAseq, Transcription factor, Zebrafish, Atlas of gene expression, Genome, Phylotypic stage
Introduction
Vertebrate embryogenesis is believed to be crucially dependent on differential gene expression. Moreover, development is organised in a hierarchical fashion, in which, in a stepwise manner, more complex structures are derived from simpler structures laid down during earlier phases of ontogeny. It is thus assumed that the employed regulatory machinery in the developing animal becomes progressively more complex. The establishment of specific transcriptional expression programs leading to specific cell fate determination is controlled by the selective expression and/or activity of transcriptional regulators (TRs), as exemplified by the role of Myod in muscle differentiation (Weintraub et al., 1991). Among these, transcription factors (TFs) bind to DNA in a sequence-specific manner. DNA regions bound by TFs form gene regulatory elements also referred to as enhancers, repressors, silencers and promoters. Many TRs are downstream effectors of signalling pathways and integrate different signalling inputs that control cell behaviour. Although the concept of master regulators with unique transcriptional functions in the organism has been suggested (Halder et al., 1995), a growing body of evidence indicates that TFs act in a combinatorial fashion to control specific regulatory output (Davidson et al., 2002; Ravasi et al., 2010). Indeed, TFs have frequently multiple roles in multiple organs and it is the particular combination of TRs expressed or repressed at a particular time and space that dictates cellular morphology and function. Expression of certain TRs can be sufficient to drive cells into a specific differentiation programme (Vierbuchen et al., 2010) or to induce a pluripotent stem cell state (Takahashi and Yamanaka, 2006). Estimations based on the analysis of known DNA-binding domains suggest that 1500–2000 genomic loci of the mouse and human genome encode transcription factors (Tupler et al., 2001; Vaquerizas et al., 2009; Venter et al., 2001). In addition, transcription is regulated at a higher order by modification of the chromatin structure. Chromatin modifications can affect gene expression by changing the accessibility of genes to transcription factors or modifying promoter and enhancer activity, in either a positive or a negative manner. The activity and/or expression of these chromatin-modifying enzymes need to be carefully orchestrated with that of the TFs and factors of the general transcriptional machinery.
Although many systematic expression studies have been performed in various vertebrate models (Belgard et al., 2011; Fu et al., 2009; Gray et al., 2004; Hunt-Newbury et al., 2007; Ravasi et al., 2010), comprehensive genome-scale data on the spatiotemporal expression of TR genes in the developing vertebrate embryo is not available. This information is a prerequisite for a systematic elucidation of transcriptional regulatory networks during development. The zebrafish (Danio rerio) embryo represents a promising model to obtain such a genome-scale description of TR gene expression as it allows the combination of transcriptome studies with large scale in situ expression analysis. We report here a comprehensive analysis of TR gene expression in zebrafish. We profiled the relative abundance of TRs by microarray analysis over different developmental stages and adult body parts, and compiled a genome-wide analysis of gene expression states by RNA sequencing (RNAseq) during organogenesis, larval maturation and adult homeostasis. We cloned 2149 gene probes and provided a comparative atlas of 1711 TR genes, including 746 new patterns of expression in the 24 hpf (hour post-fertilization) embryo. The 24 hpf stage is of particular importance as it represents the evolutionarily conserved phylotypic stage of this model organism (Domazet-Loso and Tautz, 2010). At this stage, the embryos of all the different vertebrate subclasses look very similar. Organogenesis and the vertebrate-subclass specific elaboration of the body pattern have begun at this stage, but is far from complete. The majority of TR genes is already expressed at the phylotypic stage. For example the anlage of the telencephalon expresses more than 1100 different TR genes at this early stage. Expression of these factors is largely maintained in the adult zebrafish suggesting roles of TR genes in tissue and body homeostasis. Quite unexpectedly, we find that 55% TR genes are expressed ubiquitously. Our comprehensive study of the TR gene expression state in the zebrafish embryos uncovers the complexity of the expression state of TR genes at the immature phylotypic stage and points at differential redeployment of TR genes and post-transcriptional modifications as fundamental regulatory processes in the further elaboration of body pattern.
Results
Characterisation of the repertoire of transcriptional regulatory genes
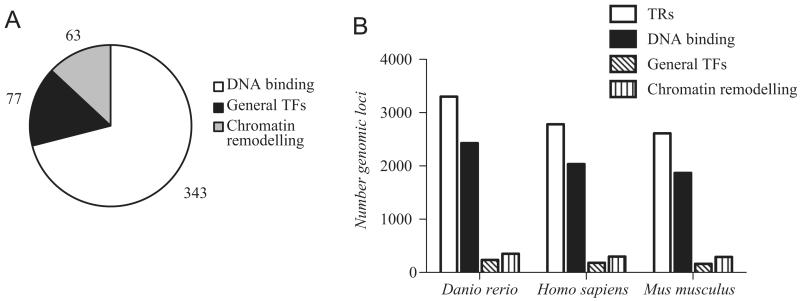
To obtain a comprehensive representation of gene loci involved in transcriptional regulation, we mined the InterPro database (Hunter et al., 2009) and the literature to systematically identify protein domain families specific to TRs. We scored 483 InterPro protein domains that fell into 3 distinct functional groups: (i) DNA-binding domains, (ii) chromatin remodelling domains and (iii) domains specific to factors of the general transcriptional machinery (Fig. 1A, Supplementary Table T1).
Fig. 1.
Gene loci encoding transcriptional regulators. (A) Categorisation of InterPro domains into distinct functional groups specific to transcriptional regulators. The number of protein domains belonging to each group is indicated. (B) Number of genomic loci encoding transcriptional regulators in the zebrafish (Zv9), human (GRCh37.p2) and mouse genome (NCBIM37). The categorisation into families is based on their predicted protein domains.
We searched the zebrafish genome (Zv9) for loci encoding proteins with at least one of these domains. We additionally mined 24,386 zebrafish Refseq transcripts (Refseq, NCBI, Nov 2010) with InterProscan (v4.6) (Zdobnov and Apweiler, 2001). We identified 3302 unique genomic loci encoding potential TRs, representing 11,6% of the 28,491 genes annotated in the zebrafish genome (Fig. 1B, Supplementary Table T2). When sorted according to potential function, 2677 (81%) of the zebrafish TR genes encode TFs with a DNA-binding domain, and 488 (15%) genes code for proteins with chromatin remodelling domains. Proteins with a putative function in general transcription are represented by 137 loci (4%). In comparison to the human (2782 genes) and mouse (2612 genes) genome, the zebrafish genome encodes more TR genes (Fig. 1B), presumably reflecting gene retained after the genome duplication at the base of the evolution of actinopterygian fish (Taylor et al., 2003).
Most transcriptional regulators are expressed throughout development
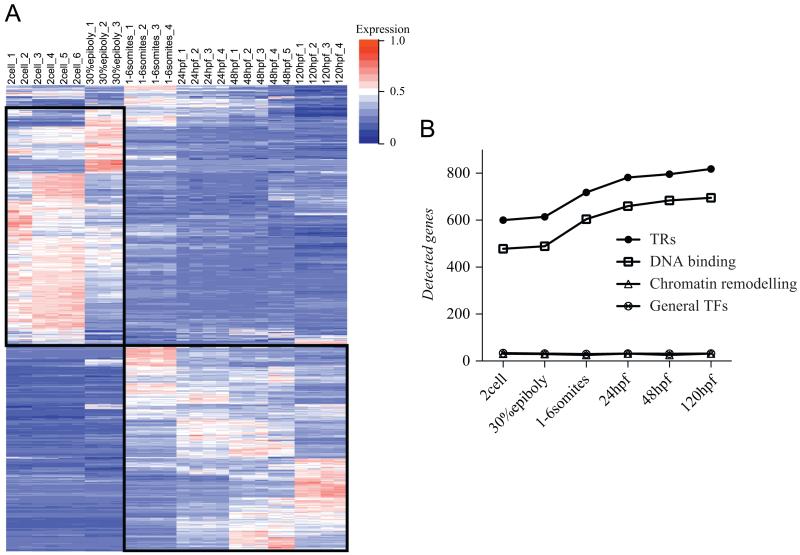
We next wished to assess the expression state of the TR genes, during embryogenesis. First, we determined the developmental profile of TR gene expression by employing a custom-designed microarray with probes representing 1565 TR genes, to which we hybridised cDNA from six different developmental stages. cDNA samples from 3 to 6 independent RNA preparations from each stage were analysed (Supplementary Fig. S1). Among the 1565 TRs genes present on the microarray, 225 are novel genes which were not included in precedent microarray analysis (Domazet-Loso and Tautz, 2010). Hierarchical clustering reveals two main clusters of genes (Fig. 2). A first cluster is composed of TRs abundant at early stages of development, before organogenesis, and low expression at subsequent stages. This group comprises known early genes such as sox32, vox, vent, gro1, gro2 (Supplementary Fig. S2). The second cluster comprises genes with prominent expression at various stages of organogenesis. It includes genes involved in somitogenesis (myod, myog, myf5, prdm1a) and neurogenesis (ascl1a, neurod, zic1) (Supplementary Fig. S2 and data not shown). These results correlate well with precedent studies on Caenorhabditis elegans (Levin et al., 2012) and ascidian embryos (Sobral et al., 2009) where early development genes and genes expressed at later stages during organogenesis were also discriminated. At the 2-cell stage prior to zygotic transcription, we detected significant levels of mRNA for 600 TR genes. By 30% epiboly after the onset of zygotic transcription, this number increased slightly. In subsequent stages, the number of expressed TR genes grew to 818 genes, levelling off by 24 hpf with a marginal increase up to 120 hpf (Fig. 2B). This overall increase in TR gene expression is exclusively due to the TF class; the number of expressed chromatin remodelling and basal transcription factors remained constant over the developmental stages examined (Fig. 2B).
Fig. 2.
TR gene expression in developing zebrafish. (A) Temporal expression profile of 1219 TRs (rows) across six developmental stages (columns) by microarray analysis. Hierarchical clustering of normalised gene expression reveals two main clusters (black rectangles) discriminating genes either expressed prior (2 cells and 30% epiboly) or during organogenesis (1–6 somites to 120 hpf larvae). Blue: low expression; white: moderate expression; red: high expression. (B) Number of TR genes detected by microarray analysis. While the number of TF genes increases, the numbers of genes encoding chromatin remodelling and general transcription factors remain constant over the stages examined.
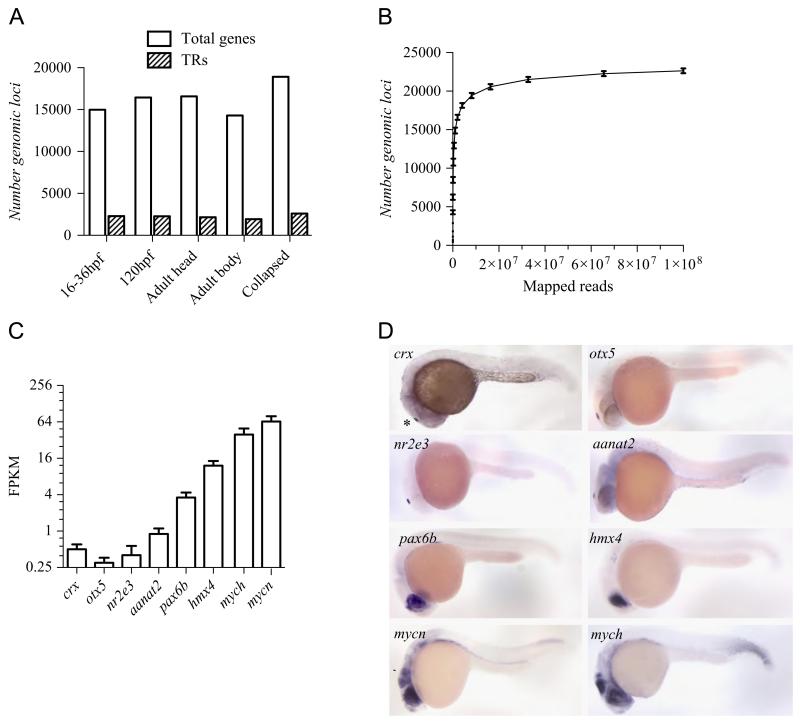
Microarray analysis has limited sensitivity and is inherently biased by the selection and specificity of the probes deposited on the chip (Marioni et al., 2008). We thus employed mRNA sequencing (RNAseq) to compare the number of TR genes expressed at 16–36 hpf and 120 hpf larvae. More than 10 million reads per condition were generated (Supplementary Table T3, Supplementary Fig. S3). The number of expressed TRs was very similar at these two developmental stages (2291 and 2273 TR genes, respectively) (Fig. 3A). Detailed comparison of the expressed TR genes at the two stages showed that 93% (2124 genes) TR genes are expressed in common at 16–36 hpf and 120 hpf. We also sequenced RNA samples derived from adult body and head to assess whether there is an additional activation of TR gene expression in adult stages. With 2163 and 1929 TR genes expressed in the adult head and the adult body, respectively, we did not find a significant increase in the total number of TR genes expressed in adult tissues (Fig. 3A).
Fig. 3.
Assessment of TR gene expression by RNAseq. (A) Number of genes (white bars) and TRs (dashed bars) expressed at two different developmental stages and two adult body parts as determined by RNAseq. The total number of TR loci quantified by transcript counting over all stages is indicated as “Collapsed”. (B) Quantification of the total number of genes detected at 24 hpf by RNAseq in function of the sequencing depth. The number of detected genes is indicated as the mean from 3 biological replicates. (C) Level of expression of transcripts represented as FPKM from a selection of genes known to be expressed in the epiphysis (crx, otx5, nr2e3, aannt2), the retina (pax6b, hmx4) or tectum and retina (mycn and mych). The relative expression is indicated as the mean of FPKM from biological triplicates. (D) RNA in situ expression data of the selected transcripts at 24 hpf. *: epiphysis; arrow head: optic tectum.
The sensitivity of detection may be limited with 10 million reads when using whole embryos, as transcripts specifically expressed in just a few cells are diluted. We thus selected one stage to further sequence the transcriptome exhaustively. We focused on 24 hpf embryos, the phylotypic stage of zebrafish, where zebrafish embryos share a very similar morphology with other vertebrate embryos and where the highest expression of conserved genes was noted (Domazet-Loso and Tautz, 2010). We generated 349 million, 76 bp long paired-end reads from three independent samples of RNA isolated from 24 hpf embryos (Supplementary Table T3). When mapped to the zebrafish genome, 77% of the reads fell into intragenic regions and 23% into intergenic regions presumably representing un-annotated transcripts. Pearson’s correlation coefficient between unfiltered RNAseq and microarray data at 24 hpf is comparable to precedent studies (r > 0.68) (Supplementary Fig. S4) (Marioni et al., 2008). To assess coverage, the number of detected transcripts was plotted over the sequencing depth. With 100 million of aligned reads the number of genes detected by at least one read in all biological triplicates reached a plateau with a mean of 22,628 genes ( ± 1.4%) (Fig. 3B). In addition, the rate of novel TRs detected increases rapidly with increasing sequencing depth until 4 million reads and then decreased slowly, showing that rare transcripts need higher coverage to be detected by RNASeq (Supplementary Fig. S5). From the 3302 loci encoding TRs in the zebrafish genome, 2488 TR gene transcripts were detected consistently in all three replicates and at significant level, in close agreement with our previous sequencing results at lower resolution. We next wished to calibrate the sequencing depth with respect to genes expressed only in restricted domains in the embryo (Fig. 3C, D). At 24 hpf, the transcripts of crx, otx5, nr2e3 and aanat2 are expressed only in the epiphysis, while pax6b, hmx4, mych and mycn have broader expression domains (Fig. 3D). The size of the expression domain correlated with the Fragments Mapped per Kilobase of transcript per Million reads sequenced (FPKM). Importantly, significant signals for the highly-tissue restricted genes crx, otx5, nr2e3 and aanat2 which were scored with 0.5, 0.3, 0.4 and 1 RPKM, respectively, were detected by the chosen sequencing depth (Fig. 3C). Hence, we scored expression of TR genes in the 24 hpf embryo with high sensitivity. Moreover, the fact that we observed only a 15% increase in the number of detected genes by increasing the depth of sequencing 34.9-fold from 10 million to 349 million reads suggests strongly that we scored efficiently the significantly expressed TR genes in the 24 hpf embryo. We confirmed the expression of 10 novel genes by qRT-PCR on 24 hpf embryos with various expression patterns (not restricted or restricted) and different expression levels (high and low expression) and find a good correlation between the RNAseq and the qRT-PCR results (R2=0.73) (Supplementary Table T7).
Different TR genes may be expressed in the embryo in comparison to the adult zebrafish. We therefore combined the lists of TR genes expressed in the embryo and the adult and found a total of 2593 genes expressed in all stages examined (Fig. 3A). Thus the 24 hpf embryo expresses detectably 75% of TR genes encoded in the genome and this number is only moderately increased in subsequent stages. This suggests that only a limited number of TR genes become activated in addition during further development and in mature tissues. Thus, the majority of TR genes remains active from the phylotypic stage into adulthood.
A library of TR clones
We next cloned the TR cDNAs. As a first strategy, we screened four normalised libraries enriched for full-length cDNAs by either hybridisation with radioactive probes specific to TRs or by direct sequencing. This library-based approach has the advantage that we could obtain predominantly full-length TR clones. In total, 196,536 clones were screened by hybridisation and 55,296 clones were directly sequenced at their 5′ and 3′ ends generating 93,279 ESTs (GenBank, EMBL accession FP104570–FP232151). The ESTs were mapped to the zebrafish genome and transcriptome with BLAT and Blast, respectively. This led to the isolation of 1242 TR cDNA clones. Genes expressed at low levels or in very restricted areas in the embryo are difficult to clone by this method. Previous deep sequencing studies in mouse indicated that transcripts detected at less than 1 FPKM correspond to genes expressed at very low levels (Mortazavi et al., 2008; Wang et al., 2009). Based on our deep sequencing data of the zebrafish transcriptome at 24 hpf, a RT-PCR screen was carried out for TR genes missed by library screening, using FPKM≥0.3 and 20 mapped reads as lower cutoff limit for candidates. An additional 907 clones were obtained, resulting in a total of 2149 TR cDNAs. This collection represents 83% of the 2593 TR transcripts detected by RNAseq over all stages and 86% of the 2488 TR transcripts expressed in the 24 hpf embryo. 72% of cloned genes are TFs followed by chromatin remodelling TRs (12%) and 5% are general transcription factors reflecting the abundance of these different classes in the genome. The remaining 11% clones represented putative TRs with less well-characterised functions in mammalian systems. This library of 2149 TR cDNAs constitutes a unique resource to study the transcriptional regulation of zebrafish development and body homeostasis.
A majority of TR genes are expressed ubiquitously in the 24 hpf embryo
We next determined the spatial expression of the TR genes by generating 1871 probes for whole mount in situ hybridisation in 24 hpf embryos, thus focusing on the phylotypic stage. We successfully obtained in situ expression patterns of 1711 TR genes. Among these, 746 (44%) are new patterns that complement existing databases of expression pattern in zebrafish (Bradford et al., 2011) (Fig. 4A). Fewer than half of the TRs assessed (768 genes, 45%) are expressed in a tissue-restricted manner. The remaining clones (55%, n=1711) showed a more or less uniform signal throughout the embryo. This together with the signal detected by RNAseq demonstrates clearly that these TRs are expressed ubiquitously. Thus, the majority of TRs have either a “housekeeping” function in many cell types, or their activity is regulated at the post-transcriptional level in a region- or stage-specific manner.
Fig. 4.
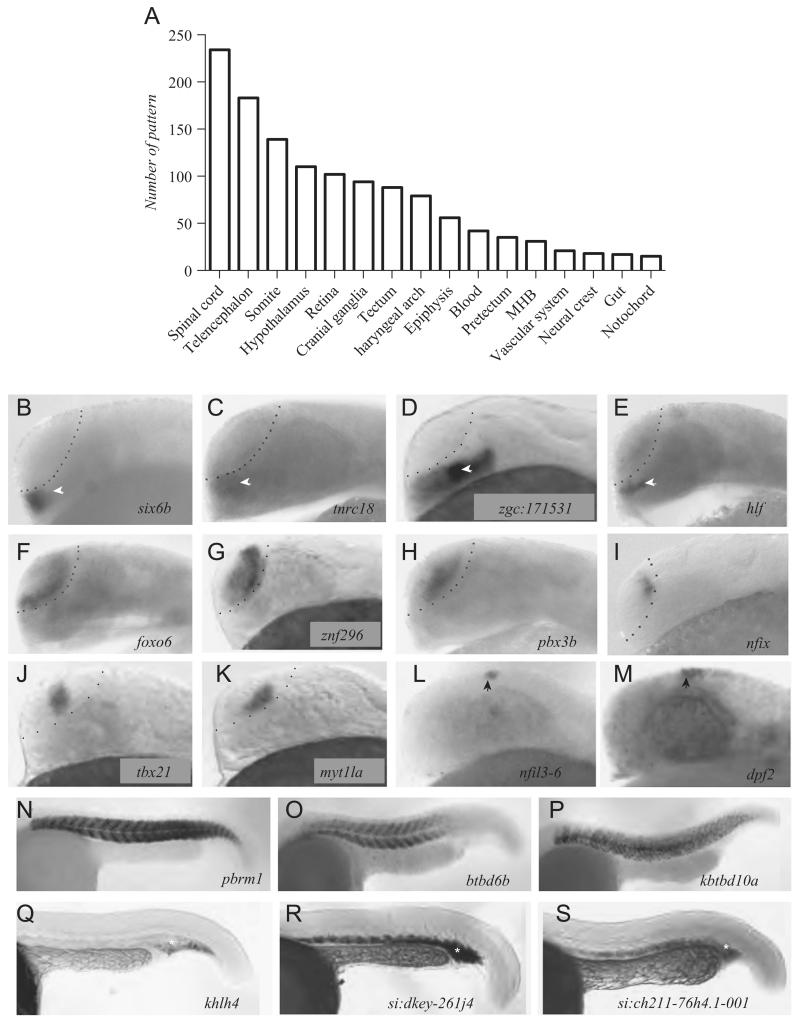
Assessment of TR gene expression patterns by in situ hybridisation. (A) Summary of in situ expression patterns of TR genes expressed in specific tissues (MHB: midbrain/hindbrain boundary). (B–M) Expression patterns of new markers of forebrain sub-domains at 24 hpf. The telencephalon border is depicted by a dashed line. White arrowheads indicate the hypothalamus and black arrowheads the epiphysis. (N–P) Expression pattern of genes expressed in somites. (Q–S) Genes expressed in the intermediate cell mass of mesoderm (*: presumptive blood precursors).
The central nervous system (CNS) and especially the spinal cord and forebrain express the highest diversity of TR genes (Fig. 4A). For instance, in the telencephalon, we detected mRNAs of 183 TR genes representing 91 InterPro families. If one includes the pan-neural (37 genes expressed in the whole neural tube) and the ubiquitously expressed genes (918 genes expressed in the whole embryo), the telencephalon of the 24 hpf zebrafish embryo expresses a total of 1138 TRs representing 67% (1138 out of 1711) of the entire transcriptional regulome analysed by in situ hybridisation. Another tissue with a high diversity of spatially restricted TRs is the somite with 139 genes from 116 InterPro families. In both somites and telencephalon, homeobox-containing TFs are the most abundant followed by TFs containing C2H2 zinc fingers (Supplementary Fig. S6). In some instances, we found also preferences for one or the other class of TRs in individual tissues. The number of tissue-restricted HMG-domain containing TFs expressed in the telencephalon was higher compared to the somites (single-sided Fisher’s exact test p<0.04). In contrast, somites express a higher, but not significant, proportion of BTB-POZ as well as SET chromatin remodelling factors when compared to the telencephalon (single-sided Fisher’s exact test p<0.09) (Supplementary Fig. S6).
The in situ expression data as well as the transcriptional profiles were compiled in a publicly accessible database (http://cassandre.ka.fzk.de/ffdb/index.php) that allows various search functions to mine the data set and to identify gene expression patterns and co-expressed genes (Supplementary Fig. S7).
Characterisation of new potential key developmental regulators
While most genes are expressed in several tissues (n=768 tissue restricted patterns), we detected 207 genes that are expressed in a single tissue in the 24 hpf embryo (Supplementary Table T5). The annotation of expression domains was based on the OBO Zebrafish Anatomy and Development Database (Supplementary Table T4). Genes expressed in a single tissue are particularly interesting as they may have unique roles in the development or function of the expressing tissue and thus may constitute putative novel key developmental regulators. For example, four TRs are expressed in the hypothalamus only. This group contains the known hypothalamic marker nkx2.1a (not shown) as well as the homeobox gene six6b, a tnrc18 homologue (LOC559514), an orthologue of mouse nkx2.4 (zgc:171531) and hlf (Fig. 4B–E). Another group of 16 genes have an expression restricted to the telencephalon only. We find patterning and differentiation genes such emx1, emx3, tbr1a and neurod6b, as well as new markers like foxo6, znf296, pbx3b, the CTF/NFI family nfix gene (zgc:136878), tbx21 and myt1la (Fig. 4F–K and Supplementary Table T5). Other genes were detectable only in the epiphysis: otx5, nr2e3, rorca, rorcb, crx, as well as the zinc finger TFs dpf2 and nfil3-6 (Fig. 4L, M and Supplementary Table T5). Restriction to single territories of expression is not confined to neural tissues: for example, 41 TR genes are uniquely expressed in the somites at 24 hpf such as the high mobility group box gene pbrm1 and two BTB-POZ containing genes btbd6b, kbtbd10a (Fig. 4N–P). We also found 14 TRs expressed exclusively in the intermediate cell mass from which blood cells develop including kelch-like 4, a new zinc finger locus (si:dkey-261j4) and an orthologue of the human AFF2 gene (ENSDARG00000052242, si:ch211-76h4.1-001) (Fig. 4Q–S and Supplementary Table T5).
The highly restricted expression patterns of these 207 novel specific markers in the 24 hpf embryo makes them prime candidates for functional studies. With 82%, TFs are overrepresented. In general, members of the TF class of TRs are more frequently tissue-restricted. Among the chromatin remodelling factors, only the BTB-POZ family shows a significant proportion of genes (38 out of the 118 genes) with restricted expression patterns mainly in the somites (Fig. 4N–P) or the central nervous system (Supplementary Table T6). Genes encoding factors of the basal transcription machinery are predominantly expressed ubiquitously.
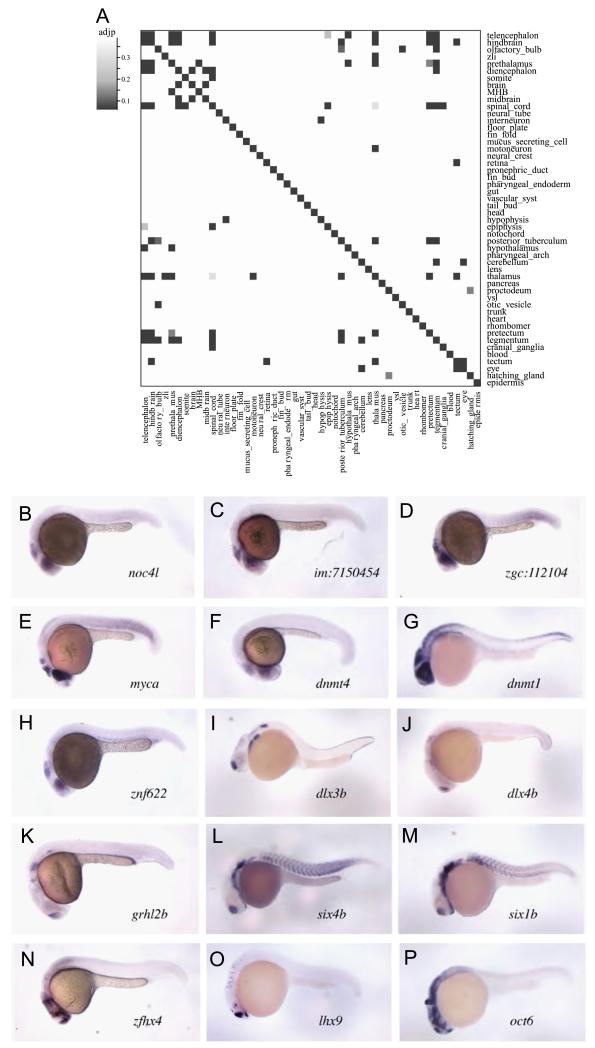
Synexpression of genes has been suggested as an indicator of functional linkage into regulatory pathways (Karaulanov et al., 2004; Niehrs and Pollet, 1999). In particular transcription factors are believed to act in a combinatorial fashion. Hence, we investigated whether tissue-restricted TR genes, whose expression is detectable in multiple tissues, are co-expressed in different tissues by a Bonferroni-corrected Pearson’s Chi-Squared test (Fig. 5A). Significant correlation is only observed for some neuronal and sensory territories. The otic placodes and olfactory bulb are part of the cranial sensory system developing from ectodermal placodes. The members of the distal-less family dlx3 and dlx4b (aka dlx7) are both required for their development (Solomon and Fritz, 2002). The expression of 10 TR genes, which include dlx3 and dlx4b as well as grhl2b, six4ba and six11b, cluster in both sensory organs (p < 1 × 10−3) at 24 hpf (Fig. 5A, I–M). We also see a correlation between forebrain structures and several diencephalic regions. For example, the telencephalon shares a significant number of TRs with the pre-thalamus (p < 10−10) and the hypothalamus (p < 10−17), as well as with more posterior regions of the midbrain like pretectum (p < 10−6), tegmentum (p < 10−5) and spinal cord (p < 6 × 10−4) (Fig. 5A). We also found that, among the 91 TRs expressed in the optic tectum, 40 (43%) TR genes are also expressed in the retina (p < 10−17). This latter finding is particularly intriguing as retina and tectum are functionally coupled by topographical projections of retinal axons into the tectum (Lemke and Reber, 2005; Polleux et al., 2007). Genes co-expressed in these two tissues like the homologous genes of yeast ncol4 and trmt1 (im:7150454), cebpz (zgc:112104), myca, the DNA-methyl transferases dnmt1, dnmt4 and the zinc finger protein znf622 (Fig. 5B–H) may thus have common roles in the two functionally linked neuronal tissues. TFs controlling generic neuronal specification like proneural genes account only partially for this correlation between different territories in the nervous system and sensory organs. For instance only five of these genes (neurog1, pou3f1, etv5b, zhfx4 and sox9b) are co-expressed in the forebrain (telencephalon, thalamus and hypothalamus) and the spinal cord (Fig. 5N–P). Together, these results show that, at a global level, there is no strong tendency towards co-expression of TR genes in different tissues. Thus TRs act mainly in a freely combinatorial fashion to specify distinct cell fate and function.
Fig. 5.
Correlation of co-expressed TRs. (A) Heat map of adjusted p-values from Pearson’s Chi-squared test showing significantly co-expressed TRs. Correlation of co-expression at 24 hpf is found mainly for neuronal regions (adjp: adjusted p-value). Example of in situ expression for TRs co-expressed in the retina and tectum (B–H), sensory placodes (I–M) or telencephalon, hypothalamus, thalamus and hindbrain (N–P). MHB: midbrain–hindbrain boundary.
Discussion
We report here a systematic characterisation of the transcriptional regulome of the zebrafish. We detected 3302 TR genes in the zebrafish genome with at least one protein domain related to transcriptional regulation including transcription factors with DNA-binding domains, chromatin remodelling proteins and factors of the general transcriptional machinery. In comparison to the mouse and human genomes, the zebrafish genome encodes a higher number of TR genes. This reflects presumably the duplication of the genome at the base of teleost evolution and the subsequent retention of some of the TR genes (Postlethwait et al., 1998). We employed microarray, deep sequencing and in situ hybridisation to assess the expression state of the TR genes. We cloned 2149 TRs and provide a comparative atlas of gene expression for 1711 genes in the 24 hpf embryo, including 746 new expression patterns. Expression of 10 genes was assessed by quantitative RT-PCR and confirmed correlation with RNAseq (R2=0.73) and in situ expression data (Supplementary Table T7). In comparison to published patterns in Zfin, 83% of the annotations are concordant. Notably 60% of the discrepancies are found among genes with non-restricted patterns, which can display higher expression levels in particular tissues and be considered as tissue restricted if the staining is not developed long enough. This work constitutes a unique resource that provides an expression pattern database and a physical library of cDNA subclones for refined expression and functional studies.
We describe the absolute expression levels of 3302 TR genes by RNAseq. Precedent microarray studies were limited by the probes deposited on the array (Domazet-Loso and Tautz, 2010) and included only a subset of these (2008 TR genes on the Agilent array G2519F, using Ensembl release 70). The 24 hpf zebrafish embryo expresses 75% of all TR loci encoded in the genome. The remaining 25% TR genes may be expressed at a different stage. However our data suggest that the number of TR genes increases only marginally over subsequent stages. We detected a similar number of expressed TR genes in 120 hpf larvae and in the adult body and head. When we count the expressed TR genes from all embryonic and adult stages, we detect 2593 expressed TR genes in approximate agreement with the 2488 genes detected by sequencing the 24 hpf embryo to saturation. The remaining TR genes encoded in the zebrafish genome may be pseudogenes or, alternatively, may comprise genes that are only activated significantly in response to specific physiological or environmental conditions that are not reproduced under standard maintenance conditions in the laboratory. In addition, some of the remaining TR genes may be expressed at such low levels that they escape our detection. Our calibration of transcript counting with in situ hybridisation of genes expressed in very few cells in the 24 hpf embryo suggests that we reached, however, a very high sensitivity. Moreover, we could not detect a strong increase of the number of expressed genes by increasing the transcript sequencing depth by 35-fold. We are confident that we provide an exhaustive evaluation of the expressed transcriptional regulome in the 24 hpf embryo. Thus, around 2500 TR genes seem to be sufficient to control the construction of a zebrafish.
Development is controlled by hierarchical decisions. It is thus assumed that new genes including TR genes are activated in the course of the elaboration of body pattern and organ function. At the phylotypic stage, vertebrate embryos share a common morphology (Haeckel, 1874; von Baer, 1828) and the body plan has been laid down but many organ systems and the vertebrate subclass specific elaboration of the body plan from the phylotypic ground state has not been completed. Moreover, although many organ primordia have formed at this stage, organs show only rudimentary functions, if any at all. It will take several further days of development before for example a functional digestive system has formed or complex behavioural traits such as hunting (from 120 hpf) will commence (Kimmel et al., 1995). Our data suggest that there is not a substantial increase in the activation of new TR genes after 24 hpf. These findings together with the fact that approximately half of the TR genes are ubiquitously expressed, underscore the importance of redeployment and posttranslational modification of TRs during subsequent organogenesis and establishment of complex organ function. Some of the TR genes such as neurod, pax6, islet1, nkx2.2, nkx6.1 and foxa3, involved in the control of neuronal differentiation in the central nervous system in the 24 hpf embryo, are redeployed for example in the differentiation of the pancreas and the liver in subsequent stages (Field et al., 2003; Grapin-Botton and Melton, 2000; Wallace and Pack, 2003). We find that ~80% of the TR genes expressed during organogenesis are also detected in differentiated adult tissues. This observation is in agreement with a precedent study in Ciona intestinalis and with a much more limited study focusing on the expression of nuclear receptor genes in zebrafish (Bertrand et al., 2007; Imai et al., 2004). This result suggests that a majority of the transcriptional regulators used to determine cellular fate during embryogenesis is still active in adult tissues to maintain the cellular differentiation state (Blau and Baltimore, 1991; Eade et al., 2012) or tissue homeostasis.
The majority of TR genes (55%, n=1711) showed ubiquitous expression as judged by deep sequencing and verified by in situ hybridisation analysis. These ubiquitously expressed genes are either constitutively active or their activity may be regulated by post-transcriptional mechanisms. Among the TR genes expressed in the 24 hpf zebrafish embryo, we found only 207 genes (12%, n=1711) expressed in a single tissue. These genes may have unique functions in the tissues, in which they are expressed in the 24 hpf embryo and are thus prime candidates for gene knock-out studies. In a precedent microarray analysis of TF expression in adult mouse tissue, 35% TFs were found to be expressed in a single tissue (Vaquerizas et al., 2009). Possibly, the poorer resolution of microarray studies to comprehensively assess the spatial expression compared to detection of gene expression by whole mount in situ hybridisation may have contributed to this discrepancy of the two studies.
Regulatory genes are frequently co-expressed in different tissues forming synexpression groups (Niehrs and Pollet, 1999). We found a number of domains in the central nervous system and sense organs that share the expression of tissue-restricted TR genes, suggesting that similar regulatory networks are operational in these domains. An intriguing pair of domains of co-expression is formed by the retina and the tectum. Neurons of the retina project axons into the tectum producing a topographic map in which the spatial relationships between the projecting axons and the target tissue are maintained (Lemke and Reber, 2005; Polleux et al., 2007). In zebrafish, the axons exit the retina at 36 hpf and invade the tectum at 46 hpf (Stuermer, 1988). The expression of these genes at 24 hpf before the axons start to find their target suggests that the retina and the tectum share regulatory mechanisms to orchestrate development of the retinotectal axonal projections. At a global level, however, there appears to be little constraint on the co-expression of TFs in other regions of the 24 hpf embryo. Although the components of regulatory cascades and other cellular processes seem to be frequently organised into synexpression groups, the TR genes appear to be much more promiscuous. This suggests little functional constraints among tissue-specific TR genes allowing high flexibility in combination of different factors. This reflects presumably the function of TRs as integrators of signalling inputs and that, as a consequence, the cooperation of TRs determines the specific regulatory output.
Materials and methods
Database of transcripts with TR protein domains
The InterPro database (release 25) was mined to select protein domains specific for each class of TR gene (described in Supplementary Table T1). The abundance of TR genes in the human (GRCh37.p2), mouse (NCBIM37) and zebrafish (Zv9 release 60) genomes was assessed by retrieving protein domain annotations form the Ensembl genome data with BioMart (Guberman et al., 2011). We found 3100 genomic loci encoding TRs in the zebrafish genome by searching BioMart with our specific set of protein domains. To ensure that all TRs were included in our study, we mined 27,580 zebrafish Refseq transcripts (NCBI, Nov 2010) with a coding sequence ≥20 amino acids, in addition. From these, 24,386 transcripts were selected with at least one predicted protein domain using InterProscan (v4.6). We then mapped these transcripts to unique genomic location with a Perl script which uses Blat (Kent, 2002) allowing a maximum distance of ±100 bp between the Refseq hit and known Ensembl exons. Alternative spliced transcripts were collapsed into a single transcriptional unit keeping the longest transcripts as reference. In this way 21,147 transcripts were mapped onto the genome. From these, 202 additional TR loci were detected giving a total set of 3302 TR genes in the zebrafish genome.
RNAseq, mapping and quantification of reads
Total RNA from wild type zebrafish (AB strain) was extracted with Trizol (Invitrogen) using a tissue homogeniser (Ultra-Turrax, Janke&Kunkel, IKA-Werk) according to the manufacturer’s protocol followed by a second round of extraction with phenol–chloroform and precipitation. Total RNA was resuspended in RNase-free water (Ambion) to reach a final concentration of 0.1–1 μg/μl RNA. RNA quality was checked using Agilent Bioanalyzer 2100 total RNA Nano series II chip (Agilent) and showed no sign of degradation (RNA index number > 8). Sequencing libraries were generated from total RNA without prior DNAse I treatment following the TrueSeq RNA (Illumina) protocol for the generation of single end (16–36 hpf, 5 dpf larvae, adult head and adult body) or paired end (24 hpf) data. Single end reads of 36 nucleotides and paired end reads (2 × 76 nucleotides) were obtained with a GAIIx (Illumina). Cluster detection and base calling were performed using the standard Illumina pipeline. Quality of reads was assessed with CASAVA v1.4 and Eland (Illumina) using the zebrafish (Zv9) genome as a reference (summarised in Supplementary Table T3). For transcript quantification, reads were mapped with the exon–exon junctions compatible mapper Tophat (version 1.4.1) (Trapnell et al., 2009) and Bowtie (version 0.12.7) against the zebrafish genome (Zv9) using known exon junctions (Ensembl, Zv9 release 60) and the options butterfly-search, coverage-search, micro-exon-search, min-anchor-length 5. The mean distance and standard deviation between read pairs were obtained from CASAVA. The total number of reads mapped with this method is 349 million reads (> 100 million reads per biological replicate, after pooling reads from technical replicates). Quantification of gene expression was performed with Cufflinks (Trapnell et al.) and HT-Seq (Anders and Huber, 2010) for computation of FPKM and raw counts respectively, keeping biological replicates as separated datasets. Data from the two quantification methods were compiled into a MySQL database using Ensembl genes numbers as unique identifier. Genes were considered as detected in a sample when RPKM (single reads) or FPKM (paired reads) were≥0.3 and number of counts≥20 in each biological replicate. Correlation of biological replicates was checked using unfiltered expression data and Pearson’s correlation coefficient r was ≥0.96 in all cases. For assessment of the sequencing depth, alignments (BAM files) obtained from Tophat were sampled and quantified with HT-Seq using either the complete list of known exon junctions (Ensembl release 60) or the selection of 3302 junctions specific to TR genes and using a threshold of detection of more than 20 read counts in each biological replicate.
Microarray design and analysis
We used a custom made microarray (Agilent #022326) composed of 35,888 probes corresponding to 2341 Refseq mRNAs and 1565 genes encoding TRs (Zv9 release 60). Briefly, sequences of Ensembl transcripts (assembly Zv7) were used to design spotted oligonucleotide probes specific for TR genes with the Agilent software eArray. As much as eight different probes were designed per transcript (31,656 probes) from the 3957 selected transcripts with at least one TR related InterPro domain (2340 genes in assembly Zv7). An additional set of 4232 probes corresponding to 529 TR genes obtained from EST databases was also synthesised resulting in a total of 35,888 different probes. An update of the array annotation was necessary to compare the microarray to next generation sequencing data. This was made using the most recent genome assembly (Ensembl zv9) as well as zebrafish cDNA databases (Refseq Nov 2010). From the original set of 35,888 probes present on the array, 27,963 probes were assigned to a new Refseq mRNA and 19,829 probes were assigned to an Ensembl gene identifier. Three to six biological replicates were produced per stage/tissue resulting in a set of 29 independent biological samples. cDNA synthesis and hybridisation to microarrays were described previously (Yang et al., 2007). Variance stabilizing normalisation (vsn) was used to correct signal variations between the different arrays and dyes, and the median of the 8 probes per transcript present on the array were computed (Bioinformatics Toolbox, MATLAB R2009b). Spearman’s correlation using unfiltered expression data was > 0.95 in all cases. The mean over the different replicates was calculated and a threshold of 5 times the background expression level used as the detection limit of the microarrays. This resulted in the selection of 1219 genes detected over the background used for expression analysis. Clustering was performed on scaled expression data. Hierarchical clustering was carried out with Pearson’s correlation and the complete-linkage method. Soft clustering was performed using the parameters c=8 and m=1.6 (Futschik and Carlisle, 2005).
Cloning of TRs genes from full-length cDNA libraries
Tissues were collected at four different developmental stages: 16 hpf to 36 hpf embryos, 120 hpf larvae, adult head and adult body. Enriched full-length cDNA libraries were produced from total RNA samples by Invitrogen (California, USA) for the 16–36 hpf library and by DNAForm (Kanagawa, Japan) for the three remaining libraries. Vector information and details on library production are available upon requests. A total of 193,356 bacterial clones were picked and gridded into separated sub-libraries. Handling, arraying, gridding, DNA-prep, sequencing and storage of the libraries were carried out following Wellcome Trust Sanger Institute (Hinxton, UK) and Genetix (Hampshire, UK) guidelines. A subset of 55,296 clones from the 16–36 hpf library were sequenced at both 5′ and 3′ ends generating 93,088 ESTs (GenBank accession FP104570–FP232151). Another set of 138,240 additional clones from a 120 hpf library, 3 months old head and body libraries, and the 16–36 hpf library were screened by hybridisation of labelled oligonucleotide probes as follows: Bacteria were gridded on Nylon filters (Performa II, Genetix) at a density of 27,648 clones/filter with a Q-Bot equipped with a 384 pins gridding head (Genetix), and grown overnight at 37 °C on LB agar plates with ampicilin (50 μg/ml). Post-gridding treatments of filters were carried out following standard protocols (Sambrook, 2001a). In total 1403 oligonucleotide probes (25mers) were designed based on Ensembl transcript predictions or Refseq (NCBI) and synthesised at 20 nmol scale (sequence information available upon request). Pools of 20–100 oligonucleotide probes were labelled with dATP γ-32P using T4-polynucleotide kinase (Fermentas) following the manufacturer’s protocol. Hybridisations of radio-labelled probes to filters were carried out in presence of tetramethylammonium chloride salts following standard protocols (Sambrook, 2001b). Filters were exposed for 3 h to X-OMAT (Kodak). Films were scanned and analysed using Photoshop7.0 and position of positive clones in respect to the gridding map determined manually. Positives clones were picked and clone identity confirmed by DNA sequencing.
Cloning of TR cDNAs by RT-PCR
Reverse transcription was performed on 1 μg total RNA extracted from 16–36 hpf embryos using SuperScript II (Invitrogen) following the manufacturer’s protocol. Pairs of PCR primers were designed with Primer3 v0.4 (Rozen and Skaletsky, 2000) using Refseq transcripts as reference. Primers are available upon request. PCR reactions were performed in 10 μl using Taq-Platinum (Invitrogen). PCR products were ligated overnight at 4 °C with pGEM-T vector (Promega) in a final volume of 5 μl following the manufacturer’s instructions, and used to transform Escherichia coli XL1Blue. Four clones for each target cDNA were screened by NcoI/SacI digestion (Fermentas). Positives clones were sequenced with T7 primer to assess identity and orientation of the inserts.
Mapping of ESTs
Mapping of ESTs to the zebrafish genome (D. rerio, assembly Zv9) was carried out using BLAT (Kent, 2002) with default parameters. A perl script was used to parse the best-hit location within a window of 1000 bp flanking mapped genes in order to allow the detection of clones with un-annotated UTR. A total of 53,712 ESTs were mapped to 3882 genomic loci. Transcript identities were further assessed by Blast using the Refseq Danio rerio repository (NCBI, Nov 2010). We assigned identity to reference transcripts when ESTs had a minimal identity of 90% over at least 100 bp and an e-value less than e−150. We successfully mapped 68,818 ESTs to 5650 independent Refseq transcripts.
RNA in situ hybridisation and statistical analysis of expression patterns
Templates for antisense DIG RNA probes were generated by cutting plasmid DNA with suitable restriction enzymes at the 5′ end of the cDNA, followed by a single step of phenol/chloroform extraction and precipitation of the linearised vector with 2.5 volume of EtOH and a final concentration of 0.3 M NaOAc. DIG RNA probes were generated with T7 (Promega) or Sp6 (New England Biolabs) RNA polymerase depending on the orientation of the insert and vector type using a DIG labelling mix (Roche). Collection of embryos, fixation and in situ hybridisation to 24 hpf embryos were carried out as described (Yang et al., 2010). The precise description of gene expression assessed by in situ RNA hybridisation is key for this study. We tried to minimise errors in annotations by using systematic annotation processes based on the anatomical description and compared our description to existing patterns in Zfin. New patterns were systematically checked twice at the level of clone identity and by repeating template and probe for a second round of in situ hybridisation. To assess the significance of co-expression, p-values of the Bonferroni-corrected Pearson’s Chi-Squared-test were computed in R and Matlab, where the sample size was >5. Adjusted p-values less than 0.01 were considered as significant.
Quantitative RT-PCR
qPCR primers were designed for 10 candidate genes using the Probe Finder software (Version: 2.49) from Roche Applied Science website. The design process involves the automatic selection of an intron spanning assay, and the following primers were selected: ENSDARG00000007812 (Fw:gcttgcacttgtccaaactg Rv:tcttctttcccatacttgaacctc); ENSDARG00000016212 (Fw:catgaggattgaagtggttgtg Rv:agtccagggaggctcgtc); ENSDARG00000032369 (Fw:tggagatctagcagaaggagaatc Rv:tcaagttcaatctcatcgctgt); ENSDARG00000009899 (Fw:tccacaacttcaatgcgatg Rv:caatgggactccaaaggtgt); ENSDARG-00000037324 (Fw:gcgctacacagaaagaaacga Rv:agcctgggcctcactctaa); ENSDARG00000075565 (Fw:tccgctgtctggaaaactaga Rv:tgcttcgtggaagaacagg); ENSDARG00000016531 (Fw:aaacctatcttcagcacaagcag Rv: tgaaactgcactcaggacaag); ENSDARG00000018619 (Fw:cagtctggaggcgttttacac Rv:agcccgctgatctcaatct); ENSDARG00000013615 (Fw:tccacatggcttgaatggt; Rv:gccttctgtaggggagatca); ENSDARG00000076251 (Fw:tggccagaccctaaaatgaa Rv:aactccagtgcggtcagattand) and beta-actin (Fw:gtgcccatctacgagggtta Rv:tctcagctgtggtggtgaag). Reverse transcription was performed with 1 μg of total RNA extracted with Trizol from two distinct pools of 24 hpf zebrafish embryos using MMLV reverse transcriptase and Oligo(dT) primers (Promega), following manufacturer’s instructions. The final RT product was diluted 5 times in water. For each gene of interest, triplicates were performed and mRNA levels determined by real-time qPCR using the StepOne Plus device (Applied Biosystems). Briefly, 2 μL of RT served as templates in the PCR reaction consisting of Go Taq qPCR master mix (Promega) and 500 nM gene-specific primers in triplicates. Ct values from biological duplicates were averaged and expression levels normalised against beta-actin. Melting curve analyses were performed to confirm correct amplification. Correlation between RNAseq and qRT-PCR expression data was made by calculating the squared correlation coefficient R2 between the log2(dCt) and log2(FPKM+1).
Data access
RNAseq data are accessible under the GEO accession number GSE39703 and microarray under the accession number GSE39728. Data on InterPro domains, EST sequences, in situ pictures, annotations, microarray and RNAseq experiments as well as links to other database (Ensembl, Refseq, Unigen and Zfin) were all integrated in a user friendly web server freely accessible at http://cassandre.ka.fzk.de/ffdb/index.php. All expression patterns will be submitted to ZFin (http://zfin.org/). cDNA clones are available upon request.
Supplementary Material
Acknowledgements
We thank M. Soysal for Grid computing (Steinbuch Centre for Computing, Karlsruhe Institute of Technology). A. Gehrlein, C. Lederer, T. Walther for excellent technical support. We thank T. Dickmeis for critical reading of the manuscript. This work was supported by the Helmholtz Association, the European commission IP ZF-MODELS LSHG-CT-2003-503496, EUTRACC LSHG-Ct-2006-037445, IP ZF-Health FP7-Health-2009-242048 and NeuroXsys Health-F4-2009 No. 223262 and Erasys Bio BMBF KZ: 0315716.
Footnotes
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.05.006.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escriva H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Baltimore D. Differentiation requires continuous regulation. J. Cell Biol. 1991;112:781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Bauer Schaper H, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Van Slyke C, Westerfield M. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science (New York, NY) 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Eade KT, Fancher HA, Ridyard MS, Allan DW. Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS Genet. 2012;8:e1002501. doi: 10.1371/journal.pgen.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, Stiles CD. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J. Neurosci. 2009;29:11399–11408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA. Endoderm development: from patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science (New York, NY) 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, Chelala C, Croft D, Cros A, Cutts RJ, Di Genova A, Forbes S, Fujisawa T, Gadaleta E, Goodstein DM, Gundem G, Haggarty B, Haider S, Hall M, Harris T, Haw R, Hu S, Hubbard S, Hsu J, Iyer V, Jones P, Katayama T, Kinsella R, Kong L, Lawson D, Liang Y, Lopez-Bigas N, Luo J, Lush M, Mason J, Moreews F, Ndegwa N, Oakley D, Perez-Llamas C, Primig M, Rivkin E, Rosanoff S, Shepherd R, Simon R, Skarnes B, Smedley D, Sperling L, Spooner W, Stevenson P, Stone K, Teague J, Wang J, Whitty B, Wong DT, Wong-Erasmus M, Yao L, Youens-Clark K, Yung C, Zhang J, Kasprzyk A. BioMart Central Portal: an open database network for the biological community. Database (Oxford) 2011;2011 doi: 10.1093/database/bar041. bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E. Anthropogenie oder Entwickelungsgeschichte des Menschen. Engelmann; Leipzig: 1874. [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science (New York, NY) 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin C, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development (Cambridge, England) 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Karaulanov E, Knochel W, Niehrs C. Transcriptional regulation of BMP4 synexpression in transgenic Xenopus. EMBO J. 2004;23:844–856. doi: 10.1038/sj.emboj.7600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lemke G, Reber M. Retinotectal mapping: new insights from molecular genetics. Annu. Rev. Cell. Dev. Biol. 2005;21:551–580. doi: 10.1146/annurev.cellbio.20.022403.093702. [DOI] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell. 2012;22:1101–1108. doi: 10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ince-Dunn G, Ghosh A. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat. Rev. Neurosci. 2007;8:331–340. doi: 10.1038/nrn2118. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Mol. Biol. (Clifton, NJ) 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sambrook J. Molecular Cloning: A Laboratory Manual. third ed. Vol. 1. CSHL Press; 2001a. 2.94. [Google Scholar]
- Sambrook J. Molecular Cloning: A Laboratory Manual. third ed. Vol. 2. CSHL Press; 2001b. 10.37. [Google Scholar]
- Sobral D, Tassy O, Lemaire P. Highly divergent gene expression programs can lead to similar chordate larval body plans. Curr. Biol. CB. 2009;19:2014–2019. doi: 10.1016/j.cub.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted action of two dlx paralogs in sensory placode formation. Development (Cambridge, England) 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Stuermer CA. Retinotopic organization of the developing retinotectal projection in the zebrafish embryo. J. Neurosci.: Off. J. Soc. Neurosci. 1988;8:4513–4530. doi: 10.1523/JNEUROSCI.08-12-04513.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali R, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science (New York, NY) 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Baer KE. Entwicklungsgeschichte der Thiere, Beobachtung und Reflexion. Bornträger; Königsberg: 1828. [Google Scholar]
- Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev. Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science (New York, NY) 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Yang L, Kemadjou JR, Zinsmeister C, Bauer M, Legradi J, Muller F, Pankratz M, Jakel J, Strahle U. Transcriptional profiling reveals barcode-like toxicogenomic responses in the zebrafish embryo. Genome. Biol. 2007;8:R227. doi: 10.1186/gb-2007-8-10-r227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rastegar S, Strahle U. Regulatory interactions specifying Kolmer–Agduhr interneurons. Development (Cambridge, England) 2010;137:2713–2722. doi: 10.1242/dev.048470. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics (Oxford, England) 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.