Abstract
Double-strand breaks (DSBs) threaten chromosome integrity. The most accurate repair of DSBs is by homologous recombination (HR), catalyzed by recombination proteins such as Rad51. Three papers (Fasching et al., 2015; Kaur et al., 2015; Tang et al., 2015) now reveal the role of three of these proteins in budding yeast: Sgs1 (BLM homolog), Top3 (TOPIIIα homolog) and Rmi1. They demonstrate several steps where all three proteins act together, and find additional functions of the Top3-Rmi1 subcomplex that are critical for the completion of meiosis.
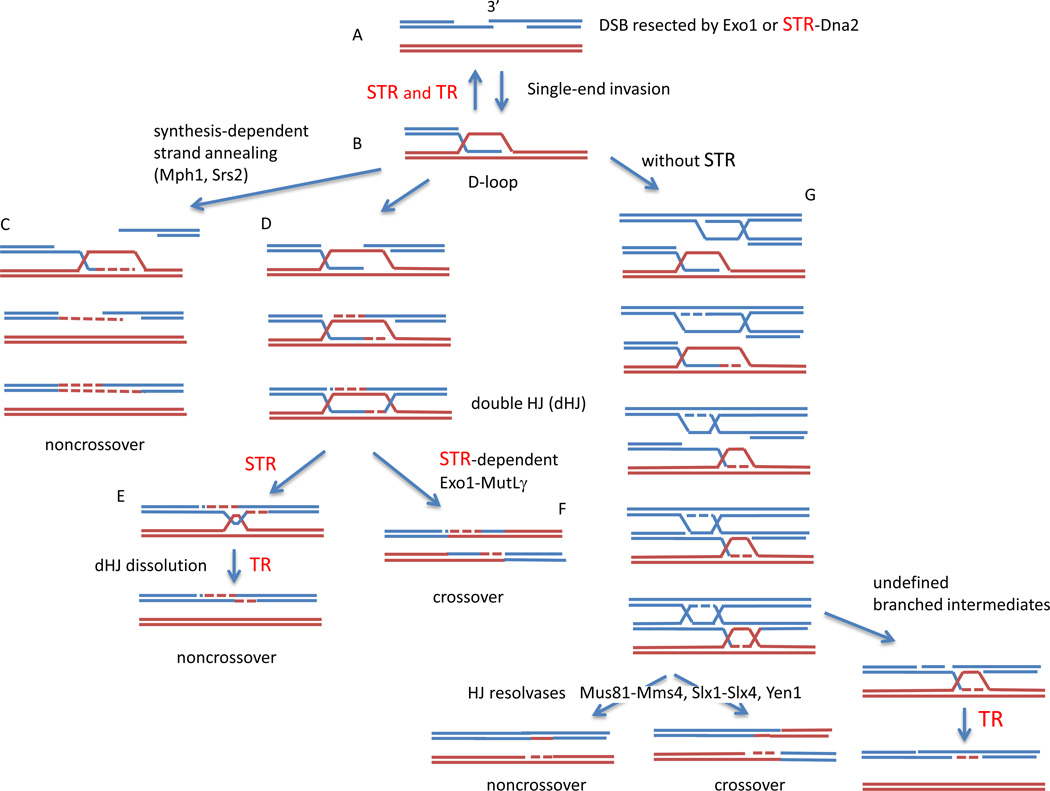
Double-strand breaks (DSBs) arise both spontaneously during DNA replication and from programmed expression of site-specific nucleases. Following the creation of a DSB, the broken ends are first resected to expose 3’-ended single-strand DNA (Fig. 1A), which organizes the assembly of a Rad51 nucleoprotein filament. The Rad51 filament is capable of searching the entire genome to locate a region of homology that can be used to patch up the broken chromosome. Homology can be located on a sister chromatid, on a homologous chromosome or in some ectopic location. Rad51 then pries open the intact double-stranded template to allow strand invasion and the formation of a three-stranded displacement or D-loop in which the single-stranded broken end base-pairs with its complementary strand of the intact duplex. (Fig. 1B) At this point the cell has several alternatives. Repair can proceed through a synthesis-dependent strand annealing (SDSA) pathway that copies the template to seal the break without an accompanying crossover (Fig. 1C). Alternatively, repair can proceed through the formation of a branched intermediate known as a double Holliday junction (dHJ) that can be cut apart by resolving enzymes (HJ resolvases) to produce crossovers between the homologs (Fig. 1D, F). Such crossovers are potentially disadvantageous in mitotic cells where an exchange between homologous chromosomes can lead to loss of heterozygosity; but in meiotic cells crossovers are necessary to generate the tension between paired homologs to assure proper disjunction of chromosomes at the first meiotic division. To avoid a crossover outcome, dHJs can also be dissolved by unwinding until a single pair of crossing-strands in a hemicatenane are removed by a topoisomerase (Fig. 1E). Also, not all strand invasions are productive, especially if the invading strand encounters a homologous sequence containing mismatches; in such cases D-loops can be dismantled to allow the broken end to search for other homologous sequences. The Sgs1-Top3-Rmi1 (STR) protein complex is involved in virtually every step along these pathways, and in mammals the orthologous mammalian complex, BTRR, involving the BLM helicase, TOPIIIα, RMI1 and an additional RMI2, protein appears to play similar roles.
Figure 1.
Roles of Sgs1, Top3 and Rmi1 in homologous recombination. A composite of steps in mitotic and meiotic recombination are shown, with the key steps requiring Sgs1-Top3-Rmi1 (STR) or Top3-Rmi1 alone (TR) shown in red. A. A double-strand break (DSB) is resected to yield 3’ ended single-strand DNA (ssDNA) tails either by the exonuclease Exo1 or by a helicase/endonuclease complex involving STR and Dna2. B. The ssDNA forms a nucleoprotein filament with Rad51 and engages in a search for homology, leading to single-end invasion and the formation of a D-loop. As shown by Fasching et al (2015) in this issue, D-loop formation can be reversed in two ways: by STR or by TR alone. C. The D-loop can be extended and the newly synthesized strand displaced, leading to DSB repair by synthesis-dependent strand annealing that yields noncrossover outcomes. D. The D-loop can be extended and result in an intermediate containing a fully ligated double Holliday junction (dHJ). The dHJ can be resolved into a noncrossover by dissolution, a process requiring STR to unwind and migrate the branched HJs and then TR to remove the remaining hemicatenane (E). Alternatively, the dHJ can be cleaved by Holliday junction resolvase. Here, the Exo1-MutLα complex that carries out this step in meiosis is shown (F). G. In the absence of STR, recombination in meiosis is greatly altered, with the appearance of multi-chromatid joint molecules (mcJMs) involving more than two of the 4 chromatids present at meiotic prophase (here three chromatids are illustrated). These mcJMs can be largely resolved by three structure-specific nucleases (Mus81-Mms4, Slx1-Slx4, or Yen1) into either crossover or noncrossover outcomes. However a minority of the intermediates formed without Sgs1 cannot be taken apart without the action of TR.
Sgs1 and BLM are members of the RecQ family of 3’ to 5’ helicases that can unwind and displace single stranded DNA from its complementary partner. People lacking BLM exhibit Bloom’s syndrome, a disease marked by a predisposition to cancer and a high level of genome instability. Top3 and its human TOPIIIα homolog are single-strand DNA cleaving enzymes that can relax supercoiled DNA, although their principal activity is in dismantling interconnected DNA molecules (Bocquet et al., 2014; Cejka et al., 2010b; Cejka et al., 2012).
As noted above, the budding yeast STR proteins play many different roles in the completion of DSB repair. In one of its guises, STR associates with the Dna2 endonuclease to promote the 5’ to 3’ resection of the DSB ends to generate long 3’-ended single-strand ends that promote Rad51 assembly and homologous recombination (Cejka et al., 2010a; Zhu et al., 2008). STR is not required for this process, as there is a parallel activity catalyzed by the Exo1 exonuclease. In mammals, BLM and Top3α, along with Rmi1 and Rmi2 promote analogous resection in mammalian cells, but in addition BLM can also act in a second pathway with EXO1 (Nimonkar et al., 2011).
STR is also implicated in the next step, when the single-stranded DNA within the Rad51 filament invades and forms sufficient base pairs with a homologous template to form a D-loop (Fig. 1B). Here STR can act to reverse this reaction, especially when the ssDNA pairs with a complementary strand containing several mismatches (Spell and Jinks-Robertson, 2004). STR also discourages annealing between two slightly mismatched ssDNA strands in the process of single-strand annealing (SSA) (Spell and Jinks-Robertson, 2004; Sugawara et al., 2004). As we will see later, removing the D-loop can occur in two ways, one requiring only Sgs1 and one needing only Top3-Rmi1(TR).
Once a D-loop has formed, repair can proceed via two major pathways: SDSA or a dHJ process. In mitotic cells Sgs1 does not appear to act in channeling repair towards SDSA, but two other 3’ to 5’ helicases, Mph1 and Srs2, are active at this step (Ira et al., 2003; Prakash et al., 2009). The dHJ pathway results in a fully ligated pair of Holliday junctions that must be resolved before chromosome segregation. In mitotic cells, where crossovers between homologous chromosomes might result in loss of heterozygosity, most dHJs are “dissolved” – producing noncrossovers (Fig. 1E) - rather than acted on by several Holliday junctions that can cleave the HJs to yield crossovers. The dissolution of dHJs requires STR in two steps, first to unwind and migrate the strands until a single hemicatenane remains and then to remove this last interconnection (Wu and Hickson, 2003). Thus, deletion of any of the STR proteins leads to a marked increase in crossovers in mitotic yeast cells during repair of a site-specific DSB. Most likely the dramatic increase in crossovers seen between sister chromatids in humans with Bloom’s syndrome (lacking BLM) reflects an analogous absence of dHJ dissolving activity.
In meiosis, the basic mitotic repair machinery becomes overlaid with a number of proteins that serve to ensure that as many as half of the recombination events, initiated by DSBs created by the Spo11 enzyme, will culminate in crossovers between homologous chromosomes. These exchanges are of course important in generating genetic diversity among germ cells but they also serve the critical function creating the necessary interconnections between chromatids that assure proper chromosome segregation. Many of these steps also counteract or modify STR function. First, STR doesn’t seem to play much of a role in promoting the much more limited 5’ to 3’ resection of DSB ends; only Exo1 seems to drive this process (Zakharyevich et al., 2010). Second, the dissolving of dHJs is blocked by the “ZMM” proteins (Lynn et al., 2007), which include the Msh4-Msh5 proteins that can bind to and apparently stabilize dHJs and thus prevent STR from dissolving them. If STR isn’t important for resection in meiosis and is thwarted in dHJ dissolution, one might think that removing STR from meiotic cells would have little consequence. But in fact the absence of STR prevents orderly progression through the normal pathways and creates novel meiotic phenotypes that dramatically change how intermediates of recombination are formed and processed.
The level of DSB formation at meiotic hotspots is sufficiently high to be able to identify and follow the kinetics of formation of a number of key molecular intermediates in meiotic recombination, including single-end invasion (i.e. the formation of a D-loop), the formation of dHJ intermediates and the appearance of both noncrossover and crossover outcomes (Oh et al., 2009). A detailed examination of the kinetics of repair revealed that noncrossovers appear earlier than crossovers and that the major pathway for resolution of the dHJs involves a noncatalytic function of Exo1 and the Msh2-Msh3 (MutLγ) mismatch repair proteins, rather than any of the 3 identified HJ resolvases: Mus81-Mms4, Slx1-Slx4, or Yen1 (Zakharyevich et al., 2010; Zakharyevich et al., 2012). Surprisingly, although crossovers appear in the absence of Sgs1, they no longer require Exo1-MutLγ.
In the absence of Sgs1, there are dramatic changes in the pattern of molecular intermediates (Jessop et al., 2006; Oh et al., 2007). Normally joint molecules (JMs) form between two of the 4 chromatids, predominantly between nonsister DNA molecules; but in the absence of Sgs1, there are much more complex JMs, involving 3 and sometimes all 4 chromatids (Fig. 1G). These results suggest that Sgs1 prevents these promiscuous strand invasions, possibly by reversing D-loop formation at one DSB end. The rejection of strand invasion is reminiscent of STR’s mitotic role in rejecting heteroduplex DNA formed during strand invasion between mismatched substrates, but in meiosis these rejections occur between identical sequences. Instead it would seem that Sgs1 is needed to assure that both ends of a DSB engage the same homologous target. A similar role for Sgs1 has been suggested in establishing a Recombination Execution Checkpoint in mitotic cells that delays recombination when the two ends of a DSB engage different partners (Jain et al., 2009).
Another surprising result that emerged from studying meiosis in the absence of Sgs1 is that the appearance of noncrossovers (NCOs) no longer precedes the advent of crossovers as seen in wild type meiotic cells. The coincident and late appearance of both NCOs and COs suggests they could arise by alternative resolution of a dHJ and that the normally predominant SDSA and dHJ dissolution pathways that lead to NCOs are absent. Consistent with these findings, the major dHJ resolution pathway, using Exo1 and MutLγ, is absent; instead crossovers – and noncrossovers – depend on the three other HJ resolvases, primarily Mus81-Eme1 and Slx1-Slx4, with Yen1 playing some sort of backup role just prior to the first meiotic division (De Muyt et al., 2012; Jessop and Lichten, 2008; Oh et al., 2008). Why the normal ZMM crossover pathway deploying Exo1-MutLγ cannot deal with the multiple JMs is still unclear. Perhaps the ZMM proteins and its associated resolvase cannot find the right DNA conformations to function when there are 3 or 4 chromatids engaged in a complex intermediate; perhaps Sgs1 plays a more direct role in creating the specific geometry for ZMM proteins to act.
The two new papers from the Hunter and Lichten labs build on this foundation and uncover an unanticipated role for a subcomplex of Top3 and Rmi1 (TR), independent of Sgs1 (Kaur et al., 2015; Tang et al., 2015). These studies show that inactivating either Top3 or Rmi1 in meiosis leads to the same dramatic appearance of JMs containing 3 or 4 chromatids as seen for the absence of Sgs1. However, whereas meiosis in the absence of Sgs1 is surprisingly complete, with quite good spore viability, the absence of either Top3 or Rmi1 results in very poor viability and the persistence of some JMs. Thus, the actions of the three HJ resolvases are insufficient to remove all the complications arising in the absence of STR. Some branched structures remain that prevent normal chromosome segregation. Thus the TR complex has an unexpected additional role in resolving branched molecules that apparently escaped the attention of the 3 HJ resolvases. These results may also account for the original characterization of Sgs1 mutations as suppressors of the slow growth of top3Δ: in mitotic cells there must also be some Sgs1-dependent (and possibly TR-dependent) branched DNA structures that require TR for their removal.
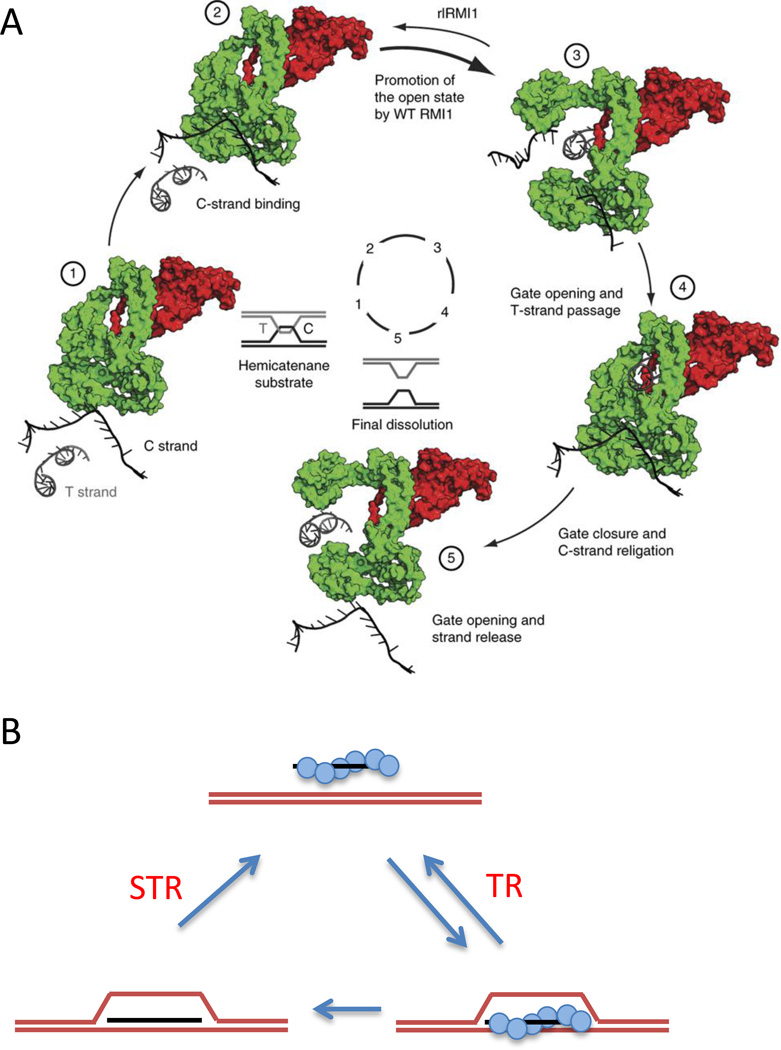
The exact nature of the refractory structures in meiosis remains to be elucidated but we can infer from the elegant biochemistry that has been carried out on Top3-Rmi1 that it must involve its ability to remove single-strand interconnections between different chromatids. Top3 belongs to a superfamily of enzymes that can cleave one DNA strand, which remains covalently attached via tyrosine, and pass though the other; but it differs from Top1 or bacterial homologs in that it is not efficient in relaxing supercoiled DNA by a series of rotations of the transiently broken strand. Instead, Top3 acts preferentially on structures that have some single-stranded DNA character, such as hemicatenanes. Recently Nicolas Thoma’s lab, in collaboration with those of Steven Kowlaczykowski, Peter Cejka and Ian Hickson, have provided a detailed model of the steps in this process, based on a high-resolution X-ray crystallographic study of the human TOPIIIα-RMI1 complex (Fig. 2A (Bocquet et al., 2014). TOPIIIα-RMI1 binds and cleaves the C (cut) strand and opens up to accommodate the binding of the T (transfer) strand and then closes again after religating the C strand, accomplishing strand passage. Mammalian TOPIIIα and budding yeast Top3 resemble prokaryotic relaxases in overall structure, but they lack a distinctive loop that has been implicated in the decatenation process. However, in the eukaryotic Top3 enzymes this loop is provided by Rmi1 (Fig. 2A). The presence of Rmi1 inhibits Top3’s supercoil relaxing activity and markedly increases its decatenation activity that is key in dissolving dHJs and, as shown below, in dismantling D-loops.
Figure 2.
Roles of STR and TR in reversing the formation of D-loops. A. Mechanism of strand passage carried out by mammalian TOPIIIα-RMI1 as illustrated by (Bocquet et al., 2014). TopIIIα cleaves the C strand and undergoes a conformational change that allows the transfer strand (T) to pass through, after which the C strand is re-ligated and the gate closes, with release of the C strand. This action is stimulated by a loop of RMI1 that is part of the active site. Figure reused with permission from Bocquet et al., NSMB 2014 Figure 5. B. Rad51 (blue circles) coating single-stranded DNA (ssDNA) facilitates strand invasion and the formation of a D-loop in the presence of the ssDNA binding protein complex, RPA, and Rad54. When the D-loop is protein-free, Sgs1 alone, or STR, can take apart the D-loop, but Sgs1 alone cannot dismantle the protein-bound. This protein-bound form can be taken apart by the Top3-Rmi1 complex acting alone.
The reversal of D-loops is the subject of the paper in this issue from Heyer’s lab in collaboration with Cejka and Kowalczykowski (Fasching et al., 2015). Artificial D-loops can be created by in vitro recombination, either bound to RPA or to Rad51 and its associated chromatin remodeler, Rad54, after which they can be purified to be protein-free. In keeping with previous results, Sgs1 by itself can dismantle a protein-free D-loop, but it fails to act on protein-bound structures (Fig. 2B). In contrast, yeast TR (and STR) can take apart protein-bound D-loops, through its strand passage activity. But, surprisingly, yeast TR won’t act on protein-free D-loops. In fact, yeast TR is quite fastidious, it will not work on D-loops created with human RPA or human Rad51 or Rad54.
(Fasching et al., 2015) also investigated the D-loop activity of the human BTRR and TRR complexes and found that they have similar activities but are much less picky about the species origin of either RPA or Rad54. Moreover, human TRR will dismantle protein-free D-loops. It will be interesting to see how mutations of TRR affect the resolution of meiotic chromosomes in mouse models to see if some of these differences will be reflected in their in vivo phenotypes. Currently little is known about how TOPIIIα mutants affect meiotic recombination and chromosome segregation, but the absence of BLM appears to reflect many of the defects seen for sgs1Δ in yeast (Holloway et al., 2010). There is no obvious defect in the early steps of recombination but there are aberrant chromosome pairings that are reminiscent of the multichromatid JMs seen in yeast.
The biochemical studies of Top3-Rmi1 reveal a strand-passage and decatenation mechanism that can explain why TR is required for the removal of some meiotic intermediates that are left behind in the absence of Sgs1, even though there are three HJ resolvases present. But whether these intermediates are extended D-loops or some other branched structure remains to be determined. Further experiments should soon get this STRaight.
Acknowledgments
JEH is supported by grants from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID, et al. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat Struct Mol Biol. 2014;21:261–268. doi: 10.1038/nsmb.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010a;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010b;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: a mechanism for disentangling chromosomes. Mol Cell. 2012;47:886–896. doi: 10.1016/j.molcel.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer W-D. Top3-Rmi1 dissolve Rad51-mediated D-loops by a toposiomerase-based mechanism. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Molecular cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, DeMuyt A, Lichten M. Top3-Rmi1 DNA single-stranded decatenase is integral to the formation and resolution of meiotic recombination intermediates. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Jessop L, Lao JP, Allers T, Lichten M, Hunter N. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods in molecular biology. 2009;557:209–234. doi: 10.1007/978-1-59745-527-5_14. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM Ortholog, Sgs1, Prevents Aberrant Crossing-over by Suppressing Formation of Multichromatid Joint Molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci U S A. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wu MKY, Zhang R, Hunter N. Pervasive and essential roles of Top3-Rmi1 decatenase orchestrate recombination iasnd facilitate chromosome segregation in meiosis. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]