Abstract
The human genome encodes a family of nine protein arginine methyltransferases (PRMT1-9), which members can catalyze three distinct types of methylation on arginine residues. Here, we identify two spliceosome-associated proteins – SAP145 and SAP49 – as PRMT9 binding partners, linking PRMT9 to U2snRNP maturation. We show that SAP145 is methylated by PRMT9 at arginine 508, which takes the form of monomethylated arginine (MMA) and symmetrically dimethylated arginine (SDMA). PRMT9 thus joins PRMT5 as the only mammalian enzymes capable of depositing the SDMA mark. Methylation of SAP145 on Arg508 generates a binding site for the Tudor domain of the Survival of Motor Neuron (SMN) protein, and RNA-seq analysis reveals gross splicing changes when PRMT9 levels are attenuated. These results identify PRMT9 as a non-histone methyltransferase that primes the U2snRNP for interaction with SMN.
INTRODUCTION
Protein arginine methylation is an abundant posttranslational modification, with about 0.5% of all arginine residues present in the methylated state in mouse embryonic fibroblasts1. Arginine methylation is enriched on RNA binding proteins2,3. Indeed, over 50% of the arginine methylation found in mammalian cells is concentrated on heterogeneous nuclear ribonucleoproteins (hnRNPs)4. In addition, a number of well-characterized methylation sites are found on histone tails5 and splicing factors6. Three distinct types of methylated arginine residues occur in mammalian cells. The most prevalent is omega-NG,NG-dimethylarginine7. In this case, two methyl groups are placed on one of the terminal nitrogen atoms of the guanidino group; this derivative is commonly referred to as asymmetric dimethylarginine (ADMA). Two other methylarginine derivatives occur; namely the symmetric dimethylated derivative (SDMA), where one methyl group is placed on each of the terminal guanidino nitrogens and the omega-monomethylated derivative (ω-MMA). These three types of arginine methylation are catalyzed by the nine members of the family of protein arginine methyltransferases (PRMTs)8. The PRMTs are classified according to their methylation products9. Type I, II and III are able to generate MMA. Type I enzymes include PRMT1, PRMT2, PRMT3, PRMT4/CARM1, PRMT6, and PRMT8, and they all perform a second methylation step to generate the ADMA mark. PRMT5 is the lone Type II enzyme described to-date, generating the SDMA mark. PRMT7 is a Type III enzyme and only generates a MMA mark10. PRMT9 has not yet been characterized, and it is the subject of this study.
PRMT9 has two distinguishing features: 1) Like PRMT7, it contains an ancestral amino acid sequence duplication that harbors a second putative AdoMet-binding motif, and 2) unique among the PRMTs, it has three N-terminal tetratricopeptide repeats (TPR). PRMT9 was identified as a potential human protein arginine methyltransferase at the same time as we described PRMT811. In that study, we noted the presence of a gene on chromosome 4q31 encoding a protein most similar to PRMT7 that was relatively well expressed in different tissues and designated it as PRMT911. In the past, the PRMT9 designation has also been used for the product of the human FBXO11 gene on chromosome 2p1612, although FBXO11 is unlikely to be a bona fide PRMT13. In some literature and protein databases, the gene on human chromosome 4q31 has previously also been referred to as PRMT10, although PRMT9 is the approved symbol and recommended gene name by the HUGO Gene Nomenclature Committee. The characterization of the PRMT9 protein (Q6P2P2 in the UniProt database) has been elusive, mainly because well-known PRMT substrates like histones and glycine-arginine rich (GAR) motif-containing proteins are not recognized (or poorly recognized) by the enzyme. Fortuitously, we found that PRMT9 can monomethylate and symmetrically dimethylate a protein that it interacts with, the spliceosome-associated protein, SAP145 (SF3B2). Thus, PRMT9 joins PRMT5 as the only mammalian Type II enzymes. SAP145 is a component of the U2 snRNP that is recruited to the branch region located near the 3′ splice site, and plays a critical role in the early stages of splicing. We were able to functionally link PRMT9 levels to the regulation of alternative splicing. Thus, we identified PRMT9 as a modulator of the SAP145/SAP49 protein complex that likely plays an important role in small nuclear ribonucleoprotein (snRNP) maturation in the cytoplasm.
RESULTS
PRMT9 identification and primary sequence features
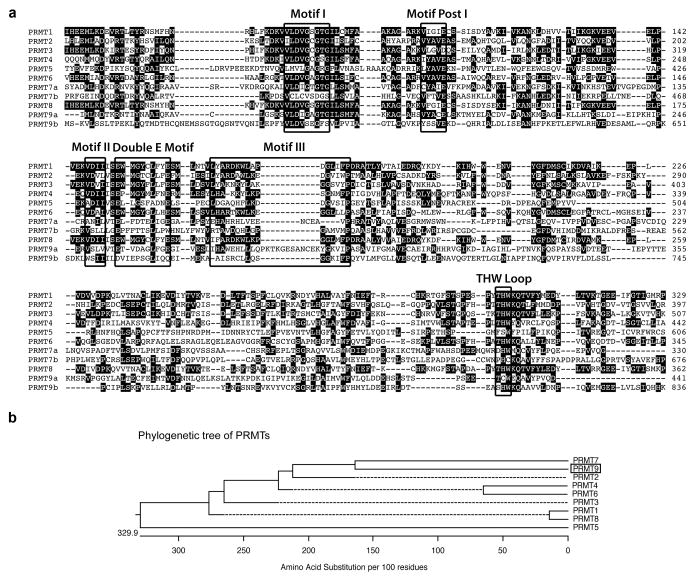
The gene encoding PRMT9 was identified a number of years ago11. A scan of the PRMT9 amino acid sequence for protein domains identified three tetratricopeptide repeats (TPRs) at its N-terminus (Supplementary Fig. 1). TPRs are helical features that often mediate protein–protein interactions14. In addition, like PRMT7, PRMT9 harbors two putative S-adenosylmethionine (AdoMet)-binding domains with a clear duplication and partial conservation of most of the six signature PRMT motifs present (Fig. 1a). A phylogenetic tree analysis of the encoded amino acid sequences of all nine members of the PRMT family reveals that it is most closely related to PRMT7 (Fig. 1b). The PRMT9 protein, with its defining TPR motifs and ancestrally duplicated sequence, is highly conserved among vertebrates, and is occasionally present in invertebrate animals, but has no orthologs in fungal species or prokaryotes (Supplementary Fig. 1).
Fig. 1. Amino acid sequence alignment of human PRMTs.
(a) The amino acid sequences from the catalytic domain of PRMTs are compared using ClustalW. The signature sequence motifs are boxed with black squares, including the motifs common to seven beta strand enzymes (Motif I, Motif Post I, Motif II, and Motif III) as well as the Double E Motif and THW Loop motifs that are specific to PRMTs. The number on the right indicates the positions of amino acid of individual PRMT, starting at the initiator methionine. Human PRMT sequences used included PRMT1: NP_001527.3; PRMT2: NP_996845.1; PRMT3: NP_005779.1; PRMT4: NP_954592.1; PRMT5: NP_006100.2; PRMT6: NP_060607.2; PRMT7: NP_061896.1; PRMT8: NP_062828.3; and PRMT9: NP_612373.2.
(b) A straight branches phylogenetic tree of all human PRMTs (shown in phenogram) was generated using ClustalW to demonstrate the evolutionary relationships among these enzymes. Protein sequence accession numbers used in the analysis are listed above.
PRMT9 associates with the SAP145 and SAP49 splicing factors
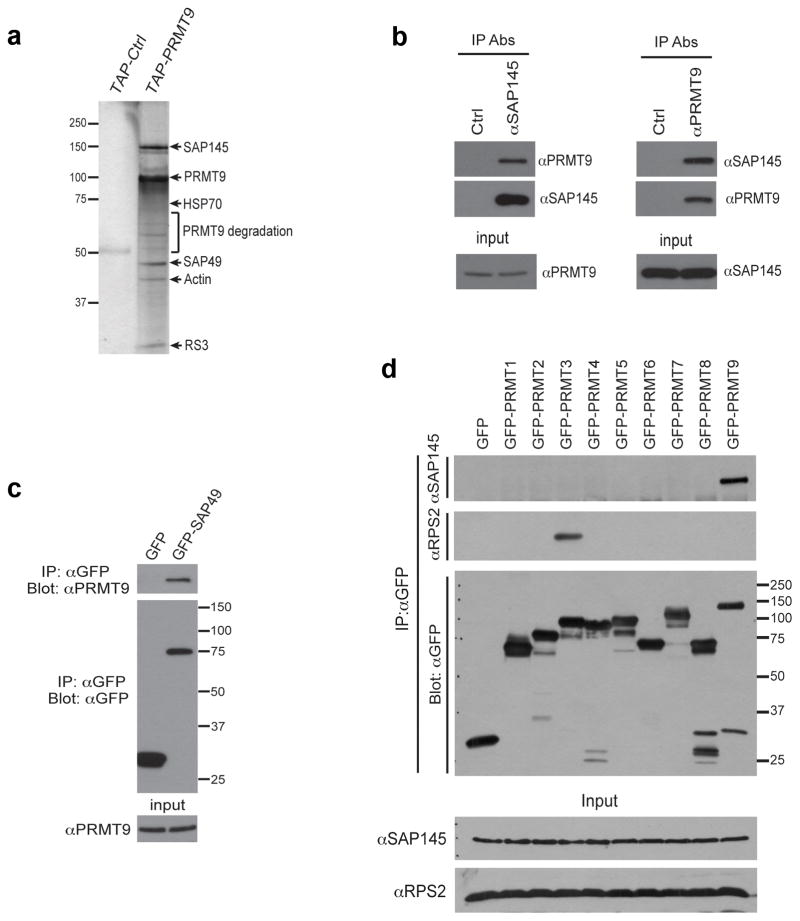
To help establish what biological processes PRMT9 may be associated with, we identified proteins that co-purified with PRMT9 expressed in HeLa cells. We performed tandem affinity purification of a TAP-tagged PRMT9 fusion, followed by tryptic digestion and mass spectrometry of the major interacting bands to identify the primary PRMT9 protein complex. The two major interacting proteins were identified as SAP145 and SAP49 (Fig. 2a). These splicing factors, also known respectively as SF3B2 and SF3B4, are tightly associated with each other15. We next developed a monoclonal antibody that specifically recognized the human PRMT9 protein (Supplementary Fig. 2a-f). Subcellular localization studies, using this antibody, revealed that PRMT9 is mainly a cytoplasmic protein (Supplementary Fig. 2d,e). Using this αPRMT9 antibody and antibodies against endogenous SAP145, we found that PRMT9 and SAP145 were reciprocally coimmunoprecipitated from HeLa cells (Fig. 2b). Similarly, GFP-tagged SAP49 coimmunoprecipitated with endogenous PRMT9 (immunoprecipiating antibodies to endogenous SAP49 are unavailable) (Fig. 2c). To determine whether the interaction between PRMT9 and SAP145 is specific for this arginine methyltransferase, we transfected the complete set of GFP-tagged PRMTs into HeLa cells and immunoprecipitated them. Only GFP-PRMT9 was able to coimmunoprecipitate SAP145 (Fig. 2d). We have previously shown that PRMT3 interacts strongly with its substrate, RPS216,17, and this interaction serves as a control here.
Fig. 2. PRMT9 interacts with SAP145 and SAP49.
(a) TAP-tag purification of PRMT9 protein complex from HeLa cells. HeLa cells were transiently transfected with either empty TAP-tag vector (TAP-Ctrl) or TAP-tag PRMT9 (TAP-PRMT9). A standard TAP procedure was applied. The eluted protein complex was separated by SDS-PAGE and silver-stained. The indicated gel slices were processed for protein identification using mass spectrometry. Actin, heat shock protein 70 (HSP70), and 40S Ribosomal Protein S3 (RS3) are likely non-specific interacting proteins.
(b) PRMT9 and SAP145 co-immunoprecipitate. Reciprocal Co-IP was performed in HeLa cells. Total cell lysates were immunoprecipitated with rabbit control IgG, αSAP145 antibody (left), and mouse control IgG, αPRMT9 antibody (right). The eluted protein samples were detected by western blotting with αPRMT9 and αSAP145 antibodies. The input samples were detected with αPRMT9 and αSAP145 antibodies, respectively.
(c) PRMT9 and SAP49 co-immunoprecipitate. HeLa cells were transiently transfected with either GFP control vector or GFP-SAP49 plasmids. Total cell lysates were immunoprecipitated with αGFP antibody. The eluted protein samples were detected by western blotting with αPRMT9 and αGFP antibodies. The input samples were detected with αPRMT9.
(d) Among all the PRMTs, only PRMT9 interacts with SAP145. HeLa cells were transiently transfected with control GFP vector and GFP-PRMTs (1 through 9). The total cell lysates were immunoprecipitated with αGFP antibody. The eluted protein samples were detected by western blotting with αSAP145, αRPS2 and αGFP antibodies. The input samples were detected with αSAP145 and αRPS2 antibodies.
Interaction of PRMT9 with SAP145
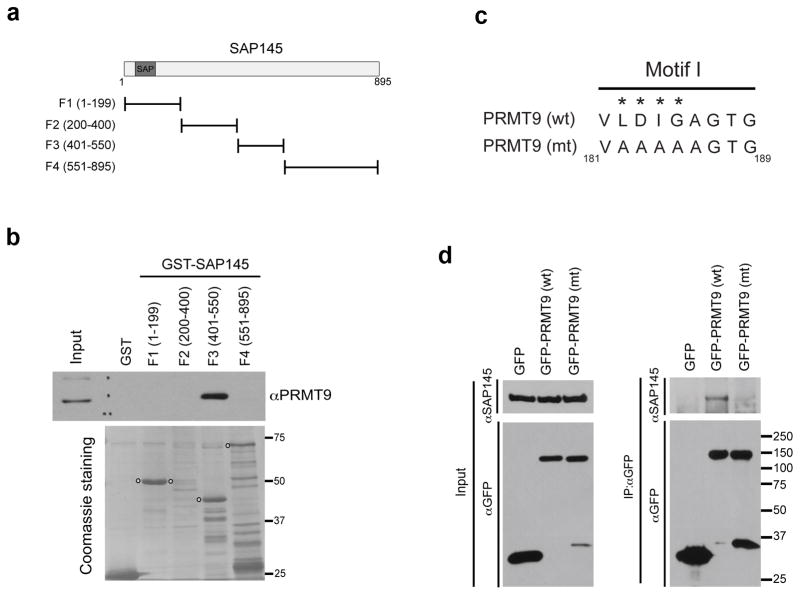
Next, we mapped the region of SAP145 that interacts with PRMT9. To do this, we broke the SAP145 protein into four roughly equal fragments (F1–4) and expressed them as GST fusion proteins (Fig. 3a), which we then used to perform a GST pull-down assay with HeLa cell lysates. Western blot analysis revealed that cellular PRMT9 interacts with fragment F3 of SAP145, which encodes for amino acids 401–550 (Fig. 3b). This fragment has no distinctive structural features that could help explain the specificity of PRMT9 for this region. We also tested whether the recombinant GST-SAP49 can pull-down endogenous PRMT9 using the similar experimental strategy. Interestingly, GST-SAP49 does not interact with PRMT9, suggesting that PRMT9 resides in SAP145/SAP49 complex through an interaction with SAP145 (Supplementary Fig. 3a). We then attempted to perform reciprocal mapping of this interaction to establish which region of PRMT9 associates with SAP145. To do this, we generated a series of flag-tagged PRMT9 deletion constructs (Supplementary Fig. 3b). We were particularly interested in the possibility that the TPR motifs would interact with SAP145. However, these motifs proved not to be sufficient for SAP145 binding. Indeed, none of the PRMT9 deletion constructs could interact with SAP145 (or SAP49) (Supplementary Fig. 3c), even though the full-length tagged enzyme retained its binding abilities. This suggests that complete structural integrity of PRMT9 is required to maintain the PRMT9/SAP145/SAP49 protein complex. Supporting this idea, we found that the catalytically inactive form of PRMT9 lost its ability to interact with SAP145 (Fig. 3c,d). Additionally, while SAP145 and SAP49 were associated with wild type GFP-PRMT9 expressed in HEK293 cells, neither splicing factor was found associated with a catalytically inactive GFP-PRMT9 similarly expressed (data not shown). These results suggest that the integrity of the AdoMet-binding domain of PRMT9 is critical for its interaction with SAP145, and that PRMT9 may release SAP145 after substrate methylation, once it takes on its AdoHcy-bound form. Thus, to test whether PRMT9 bound to its product inhibitor S-adenosylhomocysteine (AdoHcy) would also not interact with SAP145, we treated HeLa cells with adenosine-2,3-dialdehyde (AdOx), which results in elevated cellular AdoHcy levels18. We then performed a co-IP experiment to evaluate the impact of the AdoMet/AdoHcy switch on the PRMT9/SAP145 interaction. Interestingly, AdOx treatment did not affect the PRMT9/SAP145 interaction, indicating that the AdoHcy-bound form of PRMT9 still likely interacts with its substrate (Supplementary Fig. 3d).
Fig. 3. Mapping the interaction regions of PRMT9 and SAP145.
(a) A series of GST-fusion truncations of SAP145 were generated. The location of the SAP (SAF-A/B, Acinus and PIAS) domain is indicated.
(b) PRMT9 interacts with the amino acids 401–550 of SAP145. GST pull-down experiment was performed by incubating HeLa cell total lysates with purified GST or GST-tag SAP145 fragments described in (A). The pull-down samples were eluted and detected by western blotting using αPRMT9 antibody. The loading of the proteins was visualized by coomassie staining of the membrane. The open circles indicate the individual GST-tag SAP145 fragments.
(c) Generation of enzymatic mutant PRMT9. The amino acids (LDIG) located within the conserved Motif I of PRMT9 were mutated to AAAA, causing the loss of methyltransferase activity of enzyme. The numbers below indicate the location of the amino acids on the protein.
(d) Wild type, but not the enzymatic mutant, PRMT9 interacts with SAP145. Co-immunoprecipitation was performed in HeLa cells transiently transfected with GFP control vector, GFP-PRMT9 (wt) and GFP-PRMT9 (mt), as described in (c). Total cell lysates were immunoprecipitated with αGFP antibody. The eluted protein samples were detected by western blotting with αSAP145 and αGFP antibodies. The input samples were detected with αSAP145 and αGFP antibodies as well.
PRMT9 is a Type II PRMT that methylates SAP145
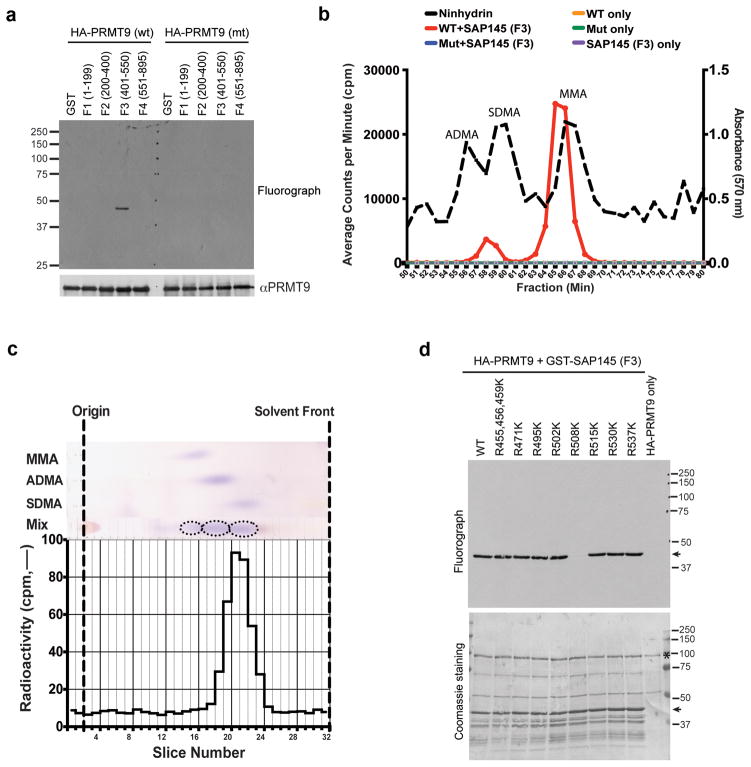
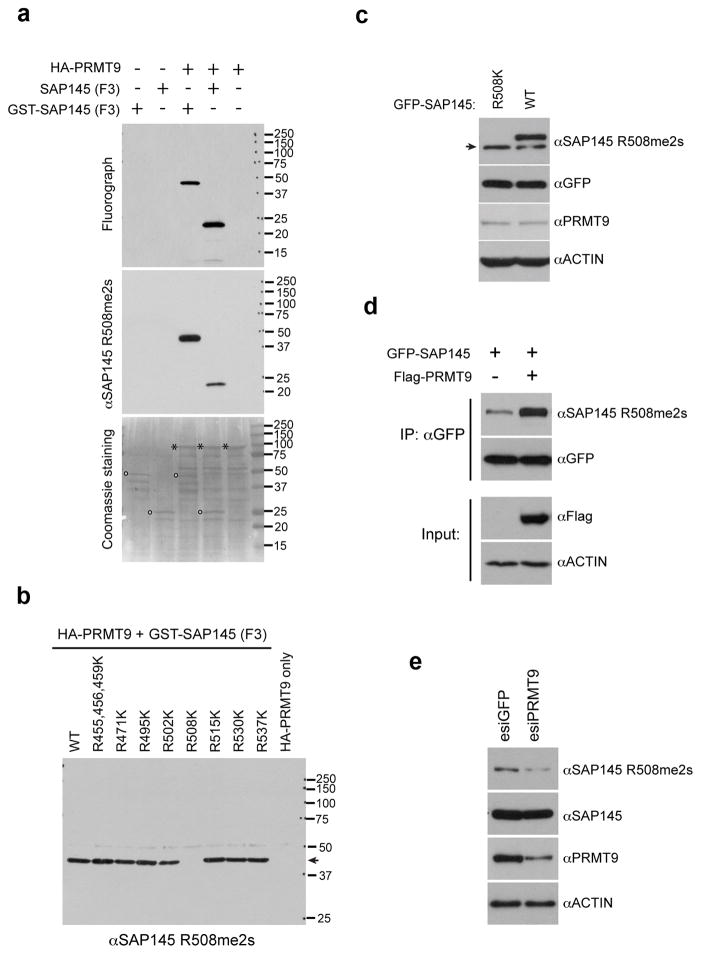
Using in vitro methylation assays with insect cell expressed HA-PRMT9 and the four fragments of SAP145 as potential substrates, we found that only the F3 fragment that physically interacted with PRMT9 (Fig. 3b) was also a good methyl-acceptor for the enzyme (Fig. 4a). The F2 fragment could not be expressed well, and thus cannot be excluded as a possible substrate. No methylation of F3 fragment was seen in a similarly expressed PRMT9 enzyme that was mutated in the AdoMet binding site (Fig. 4a). We find that PRMT9 has little or no activity on the typical substrates of other PRMTs including core histones or GAR motif-containing proteins (data not shown). To determine the methylated arginine products of PRMT9, the in vitro 3H-methylated F3 fragment of SAP145 was subjected to amino acid analysis. Mammalian expressed GFP-tagged PRMT9 was found to catalyze the formation of both MMA and SDMA only in the presence of the SAP145 fragment (Fig. 4b). No methylation was observed in control experiments with the PRMT9 fusion protein mutated in the AdoMet binding site, demonstrating that the observed activity is intrinsic to tagged PRMT9 and not to another enzyme that might be co-purifying with it (Fig. 4a,b). To confirm the formation of SDMA, the radioactive peak from the cation-exchange amino acid analysis column (corresponding to fractions 57–60) was subjected to thin layer chromatography under conditions that separated MMA, SDMA, and ADMA by their hydrophobic properties. We found that all of the radioactivity comigrated with SDMA (Fig. 4c), demonstrating that PRMT9 is a bona fide type II PRMT and joins PRMT5 as only the second enzyme of this type in mammals. A time course of in vitro methylation over a period of 20 hours showed a steady accumulation of both MMA and the final product, SDMA (Supplementary Fig. 4a,b).
Fig. 4. PRMT9 catalyzes symmetrical dimethylation of SAP145 at Arginine 508.
(a) PRMT9 methylates SAP145 fragment F3 (a.a. 401–550). The in vitro methylation was performed by incubating either wild type or enzymatic mutant recombinant HA-PRMT9 (purified from Sf21 cells) with GST or GST-tag SAP145 fragments (F1–F4, as described in Fig. 3a and b). The loading of PRMT9 was detected by western blotting using αHA antibody.
(b) PRMT9 symmetrically dimethylates SAP145 as detected by amino acid analysis. Amino acid analysis of in vitro methylation products from wild type and enzymatic mutant GFP-PRMT9 as enzymes and GST-SAP145 (401–550) fragment as substrate. Black dashed line indicates elution of nonradiolabeled standards. The radioactive peaks elute 1–2 min before the nonradiolabeled standards due to a tritium isotope effect39.
(c) PRMT9 symmetrically dimethylates SAP145 as detected by thin layer chromatography (TLC). SDMA fractions from cation-exchange chromatography of the in vitro 3H-methylation reaction were separated using thin layer chromatography (TLC). Fractions were spotted on a cellulose plate along with the addition of 5 nmol MMA, 15 nmol ADMA, and 5 nmol SDMA internal standards. Individual standards were also spotted in adjacent lanes to determine the migration distance of each methylated arginine derivative. The origin is indicated by fraction 2. The solvent front was run near the end of the plate to fraction 32. The plate was air-dried and each lane was subsequently sliced in 5 mm fractions and counted for three 30-min counting cycles. The radioactive peaks elute 1–2 min before the nonradiolabeled standards due to a tritium isotope effect39. This experiment was repeated 3 times and similar migration patterns were observed.
(d) PRMT9 methylates SAP145 at R508 in vitro. The in vitro methylation assay was performed by incubating recombinant HA-PRMT9 with a series of Arg to Lys (R to K) mutants of SAP145 fragment F3 (see Fig. 3a and b for description) for 1 h at 30° C. After exposure at −80 °C for 3 days, the membrane was stained with Coomassie blue to check the protein loading. Arrows indicate the positions of the substrates and stars indicate the positions of the recombinant HA-PRMT9.
To localize the site or sites of methylation by PRMT9 on SAP145, each of the ten arginine residues in the F3 fragment was replaced with a lysine residue. The F3 fragments containing lysine residues at nine of these sites were equally good methyl-acceptors. However, when R508 was mutated to lysine, methylation was greatly diminished, indicating the specificity of PRMT9 for this residue (Fig. 4d).
Importantly, PRMT9 is an active enzyme when purified from insect cell (Fig. 4a) and from mammalian cells (Supplementary Fig. 5a), but shows little or no activity when isolated as a GST fusion protein from bacteria (Supplementary Fig. 5b). We next tested the ability of a set of eight GST-PRMTs to methylate SAP145-F3. We found that PRMT6 could also readily methylate the F3 fragment, but that no methylation was found with the other enzymes under these conditions (Supplementary Fig. 5b). Furthermore, we tested the ability of Myc-PRMT5 (GST-PRMT5 is inactive and was thus not tested in Supplementary Fig. 5b) to methylate SAP145-F3, and we found that it was unable to do so (Figure S5C). Importantly, further analysis using the mutated SAP145-F3 fragments as substrates revealed that PRMT6 mainly methylates the R515 site (Supplementary Fig. 5d), so PRMT9 and PRMT6 modify distinct sites on SAP145. Thus, it appears that the SDMA mark at the R508 site of SAP145 is only deposited by PRMT9, and no other Type I or Type II PRMT modifies this site.
Reduced PRMT9 levels impact the methylation state of SAP145
To confirm the existence of the SAP145 R508me2s mark in cells, we generated a methyl-specific antibody against this site. Initially, we performed an in vitro characterization of this antibody and we showed that recombinant SAP145-F3 can only be recognized by the αSAP145 R508me2s antibody when it is subjected to prior methylation by PRMT9 (Fig. 5a). For immunoreactivity, the methyl-specific antibody also requires the presence of the R508 site. Thus, this antibody is indeed methyl-specific and site-specific (Fig. 5b). To establish whether this site is arginine methylated in cells, we transfected HeLa cells with wild type or mutant GFP-SAP145 (R508K), and performed Western analysis with the αSAP145 R508me2s methyl-specific antibody (Fig. 5c). The antibody clearly detected the wild-type form of the ectopically expressed fusion protein, but not the mutant form. The endogenous untagged SAP145 protein is also detected as a band that migrates slightly faster than the tagged form. When we repeated this experiment, but co-transfected cells with both SAP145 and PRMT9, we observed a dramatic increase in the levels of the SAP145 methylation as detected with the methyl-specific antibody (Fig. 5d). Endogenous SAP145 methylation cannot be further enhanced by PRMT9 overexpression (data not shown), suggesting that endogenous SAP145 is fully methylated at the R508 site. This is not a unique phenomena for a PRMT substrate, because we have previously shown that poly(A)-binding protein 1 (PABP1) is fully methylated by CARM1, and overexpression of CARM1 cannot further increase the PABP1 methylation level19. If this is the case, then the knockdown of PRMT9 should correlate well with the decrease in SAP145 methylation levels, and this is indeed what we observe; endoribonuclease-prepared siRNA (esiRNA) mediated knockdown of PRMT9 concomitantly reduces SAP145 methylation level as detected by αSAP145 R508me2s methyl-specific antibody (Fig. 5e).
Fig. 5. PRMT9 symmetrically methylates SAP145 at Arg 508 in vivo.
(a) SAP145 R508 methylation specific antibody detects in vitro methylated SAP145. The in vitro methylation assay was performed using recombinant HA-PRMT9 as enzyme, either GST-tag SAP145 fragment F3, or SAP145 fragment F3 without GST-tag (cleaved by PreScissionTM Protease) as substrates. The samples were run on a SDS-PAGE gel followed by exposure to x-ray film for 3 days. After exposure, the membrane was stripped and detected using SAP145 R508 methylation specific antibody (αSAP145 R508me2s) by western blotting. The membrane was then stained with Coomassie blue to check the protein loading. Open circles indicate the positions of the substrates and stars indicate the position of the recombinant HA-PRMT9.
(b) Arg to Lys mutation at R508 site of SAP145 abolishes the recognition of αSAP145 R508me2s antibody. The same membrane, described in Fig. 4 (d), was stripped and detected using αSAP145 R508me2s antibody by western blotting. The arrow indicates the position of specific signal detected by the antibody.
(c) Arg 508 of SAP145 is symmetrically dimethylated in the cells. HeLa cells were transiently transfected with either GFP-SAP145 or GFP-SAP145 (R508K). 30 μg of total cell lysates were subjected to western blotting detection with αSAP145 R508me2s, αGFP, αPRMT9 and αActin antibodies. The arrow indicates the endogenous SAP145 methylation signals.
(d) Overexpression of PRMT9 increases SAP145 R508 symmetrical dimethylation. HeLa cells were transfected with either GFP-SAP145 alone, or GFP-SAP145 and Flag-PRMT9. The total cell lysates were immunoprecipitated with αGFP antibody followed by western blotting with αSAP145 R508me2s and αGFP antibodies. The input samples were detected with αFlag and αActin antibodies.
(e) Knockdown of PRMT9 decreases SAP145 R508 symmetric dimethylation. HeLa cells were transiently transfected with either control esiGFP (esiRNA targeting GFP) or esiPRMT9 (esiRNA targeting PRMT9). 30 μg of total cell lysates were subjected to western blotting and detected with αSAP145 R508me2s, αSAP145, αPRMT9 and αActin antibodies.
The SMN interacts with methylated SAP145
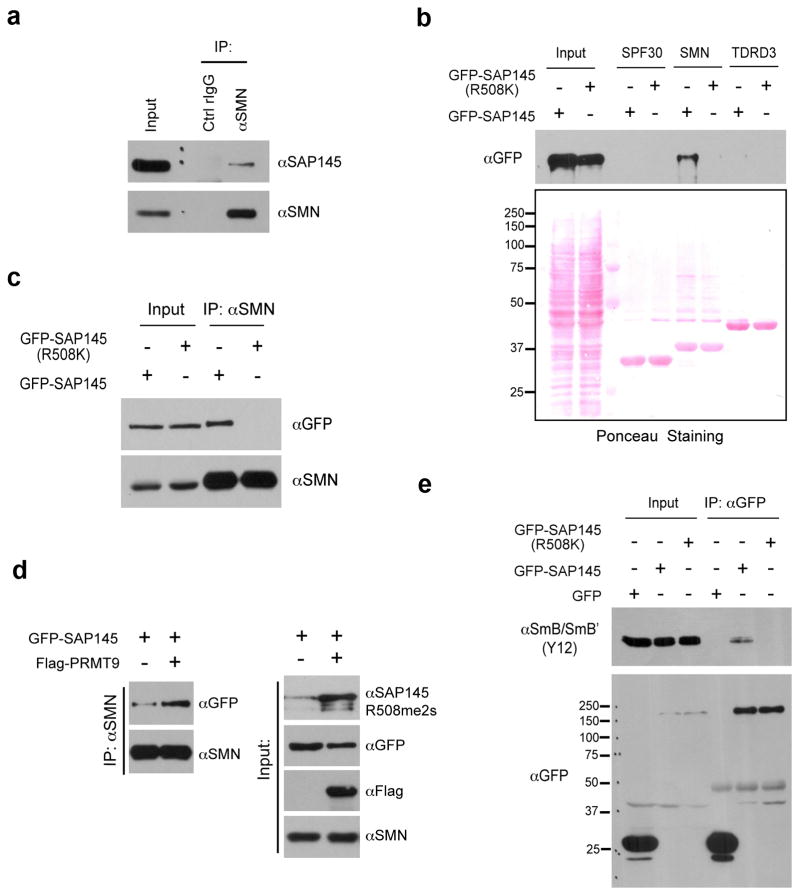
PRMT9 seems to be a rather specific enzyme, and even though many methylated proteins can be detected in HeLa cell extract with pan-MMA and pan-SDMA antibodies only SAP145 methylation is reduced when PRMT9 levels are lowered (Supplementary Fig. 6a). The SAP145 site (R508), which is the target of PRMT9 methylation, is highly conserved in vertebrates and invertebrates (Supplementary Fig. 6b), suggesting that it is a critical node of functional control, possibly through the regulation of protein-protein interactions. We first investigated whether methylation of this site regulated the interaction between SAP145 and SAP49. Overexpression of both PRMT9 and SAP145, which causes a dramatic increase in the R508me2s mark (Fig. 5d), does not alter the ability of SAP49 to interact with this protein complex (Supplementary Fig. 6c). Furthermore, disruption of R508 methylation site does not affect the assembly of the SAP145/SAP49/PRMT9 complex (Supplementary Fig. 6d), suggesting that other protein-protein interactions may be regulated by this mark. Indeed, arginine methylation motifs create docking sites for the recognition by Tudor domain containing proteins20. There are three Tudor domain-containing proteins that are well characterized as effector proteins for methyl-arginine marks, SPF30, SMN and TDRD35. SMN has been extensively studied and is involved in the assembly of ribonucleoproteins complexes, an important step for snRNPs maturation, which regulates mRNA splicing21. When we performed a co-IP experiment, we observed the endogenous interaction of SMN with SAP145 (Fig. 6a). To address whether the Tudor domain of the SMN protein mediates this interaction, we performed a GST pull down assay using the Tudor domains from three different proteins (SPF30, SMN and TDRD3). We incubated the GST-tagged recombinant proteins with HeLa cell lysates that were transiently transfected with either wild type GFP-SAP145 or GFP-SAP145 (R508K) mutant. After pull-down and Western analysis with αGFP antibody, we observed an interaction between the Tudor domain of SMN and ectopically expressed SAP145, but not with the methylation deficient SAP145 (R508K) (Fig. 6b). To further confirm this interaction in the cells, we performed coimmunoprecipitation of endogenous SMN with exogenous expressed GFP-SAP145 or GFP-SAP145 (R508K) mutant. Similar to what we observed in the pull-down experiment, endogenous SMN interacts with GFP-SAP145, but not the methylation deficient mutant (Fig. 6c). To address whether SMN-SAP145 interaction is regulated by PRMT9, we transfected HeLa cells either with GFP-SAP145 alone or with GFP-SAP145 and Flag-PRMT9, followed by a coimmunoprecipitation assay to assess the influence of PRMT9 overexpression on the SMN-SAP145 interaction. Overexpression of PRMT9 dramatically increases the levels of the GFP-SAP145 methylation as detected with the methyl-specific antibody (Fig. 6d, right panel). Concomitantly, the interaction between SMN and GFP-SAP145 was greatly enhanced (Fig. 6d, left panel). These results demonstrate that SAP145 R508 methylation by PRMT9 creates a recognition motif for Tudor domain of SMN, thus facilitates the assembly of the SMN-SAP145 complex. We next investigated whether the SMN-SAP145 interaction is important for U2 snRNP maturation. To do this we transfected HeLa cells with either GFP-SAP145 or GFP-SAP145 (R508K), and then performed a αGFP immunoprecipitation. An intact methylation site was required for SmB/B′ association with GFP-SAP145, strongly suggesting that the PRMT9 methylation site is critical for SMN-mediated Sm core assembly of the U2 snRNP (Fig. 6e).
Fig. 6. Methylation of Arginine 508 is required for SAP145-SMN interaction.
(a) SMN interacts with SAP145 in vivo. Endogenous Co-IP was performed by immunoprecipitating HeLa cell nuclear extracts using αSMN antibody, followed by western blotting detection using αSAP145 antibody.
(b) Tudor domain of SMN interacts with wild type but not R508K mutant form of SAP145. HeLa cell were transfected with either GFP-SAP145 or GFP-SAP145 (R508K). GST pull-down experiment was performed by incubating transfected cell lysates with recombinant GST-Tudor domains of SPF30, SMN and TDRD3. Eluted samples were detected by western blotting with αGFP antibody. Ponceau S staining displays the loading of total cell lysates and recombinant proteins
(c) SMN interacts with the wild type but not the R508K mutant form of SAP145. HeLa cells were transfected with either GFP-SAP145 or GFP-SAP145 (R508K). The nuclear extracts were immunoprecipitated with αSMN antibody. The eluted samples and input samples were detected with the indicated antibodies.
(d) Overexpression of PRMT9 enhances the interaction of SMN with GFP-SAP145. HeLa cells were transfected with the indicated plasmids. The nuclear extracts were immunoprecipitated with αSMN antibody. The eluted samples and input samples were detected with the indicated antibodies.
(e) An intact SAP145 R508 methylation site is required for U2 snRNP maturation. HeLa cells were transfected with either GFP-SAP145 or GFP-SAP145 (R508K). Cell lysates were immunoprecipitated with αGFP antibody. The eluted samples and input samples were detected with αSmB (Y12) and with αGFP antibodies.
PRMT9 is required for efficient alternative splicing
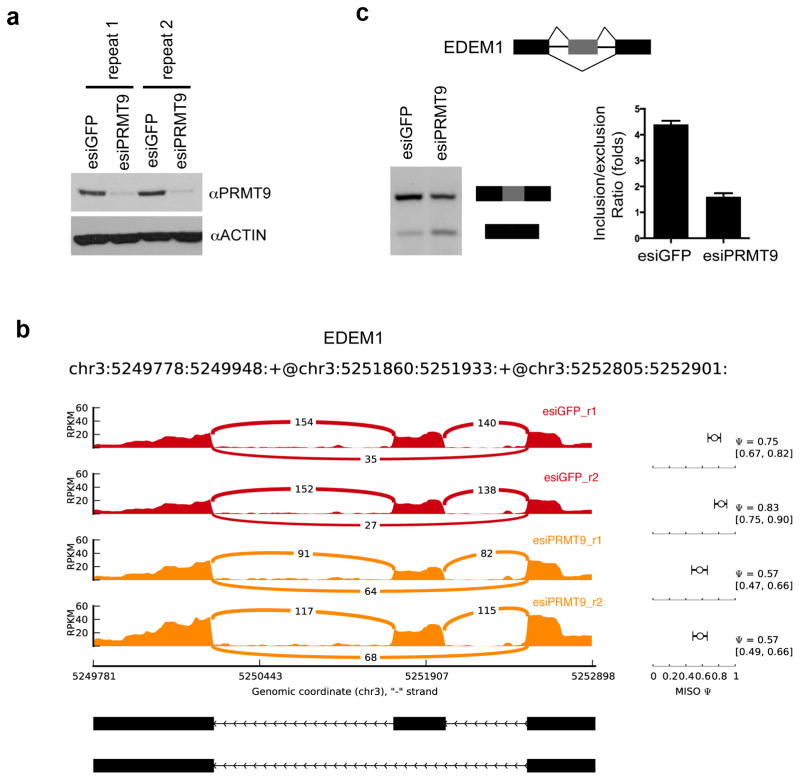
The robust and specific interaction between PRMT9 and SAP145/SAP49 (Fig. 2), the ability of PRMT9 to methylate SAP145 (Fig. 4), and the regulation of methylation dependent SMN-SAP145 interaction by PRMT9 (Fig. 6) strongly suggest that PRMT9 is involved in the regulation of mRNA splicing. To investigate this possibility, we extracted total RNA from control knockdown and esiRNA mediated PRMT9 knockdown HeLa cells and subjected them to RNA-seq analysis, particularly focusing on changes in alternatively spliced events. To reduce the experimental variation, two sets of biological replicates were prepared. Western blot analysis demonstrated the efficient knockdown of endogenous PRMT9 in both replicate samples (Fig. 7a). For RNA-seq, we performed 75-nt paired-end sequencing and achieved about 220 million and 120 million unique mapped reads for control knockdown and PRMT9 knockdown, respectively. We used mixture-of-isoforms (MISO) model 2 to compute and quantify alternative splicing events. We were able to identify five types of alternative splicing events, which were significantly altered. These events included 34 skipped exons (SE), 13 retained introns (RI), 9 alternative 3′ splice sites (A3SS), 9 alternative 5′ splice sites (A5SS) and 3 mutually exclusive exons (MXE) (Supplemental Table 1). One example of the MISO output landscape is displayed in Fig. 7b, showing RNA-seq read coverage across EDEM1 exon 8 to exon 10 from esiGFP control and esiPRMT9 knockdown HeLa cells. To validate the alternative splicing called by MISO, RT-PCR was performed using the primers located within the flanking exons of the EDEM1 gene. Knockdown of PRMT9 using esiRNA reduces the inclusion/exclusion ratio to about 2.5-fold (Fig. 7c). At another gene loci (NGLY1), knockdown of PRMT9 expression increases the inclusion/exclusion ratio to about 2.5-fold (Supplementary Fig. 7a,b). It should be noted that esiRNA mediated knockdown of PRMT9 only causes about a 50% decrease in SAP145 R508 methylation level (Fig. 5e). A small amount of enzyme is still capable of carrying out substantial methylation. This may explain why we can only capture a small group of altered splicing events. Total removal of PRMT9 may have more profound effects on alternative splicing. Thus, PRMT9 is impacting alternative splicing in a number of different ways, possibly through its ability to methylate SAP145 and promote the interaction between SAP145 and SMN.
Fig. 7. PRMT9 regulates alternative splicing.
(a) Two sets of biological replicated samples for RNA-seq. HeLa cells were transiently transfected with esiGFP control or esiPRMT9. The total RNA was extracted 72 hours after transfection. Western blotting confirms the knockdown of PRMT9 protein using αPRMT9 and αActin antibodies.
(b) RNA-seq read coverage across EDEM1 exon 8 to exon 10 from esiGFP control and esiPRMT9 knockdown HeLa cells. MISO (Ψ) values and 95% confidence intervals were shown at right.
(c) Validation of exon exclusion of EDEM1 exon 9 in response to PRMT9 knockdown. RT-PCR experiments were performed using RNA extracted from esiGFP control and esiPRMT9 transfected HeLa cells with primers located on exon 8 and exon 10 (left panel). The PCR products were quantified by densitometric analysis. Inclusion vs exclusion ratio was calculated. Error bars represent standard deviation calculated from three independent experiments (right panel).
DISCUSSION
We have demonstrated that PRMT9 is a Type II enzyme using both amino acid analysis approaches and methyl-specific antibodies that recognize the SDMA mark on SAP145. It is unclear how many additional substrates PRMT9 might have, but it is likely not as promiscuous as PRMT5, which methylates a broad swath of GAR motif-containing proteins. PRMT5 is the founding member of the Type II class of PRMTs, and clearly the dominant enzyme in this class. When cell lysates are subjected to Western blot analysis using pan-SDMA antibodies, the majority of the bands detected in wild-type cells are lost in PRMT5 knockdown cell lines1. Quantification of the different types of arginine methylation across a large number of tissue and cell types have estimated the ratios to be roughly 1500:3:2:1 for Arg:ADMA:MMA:SDMA1,7,22. Measurements of cellular SDMA levels using PRMT5 knockout cells have yet to be performed, but this would establish what proportion of the SDMA is deposited by PRMT5 and PRMT9, respectively. Although PRMT5 and PRMT9 are both Type II enzymes, they do not display redundancy. In in vitro methylation assays these two PRMTs do not recognize each other’s substrates (Supplementary Fig. 5c & data not shown). In addition, subsets of SDMA-specific antibodies selectively recognize PRMT5 or PRMT9 substrates as validated using the respective knockdown cell extracts (data not shown). The extended seven amino acid (CFKRKYL) PRMT9 methylation motif in SAP145 (Supplementary Fig. 6b) is not found in any other protein. However, when we reduce the search to a five amino acid motif (FKRKY) we did identify eight human proteins that could possibly be substrates for this newly characterized enzyme, and they will be tested in the near future.
After we designated the gene on human chromosome 4q31 as PRMT911, this designation was also used for the FBXO11 gene on human chromosome 2p1612. FBXO11 does not contain the signature amino acid sequence motifs of the PRMT family and the description of its activity as a protein arginine methyltransferase likely was the result of contamination of the FLAG-tagged protein with PRMT512. No methyltransferase activity has been observed with the nematode homolog of FBXO1113. It has been clearly shown that the monoclonal M2 antibody used to pull down FLAG-tagged proteins will also bring down endogenous PRMT5, thus contaminating any preparation of a FLAG-tagged protein9,23. Due to the usage of PRMT9 for both of these gene products, PRMT10 has also been used to designate the 4q31 product. However, PRMT9 is now the approved symbol and recommended gene name by the HUGO Gene Nomenclature Committee for the PRMT characterized in this work.
Classic studies in the arginine methylation field first implicated this posttranslational modification in the regulation of RNA metabolism3,4. More recently, large-scale proteomic analysis of arginine methylated proteins revealed the identity of many of these RNA binding proteins 24,25. Both Type I and Type II PRMTs are involved in the regulation of splicing. The Type I enzyme, CARM1, enhances exon skipping in an enzyme-dependent manner6,26. It regulates splicing by methylating a number of splicing factors (CA150, SAP49, SmB and U1C), likely at transcriptionally active sites to which it is recruited as a transcriptional coactivator6. PRMT6 is another transcriptional coregulator that does not exclusively modulate gene expression, but also impacts alternative splicing patterns27. Indeed, transcriptome analysis of PRMT6 knockdown cells revealed a number of gene-specific exon pattern changes, including altered exon retention and exon skipping28. The Type II enzyme, PRMT5, functions as a transcriptional repressor in the nucleus by symmetrically methylating histone H4R3 and H3R829. In the cytoplasm, PRMT5 is primarily found in the 20S methylosome complex, along with MEP50, pICIn, and various Sm proteins (SmB/B′, D1–3, E, F, & G)30–32. In this complex, the Sm proteins are symmetrically dimethylated by PRMT5. In turn, the methylated Sm proteins interact with the Tudor domain of SMN33, which helps load the Sm proteins onto spliceosomal snRNAs (U1, U2, U4, U5, U11 & U12)21,34. Thus, PRMT5 plays a critical role in spliceosomal U snRNP (small nuclear ribonucleoprotein particle) maturation. PRMT5 has a rather central role in pre-mRNA splicing, and it is assumed that pleiotropic splicing defects will be observed upon PRMT5 loss; this is indeed the case. The conditional removal of PRMT5 in mouse neural progenitor cells results in defects in both constitutive splicing and alternative splicing35. Furthermore, in Arabidopsis, PRMT5 is essential for proper pre-mRNA splicing36,37.
The splicing factors SAP145 and SAP49 are tightly associated with each other15. SAP145 and SAP49 are dedicated components of the nuclear U2snRNP. The region of SAP145 that interacts with SAP49 has been mapped, and it lies adjacent to the R508me2s site38. Methylation of R508 does not impact the SAP145/SAP49 interaction (Supplementary Fig. 6). The PRMT9/SAP145/SAP49 complex likely forms in the cytoplasm, where PRMT9 deposits the SAP145R508me2s mark for subsequent recognition by SMN. In mammals, newly synthesized snRNAs are first exported to the cytoplasm, where they undergo maturation (partial snRNP complex assembly) before being imported back into the nucleus to form catalytically active snRNPs21. As described above, PRMT5 plays a critical role in the biogenesis of the Sm-class of spliceosomal snRNPs. PRMT9 seems to add another level of maturation, which is specific for U2 snRNA. A fairly unique feature of PRMT9 is its ability to strongly interact with its substrate, SAP145 (Fig. 2d); most PRMTs, with the exception of PRMT3, do not do this. The PRMT9/SAP145 interaction occurs in the cytoplasm. However, copious quantities of SAP145 are observed in the nucleus38. Thus, it is likely that some signal, perhaps a posttranslational modification, regulates the dissociation of PRMT9 and SAP145 to facilitate nuclear translocation of the maturing U2 snRNP. Indeed, we observe reduced SmB/B′ association with mutant SAP145 (GFP-SAP145R508K), which is indicative of a U2 snRNP maturation defect. This flaw in U2 snRNP maturation could modify the fidelity of branch point recognition and alter a subset of alternative splicing events.
Methods
Plasmids and antibodies
Human PRMT9 cDNA (NM_138364.2) was subcloned into a pGEX-6p-1 vector (GE Healthcare Life Sciences), a pEGFP-C1 vector (Clontech), and a 3XFlag vector (Invitrogen). NTAP-PRMT9 used for tandem affinity purification was generated by cloning human PRMT9 cDNA into pCeMM NTAP(GS) vector (EUROSCARF). Human SAP49 cDNA, SAP145 cDNA and its truncated cDNAs were cloned into pEGFP-C1 vector. Both wildtype and mutant HA-PRMT9 were cloned into pBacPAK vector (Clontech) for insect cell expression. Rabbit anti-GFP polyclonal antibody was purchased from Invitrogen (Cat# A6455, used for IP, 2ul/IP). Mouse anti-HA monoclonal antibody was purchased from Covance (Cat# MMS-101P-200, used for Western blot, 1:2000 dilution). Mouse anti-Flag M2 monoclonal antibody was purchased from Sigma (Cat# A2220, used for IP, 20ul/IP). Rabbit anti-SAP145 polyclonal antibody was purchased from Novus Biologicals (Cat# NB100-79848 used for Western blot, 1:3000 dilution and IP, 5ul/IP). Rabbit anti-SAP49 polyclonal antibody was purchased from Abcam (Cat# ab66659, used for Western blot, 1:1000 dilution). Mouse anti-PRMT9 monoclonal antibody was raised, using the recombinant GST-PRMT9 as antigen, by the Antibody Facility Core at M.D. Anderson Cancer Center (used for Western blot, 1:1000 dilution and IP, 10ul/IP). The SAP145R508me2s antibody was identified in a screen for SDMA-specific antibodies (used for Western blot, 1:1000 dilution). Cell Signaling Technology generated this panel of antibodies using the XXXXRme2sXXRme2sXXXXRme2sXXXRme2sXXXX peptide as an antigen. The resulting antibodies were screened on PRMT5 knockout cells, and the majority displayed a loss of immunoreactivity. However, two of these antibodies, BL8244 and BL8245, strongly recognized a band of roughly 150 kDa, which is not lost in PRMT5 KO cells. When we tested these two antibodies on PRMT9 knockdown cells the immunoreactivity of this 150 kDa band was reduced. We thus went on to characterize these antibodies in vitro (Fig. 5). Uncropped immunoblots for each figure are shown in Supplementary Fig. 8.
Tandem affinity purification
HEK293 cells (from ATCC) were transfected with NTAP-PRMT9. A total of 1X108 cells were used for tandem affinity purification. In brief, cells were first lysed in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 125 mM NaCl, 5% glycerol, 0.2% NP-40, 1.5 mM MgCl2, 25 mM NaF, 1 mM Na3VO4 and protease inhibitors. After centrifugation at maximum speed for 10 minutes, the supernatant was incubated with rabbit-IgG Sepharose (GE Healthcare Life Sciences) at 4° C for 4 hours. The bound beads were first washed with lysis buffer and then TEV-protease cleavage buffer (10 mM Tris-HCl (pH 7.5), 100 mM NaCl and 0.2% NP-40). The bound protein complexes were eluted by addition of 50 μg TEV protease (4° C, overnight). TEV-protease cleavage products were then incubated with Streptavidin agarose (Millipore) at 4° C for 2 hours. The bound proteins were heated at 100 °C for 5 minutes in SDS sample buffer and analyzed by SDS-PAGE followed by either silver staining or SYPRO Ruby staining. After comparison with the control samples, differentially expressed bands were cut from the gel, and proteins were identified by LC-MS/MS using the Protein and Metabolite Analysis Core Facility at UT Austin.
Baculovirus expression and protein purification
HA-PRMT9 (both wild type and the enzymatic mutant as described below) in pBacPAK8 vector were cotransfected with linearized Baculovirus DNA (BD Biosciences) using FuGENE 6 transfection reagent (Roche). The expression and purification of recombinant proteins were followed according to the instruction manual of Baculovirus Expression Vector System (6th Edition, May 1999), except that HA conjugated beads and HA peptide (Sigma) were used to bind and elute the recombinant proteins, respectively. Purified proteins were then dialyzed in 1X TBS buffer (50 mM Tris-Cl, pH 7.5. 150 mM NaCl) and used for the in vitro methylation assay.
Immunoprecipitation
HeLa cells were washed with ice-cold PBS and lysed with 1 ml of co-immunoprecipitation buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2) with protease inhibitor cocktail. After sonication, insoluble materials were removed by centrifugation at maximum speed for 10 minutes. Whole cell lysates were incubated with 2 μg of immunoprecipitation antibody overnight. After incubation with Protein A/G agarose beads, the bound proteins were eluted and analyzed using Western blots.
Immunofluorescence
HeLa cells (from ATCC) were grown on glass coverslips to desired confluence before fixation. Cells were rinsed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. After blocking with 20% newborn calf serum, cells were incubated with monoclonal PRMT9 antibody (1:1000) at 4° C overnight. Cells were then stained with secondary antibody and DAPI sequentially. The coverslips were then sealed and examined under a NIKON Eclipse E800 fluorescent microscope.
GST-Pull down
GST and different recombinant proteins were expressed and purified from E.coli. The individual protein was incubated with HeLa cell lysates at 4° C overnight. The protein complex was then incubated with Glutathione Sepharose 4B resin (GE Healthcare Life Sciences) for 1 hour at 4° C. The eluted samples were loaded on SDS-PAGE gel and detected by Western blots using the indicated antibodies. Equal loading of the recombinant proteins was visualized by either Coomassie blue staining or ponceau staining.
Nuclear and cytoplasmic extraction
Cytoplasmic and nuclear fractions of HeLa cells were extracted following the protocol as described in the instructions from NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotech).
esiRNA Knockdown
esiRNA used for transient knockdown of PRMT9 was designed using the website-http://cluster-12.mpi-cbg.de/cgi-bin/riddle/search. The preparation of dsRNA was followed according to the instructions of MEGAscript® RNAi Kit (Invitrogen). dsRNA was then digested with RNase III to produce a blender of 18–30 bp siRNA mixture. After purification, the esiRNA was dissolved in H2O and tested for knockdown efficiency.
Site-directed mutagenesis
Human PRMT9 was mutated to create a catalytically inactive mutant by introducing quadruple mutations (L182A, D183A, I184A, G185A) in the conserved motif I in both the HA-PRMT9 and GFP-PRMT9 plasmid vectors. Mutant primers (IDT, San Diego, CA) were as follows: forward 5′-TGT TTG GGG TCC AAA AGT GTT GCG GCC GCT GCG GCA GGA ACT GGA ATA CTA AGC-3′ and reverse primer 5′-GCT TAG TAT TCC AGT TCC TGC CGC AGC GGC CGC AAC ACT TTT GGA CCC CAA ACA-3′ (Tm = 86.7°C). PCR reactions were set up according to the QuikChange XL Site-Directed Mutagenesis kit (Agilent Technologies, Inc.). GST-SAP145 Arginine to Lysine series mutants were generated using the same strategy. All mutations were confirmed on positive colonies by DNA sequencing.
In vitro methylation assay
In vitro methylation reactions were carried out in 30 μl of phosphate-buffered saline (pH = 7.4.) containing 0.5–1.0 μg of substrate, 3 μg of recombinant enzymes and 0.42 μM S-adenosyl-l-[methyl-3H]methionine (79 Ci/mmol from a 7.5 μM stock solution; PerkinElmer Life Sciences). The reaction was incubated at 30° C for 1 h and then separated on SDS-PAGE, transferred to a PVDF membrane, treated with En3Hance™ (PerkinElmer Life Sciences), and exposed to film for 1–3 days at −80° C.
High-resolution cation exchange chromatography
In vitro methylation reactions were set up with approximately 2 μg of enzyme protein, approximately 5 μg of substrate, and 0.7 μM S-adenosyl-L-[methyl-3H]methionine (PerkinElmer Life Sciences, 75–85 Ci/mmol, 0.55 mCi/ml in 10 mM H2SO4/EtOH (9:1, v/v)) in a final reaction volume of 60 μl. The reactions were incubated and rotated for 1 h, 5 h, or 20 h at 37° C, in a reaction buffer of 50 mM potassium HEPES, 10 mM NaCl and 1 mM DTT, pH 8.0. To quench the reaction, a final concentration of 12.5% trichloroacetic acid was added with 20 μg of the carrier protein bovine serum albumin. After a 30 min incubation at room temperature, the precipitated proteins were centrifuged at 4000 x g for 30 min and washed with cold acetone to remove excess [3H]AdoMet, and the pellet was air dried before acid hydrolysis. 50 μl of 6 N HCl was added to each dried pellet, and 200 μl of 6 N HCl was added to a Waters PicoTag Chamber. Each sample was acid hydrolyzed in vacuo at 110° C for 20 h. After acid hydrolysis, the pellets were dried using vacuum centrifugation. To prepare samples for high resolution cation-exchange chromatography, the hydrolyzed samples were resuspended in 50 μl of water and mixed with sodium citrate buffer (0.2 M in Na+, pH 2.2) and standards including 1 μmol each of ω-MMA (acetate salt, Sigma, M7033), SDMA (di(p-hydroxyazobenzene)-p′-sulfonate salt, Sigma, D0390) and ADMA (hydrochloride salt, Sigma, D4268). Samples were loaded onto sulfonated polystyrene resin and eluted using 0.35 M sodium citrate running buffer, pH 5.25, at 55 °C. 1 ml fractions were collected and the positions of the internal standards were determined using a ninhydrin assay using only 50 μl of each fraction. 950 μl of the fraction was assayed for radioactivity by the addition of 10 ml of Safety Solve Scintillation cocktail (Research Products International, 111177) and data was reported as the average of three 5-min counting cycles on a Beckman LS6500 Liquid Scintillation instrument.
Thin-layer chromatography
Fraction samples were spotted on a cellulose plate along with the addition of 5 nmol MMA, 15 nmol ADMA, and 5 nmol SDMA of internal standards as described above. Individual standards were also spotted in adjacent lanes to determine the migration distance of each methylated arginine derivative. The origin is indicated by fraction 2. The solvent front was run near the end of the plate to fraction 32. The plate was air-dried and each lane was subsequently sliced in 5 mm fractions and counted using liquid scintillation counting for three 30-min counting cycles. This experiment was repeated 3 times and similar migration patterns were observed.
Protein sequence alignment using ClustalW
The parameters for the alignment using ClustalW were set as following: Gap Penalty: 10, Gap Length Penalty: 0.2, Delay Divergent Seqs (%) 30, Protein Weight Matrix: Gonnet Series for multiple alignment parameters. For pairwise alignment, Gap Penalty: 10, Gap Length 0.1, Protein Weight Matrix: Gonnet 250.
Deep sequencing and alternative splicing analysis by MISO
Paired-end strand-specific RNA-seq reads were aligned to the human genome (hg19) using TopHat 2.0.91. We performed 75-nt paired-end sequencing and achieved about 220 million and 120 million unique mapped reads for control knockdown and PRMT9 knockdown, respectively. Based on the alignment result from TopHat, mixture-of-isoforms (miso) model2 was used to compute ‘percentage spliced in’ (PSI or Ψ) values to quantify alternative splicing, indicating the fraction of a gene’s mRNAs that include the alternative region. Five types of alternative splicing were analyzed, including skipped exons (SE), alternative 3′/5′ splice sites (A3SS, A5SS), mutually exclusive exons (MXE) and retained intron (RI). Then, change of PSI (Ψ) and Bayes factor (BF > 10) were used to identify significantly altered splicing in PRMT9 knockdown.
Supplementary Material
Acknowledgments
We thank Maria Person for mass spectrometry analysis at the “Protein and Metabolite Analysis Facility” (UT Austin) supported by RP110782 (CPRIT). MT Bedford is supported by an NIH grant (DK062248); Steven Clarke is supported by an NIH grant (GM026020). Andrea Hadjikyriacou and Cecilia Zurita-Lopez were supported by an NIH training grant (GM007185). MT Bedford is a cofounder of EpiCypher.
Footnotes
Competing Financial interests: M.T.B is a cofounder of EpiCypher and a consultant for Cell Signaling Technologies.
Accession codes: Deep-sequencing data can be accessed with GEO accession number GSE63953.
CONTRIBUTIONS
Y.Y., S.G.C. and M.T.B. conceived the project. Y.Y., A.H., S.G., D.K., C.Z-L. and R.K., carried out the experiments. Z.X. and W.L. analyzed the RNA-Seq data. A.G. developed the methyl-specific antibodies. M.T.B. wrote the manuscript and supervised the work. Y.Y. and S.G.C. provided valuable support with the writing.
References
- 1.Dhar S, et al. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci Rep. 2013;3:1311. doi: 10.1038/srep01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74:969–976. doi: 10.1016/0006-291x(77)91613-8. [DOI] [PubMed] [Google Scholar]
- 5.Gayatri S, Bedford MT. Readers of histone methylarginine marks. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Paik WK, Kim S. Natural occurrence of various methylated amino acid derivatives. John Wiley & sons; 1980. [Google Scholar]
- 8.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 9.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. The Journal of biological chemistry. 2013;288:37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 12.Cook JR, et al. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun. 2006;342:472–481. doi: 10.1016/j.bbrc.2006.01.167. [DOI] [PubMed] [Google Scholar]
- 13.Fielenbach N, et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 16.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 18.Chen DH, Wu KT, Hung CJ, Hsieh M, Li C. Effects of adenosine dialdehyde treatment on in vitro and in vivo stable protein methylation in HeLa cells. Journal of biochemistry. 2004;136:371–376. doi: 10.1093/jb/mvh131. [DOI] [PubMed] [Google Scholar]
- 19.Zeng H, et al. A TR-FRET-based functional assay for screening activators of CARM1. Chembiochem : a European journal of chemical biology. 2013;14:827–835. doi: 10.1002/cbic.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 21.Matera AG, Wang Z. A day in the life of the spliceosome. Nature reviews. Molecular cell biology. 2014;15 doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka M. Epsilon-N-methylated lysine and guanidine-N-methylated arginine of proteins. 3. Presence and distribution in nature and mammals. Seikagaku. 1972;44:364–370. [PubMed] [Google Scholar]
- 23.Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 24.Guo A, et al. Immunoaffinity Enrichment and Mass Spectrometry Analysis of Protein Methylation. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.O113.027870. O113.027870 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlmann T, et al. A method for large-scale identification of protein arginine methylation. Mol Cell Proteomics. 2012;11:1489–1499. doi: 10.1074/mcp.M112.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 27.Harrison MJ, Tang YH, Dowhan DH. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 2010;38:2201–2216. doi: 10.1093/nar/gkp1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowhan DH, et al. Protein arginine methyltransferase 6-dependent gene expression and splicing: association with breast cancer outcomes. Endocrine-related cancer. 2012;19:509–526. doi: 10.1530/ERC-12-0100. [DOI] [PubMed] [Google Scholar]
- 29.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friesen WJ, et al. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem. 2002;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- 32.Meister G, et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 33.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 34.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. The EMBO journal. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezzi M, et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes & development. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng X, et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez SE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 38.Terada Y, Yasuda Y. Human immunodeficiency virus type 1 Vpr induces G2 checkpoint activation by interacting with the splicing factor SAP145. Molecular and cellular biology. 2006;26:8149–8158. doi: 10.1128/MCB.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.