Abstract
Budding yeast Slx4 interacts with the Rad1–Rad10 endonuclease that is involved in nucleotide excision repair (NER), homologous recombination (HR) and single–strand annealing (SSA). We previously showed that Slx4 is dispensable for NER but is essential for SSA. Slx4 is phosphorylated by the Mec1 and Tel1 kinases after DNA damage on at least six Ser/Thr residues, and mutation of all six residues to Ala reduces the efficiency of SSA. In this study, we further investigated the role of Slx4 phosphorylation in SSA, specifically in regulating cleavage of 3′ non–homologous (NH) DNA tails by Rad1-Rad10 during SSA and HR. Slx4 became phosphorylated after induction of a single double–strand break (DSB) during SSA and dephosphorylation coincided approximately with completion of repair. Slx4 is recruited to 3′ NH tails during DSB repair, but this does not require phosphorylation of Slx4. However, we identified specific damage-dependent Mec1/Tel1 site of Slx4 phosphorylation, Thr 113, that is required for efficient cleavage of NH tails by Rad1–Rad10. Consistent with these data, deletion of both Mec1 and Tel1 severely reduces the efficiency of NH DNA tail cleavage during HR. These data show that phosphorylation of Slx4 by Mec1 and Tel1 plays an important role in facilitating NH DNA tail cleavage during HR.
Keywords: Slx4, Rad1, SSA, Mec1, Tel1, homologous recombination
1. Introduction
Homologous recombination (HR) is an important mechanism for repairing DNA double-strand breaks (DSBs). Resection of a DSB creates a 3′ single–stranded tail that can search for and pair with an intact homologous donor sequence, which is used as a template for repairing the break (reviewed in [1,2]). Before an invading end can prime new DNA synthesis to copy the donor template, any non-homologous sequence at the 3′ end must first be removed. Repair is then completed by gap filling and end ligation. In budding yeast, 3′ non–homologous (NH) tails are removed by the Rad1–Rad10 structure-specific endonuclease, while XPF and ERCC1, the orthologues of these proteins, perform similar functions in mammalian cells [3–6].
3′ NH tails also arise during single-strand annealing (SSA), a sub-pathway of HR that can be used to repair DSBs flanked by repeated sequences (Fig. 1A) [5]. After resection of the ends of the DSB, Rad52 and Rad59 promote the annealing of homologous sequences, resulting in an intermediate that has two non–homologous 3′–ended tails [7,8]. Work from several labs showed that Rad1–Rad10 cleaves these 3′ single–strand tails, providing a 3′ hydroxyl for filling in by DNA synthesis followed by ligation to complete repair [9–14]. This results in deletion of one copy of the repeat plus the sequence located between the repeats (Fig. 1A). In either gene conversion or SSA, tail removal by Rad1–Rad10 also requires the Msh2–Msh3 mismatch recognition complex [14–18] but it is not entirely clear why. The available data suggest that Msh2–Msh3 may recognize and stabilize the 3′ tails at the dsDNA–ssDNA junction, somehow facilitating their cleavage. Rad59 and Srs2 have also been implicated in 3′ non–homologous tail removal [14,19].
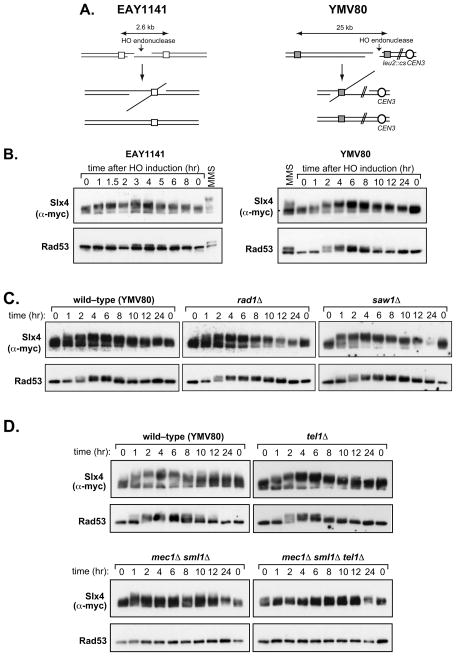
Figure 1. Phosphorylation of Slx4 during repair of a HO–induced DSB by SSA.
(a) Schematic diagram of SSA strains used in this study. (b) Strains GTY14 (EAY1141 Slx4-13Myc) and GTY27 (YMV80 Slx4-13Myc) were grown in YEP-lactate medium overnight, and HO expression was induced by the addition of galactose. Aliquots were taken at the indicated times after addition of galactose and analysed by Western blotting for Slx4 and Rad53. (c) Western blot analysis of Slx4 and Rad53 hyperphosphorylation in YMV80 Slx4-13Myc strains in which RAD1 (GTY42) or SAW1 (GTY44) were deleted. (d) Western blot analysis of Slx4 and Rad53 hyperphosphorylation in YMV80 Slx4-13Myc strains in which MEC1 (GTY37), TEL1 (GTY38), or both MEC1 and TEL1 (GTY39) were deleted.
The requirement for Rad1–Rad10 during gene conversion depends on the number and length of non–homologous tails. When both ends of a DSB have tails 30 nucleotides or longer, Rad1–Rad10 is essential for gene conversion [14–16,20]. However, when only one non–homologous tail arises Rad1–Rad10 is less critical [16,20,21]. When 3′ tails are less than 10 nucleotides long, Rad1 and Msh2–Msh3 are not required for gene conversion or SSA, and instead they are removed by the proof–reading activity of DNA polymerase δ [14]. Rad1–Rad10 is required for removal of all tails longer than 30 nucleotides whereas Msh2–Msh3 is not required in SSA when the repeat sequences are longer than 1kb [15].
Slx4, a protein with multiple functions in the DNA damage response, was recently identified as a Rad1–Rad10 interacting factor that is required for SSA but not for NER [22–24]. Human SLX4 also interacts with XPF–ERCC1 [25–27]. Although Slx4 is essential for SSA in yeast, the precise step that is controlled by Slx4 is unclear and it is not known if Slx4 is required for Rad1 activity during SSA. It is unlikely that the function of Slx4 in SSA is to recruit Rad1–Rad10 to NH tails; this is mediated instead by the novel SSA factor Saw1 [23]. At present, it is not yet clear if Slx4 is recruited to NH tails and if so, what mechanisms are at play. Slx4 is phosphorylated in a Mec1/Tel1-dependent manner after treatment with DNA damaging agents, including ionising radiation (IR), UV light and DNA alkylating agents. This is independent of the Rad53 and Chk1 kinases that are activated by Mec1 and Tel1 [28]. Previously, we identified six sites in Slx4 phosphorylated by Mec1/Tel1. Simultaneous mutation of all six Ser/Thr residues to Ala reduced the efficiency of SSA but did not affect cellular resistance to DNA alkylating agents or viability in the absence of the Sgs1 helicase, the other two known functions of Slx4 [22,24]. It is not yet known what aspect of SSA requires phosphorylation of Slx4. In this study we further characterise Slx4 phosphorylation during DSB repair and demonstrate that phosphorylation of Thr 113 of Slx4 by Mec1 and Tel1 is required for 3′ NH DNA tail cleavage by Rad1-Rad10 during SSA and HR.
2. Materials and methods
2.1 Yeast strains and plasmids
Yeast strains used in this study were derived from EAY1141 [29], YMV80 [30] or tNS1379 [31], except where otherwise indicated. All strains are listed in Table 1. Strains with deletions of SLX4, RAD1 or SAW1 were constructed using slx4::NatMX, slx4::KanMX, rad1::KanMX, rad1::HphMX, or saw1::KanMX disruption PCR products in accordance with published procedures [32]. pTAK75 (a gift from T. Usui) was used to construct tel1::TRP1 disruptions. Strains expressing Slx4-13Myc (Slx4 tagged with a C-terminal 13xMYC epitope) were constructed by a PCR-based method, using the markers KanMX, HphMX, NatMX or URA3-MX::kanMX. Strains for assaying removal of non-homologous DNA ends during HO-induced gene conversion were generated by transforming plasmids pFP122, pFP140 or pFP120 [6] into wild-type (YMN4), mec1Δ tel1Δ sml1Δ (YFD0814), slx4Δ (tNS2362) and rad1Δ (tNS2372) strains containing hmlΔ matα-inc hmrΔ. Slx4 plasmids containing individual Ser/Thr to Ala point mutations were generated by mutagenesis of the wild-type SLX4 sequence in a low-copy yeast centromeric vector, using the QuikChange site-directed mutagenesis kit (Stratagene). Presence of the desired mutations was confirmed by DNA sequencing.
Table 1.
| Strain | Genotype | Background | Reference |
|---|---|---|---|
| EAY1141 | mat::leu2::hisG hmrΔ3 thr4 leu2 trp1 THR4-ura3-A(205bp)-HOcs-URA3-A ade3::GAL10-HO::NatMX | tNS1379 | Goldfarb and Alani, 2005 |
| tNS2370 | rad1Δ:: KanMX | EAY1141 | This study |
| GTY3 | slx4::KanMX | EAY1141 | Flott et al., 2007 |
| GTY13 | SLX4(MYC13)::KanMX::slx4 | EAY1141 | This study |
| SLY1881 | rad1-D825A-3×HA::KanMX [pFH800] | tNS1379 | Li et al., 2008 |
| SLY2426 | rad1-D825A-3×HA::KanMX saw1::HphMX [pFH800] | SLY1881 | Li et al., 2008 |
| GTY48 | slx4::NatMX | SLY1881 | This study |
| GTY59 | slx4::NAT 13myc-SLX4wt::LEU2::leu2 | SLY1881 | This study |
| GTY60 | slx4::NAT 13myc-Slx4-T113A::LEU2::leu2 | SLY1881 | This study |
| GTY61 | slx4::NAT 13myc-Slx4-T113E::LEU2::leu2 | SLY1881 | This study |
| GTY62 | slx4::NAT 13myc-Slx4-mut6A::LEU2::leu2 | SLY1881 | This study |
| GTY37 | SLX4(MYC13)::Ura3MX::slx4 mec1::HphMX sml1::KanMX | YMV80 | This study |
| GTY38 | SLX4(MYC13)::KanMX::slx4 tel1Δ::TRP1 | YMV80 | This study |
| GTY39 | SLX4(MYC13)::Ura3MX::slx4 mec1Δ::HphMX sml1Δ::KanMX tel1Δ::TRP1 | YMV80 | This study |
| GTY42 | rad1Δ:: KanMX SLX4(MYC13)::HphMX::slx4 | YMV80 | This study |
| GTY44 | saw1Δ:: KanMX SLX4(MYC13)::HphMX::slx4 | YMV80 | This study |
| GTY11 | slx4Δ::KanMX | YMV80 | This study |
| GTY32 | rad1Δ:: KanMX | YMV80 | This study |
| YMV126 | sml1Δ::KanMX mec1Δ::HphMX | YMV80 | |
| GTY36 | tel1Δ::TRP1 | YMV80 | This study |
| GTY35 | mec1Δ::HphMX sml1Δ:: KanMX tel1Δ::TRP1 | YMV80 | This study |
| YMN4 | MATα-inc hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL::HO | This study | |
| YFD814 | MATα-inc hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL::HO sml1::KAN, mec1::NAT, tel1::TRP1 | YMN4 | This study |
| tNS2362 | slx4::KanMX | YMN4 | This study |
| tNS2372 | rad1::KanMX | YMN4 | This study |
| JKM179 | MatΔ hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG′ ura3-52 ade3::GAL::HO | W303 | Lee et al., 1998 |
| SLY2439 | rad52Δ | EAY1141 | This study |
2.2 HO induction and analysis of DSB repair
Yeast cultures were grown overnight in YEP-lactate at 30°C to a density of approximately 1×107 cells/ml and induced by addition of galactose to a final concentration of 2%. Aliquots were removed at the indicated time points and samples were processed for either western blotting or Southern hybridization.
2.3 Southern blot analysis
Genomic DNA was purified as previously described, digested with BglII (EAY1141 strains) or Asp718 (YMV80 strains) and analyzed by Southern hybridization with appropriate 32P-labelled probes [29,30,33].
2.4 Western blotting and antibodies
Analysis of Slx4p and Rad53p phosphorylation was performed as described previously [22], except that samples were resolved on 3–8% Tris-Acetate gels (Invitrogen) before immunoblotting for Slx4 and Rad53. Primary antibodies used in this study were anti–Myc (9E10; Roche) and anti-Rad53 (a mixture of yN–19 and yC–19; Santa Cruz).
2.5 Measurement of SSA and gene conversion efficiency
To measure SSA efficiency, cultures were grown overnight in either YEP-lactate or SC–Leu +2% raffinose, diluted and plated onto glucose- or galactose-containing solid media (YPA or SC–Leu). Plates were incubated at 30°C for 2–4 days before counting colonies. The viable fraction was derived by dividing the number of colony-forming units on galactose media by that on glucose media. Efficiency of 3′ non–homologous tail removal during gene conversion was determined as described previously [6].
2.6 Quantitative PCR assay to monitor 3′ NH tail formation and removal
Yeast cells were lysed with glass beads and genomic DNA was extracted using Qiagen’s DNeasy Blood and Tissue kit. The DNA was diluted and used as a template in SYBR green-based real-time PCR assays. Primer pairs TAIL1 and REP1 were used for the ura3 substrate in EAY1141 strains and primer pairs TailU2 and IntU2 were used for the “U2” substrate in YMV80 strains. In each case, signals from the CEN8 and PHO5 control loci were used to normalise signals from the SSA substrate.
2.7. Chromatin immunoprecipitation
Cross–linking and chromatin immunoprecipitation (ChIP) were carried out as described previously [23] with the following minor modifications. For cross-linking, 1% (v/v) formaldehyde (Sigma) was added and the reactions were incubated with rotation for 5 min prior to neutralization by 2.5 M glycine. For immunoprecipitation, 0.3 ml anti-Slx4 sera was added to cell lysates and the immunoprecipitates were subjected to real time quantitative PCR with primer sets that anneal to proximal (pJC1: GCCCAGTATTCTTAACCCAACTGCAC, pJC2: CAGCTGGCGTAATAGCGAAGAGGCCC) or distal side (pJC3: CCTTAGTAGTTGGTAACCTGACAAAGG, pJC4: CCTTCTGTTCGGAGATTACCGAATC) of the 3′ flaps and to the MAT locus. “Fold immunoprecipitate” represents the ratio of the Slx4 IP PCR signal before and after HO induction, normalized by the input signal and that of the PCR signal of the MAT control. Data represent the mean + s.d. of three or more independent experiments.
3. Results
3.1 Slx4 becomes phosphorylated during SSA repair of a HO–induced DSB
As we were interested in the role of Slx4 phosphorylation during SSA, we tested phosphorylation of Slx4 during SSA repair of a HO endonuclease-induced DSB in two different SSA strains, in which the homologous repeats flanking the DSB were separated by 2.6 kb (EAY1141; Fig 1A, left panel) or 25 kb (YMV80; Fig 1A, right panel) respectively [29,30]. To allow detection of the Slx4 protein in these SSA strains, Slx4 was C-terminally tagged with thirteen Myc epitopes; this form of Slx4 fully rescues the phenotypes of slx4Δ cells [28]. Cultures of EAY1141 Slx4-13Myc and YMV80 Slx4-13Myc were grown overnight in YEP-lactate before addition of galactose to induce the HO endonuclease. Samples were collected at different times after DSB induction, and the phosphorylation of Slx4 – and Rad53 for comparison –was judged by electrophoretic mobility shift [28]. Since 5′–3′ resection of the DSB ends proceeds at a rate of approximately 4 kb/hr, it takes approximately 1.5 hr in EAY1141, and 6 hr in YMV80, for resection to reach the distal repeat, enabling SSA to occur [5,29,30]. Slow-migrating phosphorylated forms of Slx4 were readily detectable 1 hr after DSB induction, and reached a maximum approximately 3 hr after HO induction in the EAY1141 strain, or 6 h in the YMV80 strain (Fig. 1B). Reversal of Slx4 phosphorylation appeared to coincide with completion of SSA in each strain, as judged by Southern blot analysis (data not shown). Rad53 hyperphosphorylation was also monitored as a readout of DNA damage signalling. In both SSA strains, hyperphosphorylation of Rad53 was detected 2–3 hr after DSB induction and dephosphorylation coincided approximately with completion of repair, consistent with previous observations [30].
3.2 Slx4 phosphorylation during SSA requires Mec1/Tel1 but not Rad1 or Saw1
We next examined the genetic requirements for Slx4 phosphorylation during SSA. First we asked whether Slx4 phosphorylation required Rad1, with which Slx4 associates, or Saw1 that is required for recruitment of Rad1–Rad10 to 3′ NH tails during SSA [22,23,34,35]. As seen in Fig. 1C, Slx4 became phosphorylated in both rad1Δ and saw1Δ cells with kinetics similar to wild-type cells and Slx4 phosphorylation in rad1Δ and saw1Δ cells was eventually lost by 24 hr post-induction (Fig. 1C). Rad53 hyperphosphorylation persisted until around 12 hr in these mutants.
Next, we examined Slx4 phosphorylation during SSA in cells lacking either or both of the upstream yeast DNA damage-responsive kinases, Mec1 and Tel1 that phosphorylate Slx4 after MMS [28]. Slx4 phosphorylation appeared to be unaffected by deletion of TEL1 (Fig. 1D). In contrast, phosphorylation of Slx4 was significantly compromised by the loss of Mec1, although some transient phosphorylation could be seen at early time-points after DSB induction in mec1Δ sml1Δ cells (all mec1Δ strains also carry the sml1Δ mutation which suppresses the lethality of a MEC1 deletion).
Loss of both kinases completely abolished Slx4 phosphorylation (Fig. 1D). Thus Slx4 phosphorylation in response to a HO–induced DSB during SSA requires Mec1 and/or Tel1, but not Rad1 or Saw1. Rad53 hyperphosphorylation showed a similar pattern, although it was completely lost when MEC1 was deleted. As with Slx4, deletion of TEL1 had little effect.
3.3 Slx4 is recruited to 3′–non-homologous tails in a phosphorylation–independent manner
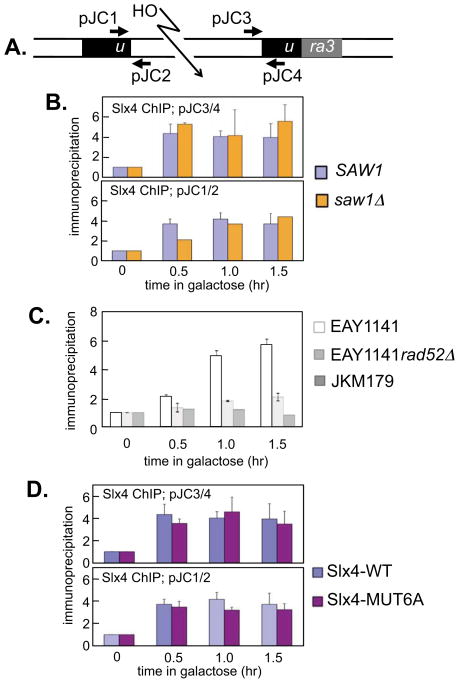
Phosphorylation of Slx4 by Mec1/Tel1 during SSA suggests that the post–translational modification of Slx4 may affect an important regulatory step. To study how phosphorylation might regulate Slx4, we first tested if Slx4 is recruited to 3′ NH tails during SSA, and if this is affected by phosphorylation. Cells were employed that express a rad1D825N mutant that retains the ability to bind DNA but is defective in DNA cleavage activity, resulting in the persistence of a HO-induced DSB in an SSA reporter cassette (Fig. 2A; strain EAY1141) [36]. As noted previously, expression of nuclease–deficient rad1 is necessary to detect Rad1 protein at the 3′ NH tails by ChIP. This is possibly due to the rapid dissociation of wild–type Rad1 from the post–cleavage complex [23]. We found that Slx4 can be detected at 3′ NH tails in cells harboring the rad1D825N mutation within half an hour of HO expression and the level remains constant until 1.5 hr post HO expression (Fig. 2B).
Figure 2. Association of the Slx4 with 3′ non–homologous DNA tails.
(a) Schematic diagram of ChIP experiment. Persistent 3′ non-homologous DNA tails are generated after HO–induced DSB induction in a strain expressing nuclease-deficient Rad1–D825A[23]. ChIP (chromatin immunoprecipitation) assays were carried out as previously described [23], using the indicated primers, to detect the accumulation of proteins at the persistent 3′ non-homologous tails in the rad1–D825A mutant. (b) ChIP assays to measure levels of Slx4 at persistent 3′ NH tails were performed in strains SLY1881 (Rad1-D825A) and SLY2426 (Rad1-D825A saw1Δ). Anti–Slx4 immunoprecipitates were subjected to real time quantitative PCR with primer sets that anneal to proximal or distal side of the 3′ flaps and to the MAT locus. Fold immunoprecipitate represents the ratio of the PCR signal in Slx4 immunoprecipitates before and after HO induction, normalized by the PCR signal of the MAT control. Data represent the mean + s.d. of three or more independent experiments. (c) ChIP assays to measure levels of Slx4 wild–type (WT) or Slx4–MUT6A bound at persistent 3′ non-homologous tails were performed in strains that express 13Myc–tagged wild–type Slx4 (GTY59) or 13Myc–tagged Slx4–MUT6A (GTY62). Anti–Slx4 antibodies, not Myc antibodies were used to immunoprecipitate Slx4. (d) Slx4 ChIP in strains EAY1141 rad1D825A-3HA, EAY1141 rad1D825A-3HA rad52Δ and JKM179 with primer sets proximal to HO-induced DSBs. In both panels, for ChIP in strains with EAY background, primer set DJP1/2 was used for real-time PCR. Right panel: for ChIP in JKM179, a PCR primer set L6-2317-5′ (CTTCAATATTATTCGACCACTCAAG) and L6-2398-3′ (GTGAATTTGGATTTTATTTAAAAATCG) was used. For ChIP in JKM179, a PCR primer set R6-2821-5′ (GTTCCCAATGTTTGTTTATACACTC) and L6-2904-3′ (CAAATACATAGACATAAACAAAAGA) was used.
From this experiment it was not clear if Slx4 recognises the DSB, ssDNA tails created by resection or NH tails found after annealing of the repeats. To address this we took strain EAY1141, in which a HO–induced DSB is repaired by SSA, and deleted RAD52. In the resulting cells, the HO–induced DSB is resected but cannot be repaired. In these cells, Slx4 recruitment to the vicinity of the HO–induced DSB is greatly reduced compared with cells that express Rad52 (Fig. 2C). We also examined Slx4 recruitment in cells expressing HO endonuclease in a strain (JKM179) that was also deleted for HML and HMR sequences such that induction of HO in these cells results in generation of a single DSB at the MAT locus that cannot be repaired by gene conversion [37,38]. Slx4 was not recruited to the vicinity of the HO–induced DSB in these cells (Fig. 2C). Taken together, these data imply that Slx4 does not simply bind to a DSB or to ssDNA tails created by DSB resection. Instead it is more likely that Slx4 binds to NH DNA tails or another structure that arises during HR.
Previously, we showed that the recruitment of Rad1–Rad10 complex to 3′ NH tails depends on the Saw1 protein and deletion of SAW1 abolished the Rad1–Rad10–dependent 3′ tail removal and SSA [36]. We thus tested if the association of Slx4 with the 3′ tail DNA relies on Saw1. As shown in Fig. 2B, saw1Δ cells showed efficient Slx4 recruitment at 3′ NH tails. The results suggest that binding of Slx4 to the 3′ NH tails does not require Saw1 even though recruitment of Rad1–Rad10 does require Saw1. Six S/T-Q motifs within Slx4 have been found to be phosphorylated by Mec1/Tel1 in vitro and in vivo and mutation of all six reduces the efficiency of SSA [22] although the mechanistic basis for this defect is not known. Most importantly, the slx4–MUT6A mutant [22], in which the six known Mec1/Tel1 phosphorylation sites are mutated to Ala, showed levels and kinetics of association at the 3′ NH tails indistinguishable from that of wild–type Slx4 (Fig. 2D), suggesting that the phosphorylation by Tel1/Mec1 does not regulate the recruitment of Slx4 at 3′ NH tails.
3.4 Slx4 phosphorylation on Thr113 is important for 3′ non-homologous tail removal during HR
We wished to investigate which step of SSA/HR is regulated by Slx4 and, in particular, if Rad1–Rad10 activity is affected. To examine the role of Slx4 phosphorylation in 3′ NH tail removal, we developed a quantitative PCR assay to monitor formation and removal of 3′ NH tails generated during SSA repair of a site-specific DSB in vivo (Fig. 3A). In our experimental system, a DSB is induced at an HO restriction site flanked by homologous ura3 205 bp repeat sequences. The DSB is subjected to 5′–3′ resection of the DSB ends followed by annealing of flanking repeats, NH tail removal, repair synthesis and end-ligation. This results in deletion of the sequence between the ura3 repeats, as well as one of the repeats [29]. Primer pairs TAIL1 and REP1, which amplify regions within the non–homologous tail and the homologous repeats respectively (Fig 3A, upper panel), were used for quantitative PCR on genomic DNA extracted from cells after HO induction.
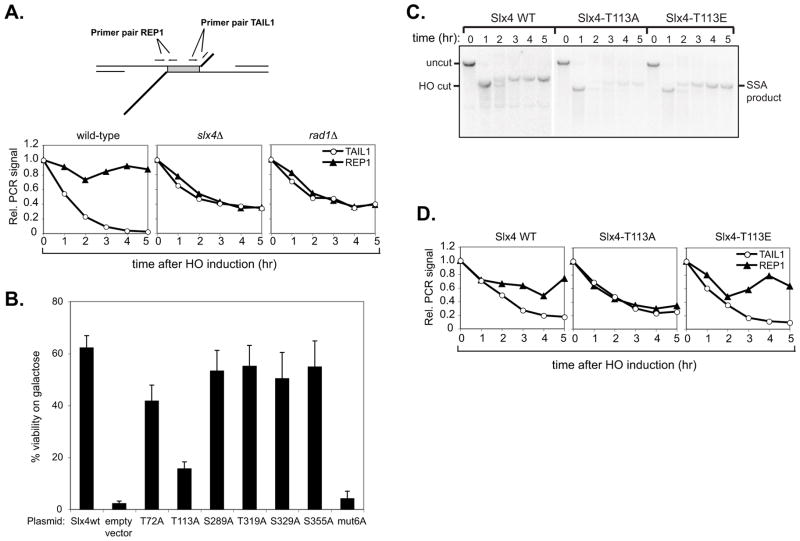
Figure 3. Slx4 phosphorylation on Thr113 is required for cleavage of 3′ NH tails.
(a) Detection of 3′ non-homologous tail formation and removal by qPCR. Upper panel: Quantitative PCR (qPCR) assay to monitor 3′ non–homologous tail formation and removal at a double flap structure created during SSA repair of a DSB between 205bp ura3 direct repeats. Primer pairs TAIL1 and REP1 are used to detect the presence of the single-stranded non-homologous tail and repeat regions. Lower panel: 3′ non-homologous tail formation and removal at the 205bp ura3 SSA substrate was monitored in wild-type (EAY1141), rad1Δ (tNS2370) and slx4Δ (GTY003) strains. Graphs represent normalised PCR signals from primer pairs TAIL1 and REP1. Values obtained for all time points were normalised to the 0 hr value. Signals from CEN8 and PHO5 control loci were averaged and used to normalise the ura3 SSA substrate values. (b) EAY1141 slx4Δ cells (GTY03) were transformed with low-copy plasmids expressing 13Myc–tagged wild–type Slx4 or individual Slx4 S/T-Q to A-Q point mutants. SSA efficiency in these cells was determined by plating on glucose or galactose media, as described in Materials and Methods. Error bars represent standard error of the mean (SEM) from three separate experiments. (c) Southern blot analysis of SSA in tNS2402, tNS2405, tNS2447 (EAY1141 slx4Δ cells transformed with SLX4–wt, slx4–T113A or slx4–T113E plasmids, respectively). (d) 3′ non-homologous tail formation and removal was monitored in EAY1141 slx4Δ (GTY03) cells transformed with SLX4–wt, slx4–T113A or slx4–T113E plasmids by qPCR, using primer pairs TAIL1 and REP1.
During SSA repair in the wild-type EAY1141 strain (Fig. 3A, lower panel), the relative signal from primer pair TAIL1 decreases from 1 to almost 0 over a 5 hr period as resection creates single-stranded tails, which are then removed. The relative signal from primer pair TAIL1 did not decrease to 0 in either mutant (slx4Δ = 0.34, rad1Δ = 0.39), indicating that the ssDNA tails were still present up to 5 hr after HO induction. The decrease of signal to about 0.5 is expected since one strand of the tail region will be removed by resection and quantitative PCR will begin with only 50% of the starting template. In the wild-type strain, the relative signal from primer pair REP1 decreases transiently as the corresponding region becomes single-stranded and then is restored to approximately 0.85 of the initial value as the region is repaired to double-stranded DNA. In the mutants, the REP1 signal decreases to approximately 0.35 in slx4Δ cells and 0.37 in rad1Δ cells and does not recover, indicating that the target region has not been repaired to dsDNA by 5 hr post HO-induction. These results are in good agreement with the kinetics of SSA product formation, monitored by Southern blotting (data not shown).
Mutation of the six known Mec1/Te1l phosphorylation sites in Slx4 reduces the efficiency of SSA. The mechanistic basis for this defect is not known and the contribution of individual phosphorylation sites has not been examined [22]. To investigate whether individual sites are required for the SSA-related function of Slx4, we individually mutated the Ser/Thr residues in each of the six phosphorylated S/T–Q motifs in Slx4 – T72, T113, S289, T319, S329 and S355 – to Ala and tested the ability of the single mutants to support SSA. EAY1141 slx4Δ cells bearing low-copy plasmids for expression of wild-type Slx4 or Ser/Thr to Ala point mutants were plated on glucose- or galactose-containing media to determine viability in the presence of HO endonuclease. As shown in Fig. 3B, SSA efficiency was reduced in the Slx4-Thr113Ala (T113A) mutant to approximately 30% of wild-type levels. Other single Ala point mutants, apart from T72A which showed a slight reduction, did not show significantly reduced viability on galactose, whereas the slx4-MUT6A mutant lacking all six Slx4 Mec1/Tel1 phosphorylation sites was severely defective. Consistent with an important role for Slx4 Thr113 in SSA, Southern blot analysis revealed a defect in SSA product formation in slx4Δ cells transformed with the Slx4-T113A mutant plasmid (Fig. 3C). In contrast, slx4Δ cells expressing wild-type Slx4 or, intriguingly, an Slx4-Thr113Glu (T113E) phospho–mimetic mutant, were proficient for SSA (Fig. 3C). Taken together, these results suggest that full Slx4 function in SSA requires Thr113, along with one or more of the other phospho–sites.
We next measured 3′ tail removal in slx4Δ cells expressing wild-type Slx4, the Slx4-T113A mutant, or the Slx4-T113E phospho–mimetic mutant, from low-copy plasmids (Fig. 3D). The Slx4-T113A mutant showed a partial defect in tail removal (approximately two-fold relative to the wild-type at 5 h post-HO induction), consistent with the observed SSA defect. Interestingly, the Slx4-T113E phospho–mimetic mutant appeared proficient in tail removal, as the signal from the REP1 control primer pair was restored to similar levels as in the wild-type (Slx4-T113E = 0.63, Slx4 wt = 0.74), indicating successful completion of repair.
3.5 Mec1 and Tel1 are required for 3′ NH tail cleavage
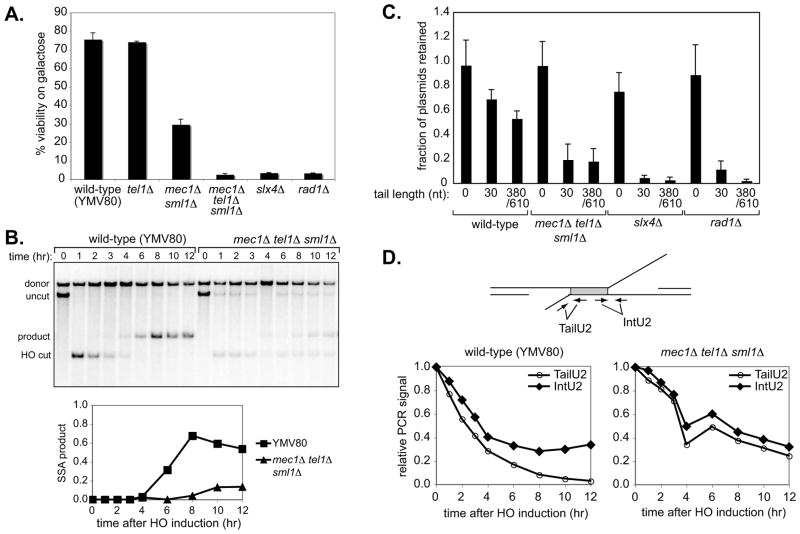
Given the dependence of Slx4 phosphorylation on Mec1 and Tel1, we examined the effect of MEC1 and/or TEL1 deletion on SSA. YMV80 (wild-type), mec1Δ sml1Δ (YMV126), tel1Δ (GTY36), or mec1Δ tel1Δ sml1Δ (GTY35) cultures were grown overnight in YEP-lactate, diluted and plated to glucose- or galactose-containing media to determine SSA efficiency. Like the wild-type YMV80 strain, the tel1Δ mutant was proficient in SSA. In contrast, SSA efficiency was severely decreased in the mec1Δ sml1Δ mutant judged by viability on galactose, and nearly abolished in the mec1Δ tel1Δ sml1Δ mutant (Fig. 4A). YMV80 slx4Δ (GTY11) and rad1Δ (GTY32) mutants, which are defective in SSA, were included as controls. We also monitored SSA in the mec1Δ tel1Δ sml1Δ strain by Southern blotting and observed little SSA product formation even after 12 hr (Fig. 4B).
Figure 4. Mec1/Tel1 are required for 3′ non–homologous tail removal during HR.
(a) The efficiency of SSA repair in wild-type (YMV80) or mec1Δ sml1Δ (YMV126), tel1Δ (GTY36), mec1Δ tel1Δ sml1Δ (GTY35), slx4Δ (GTY11) and rad1Δ (GTY32) strains was determined by plating on glucose or galactose media, as described in Materials and Methods. Error bars represent standard error of the mean (SEM) from three separate experiments. (b) Upper panel: Southern blot analysis of SSA in wild-type (YMV80) and mec1Δ tel1Δ sml1Δ (GTY35) strains. Lower panel: quantitation of SSA product formation in YMV80 and GTY35. (c) Gene conversion efficiency was measured in wild-type (YMN4), mec1Δ tel1Δ sml1Δ (YFD814), slx4Δ (tNS2362) and rad1Δ (tNS2372) strains containing hmlΔmatα-inc hmrΔ and transformed with the plasmids pFP122, pFP140 or pFP120. Mean fractions of plasmids retained ± 95% confidence interval (CI) are shown. (d) 3′ non-homologous tail formation and removal was assayed in wild-type (YMV80) and mec1Δ tel1Δ sml1Δ (GTY35) strains by qPCR, using primer pairs TailU2 and IntU2 to detect the non-homologous tail and internal control (repeat) regions as indicated.
We then asked whether Mec1 and Tel1 are required for 3′ tail removal in the context of gene conversion. We used plasmids containing inverted repeats of E. coli lacZ sequences, one of which lacks a promoter while the other is interrupted by an HO cleavage site [6]. Plasmids in which HO cleavage produces no tails (pFP122), or NH tails of 30 nt (pFP140) and of 308/610 nt (pFP120) after HO induction were transformed into hmlΔ matα-inc hmrΔ wild-type (YMN4), mec1Δ tel1Δ sml1Δ (YFD0814), slx4Δ (tNS2362) and rad1Δ (tNS2372) strains, to test their ability to perform gene conversion in the presence of NH DNA tails of various lengths [6]. Both SLX4 and RAD1 were strictly required for repair of the HO-induced DSB when tails of 30 nt or 308/610 nt, were present but were not required when there were no NH tails (Fig. 4C). In cells lacking both Mec1 and Tel1, plasmid retention in the presence of short (30 nt) or long (380/610 nt) tails was also greatly reduced (Fig. 4C), indicating a defect in tail removal, whereas gene conversion in the absence of 3′ NH tails was normal in these cells (Fig. 4C). These results suggest that Mec1 and Tel1 are required for NH tail removal during SSA and HR.
To directly test the role of Mec1/Tel1 in 3′ NH tail removal, we employed a quantitative PCR tail-cleavage assay (Fig. 4D, upper panel). Using the primer pairs TailU2 and IntU2, we monitored removal of the long NH tail formed after DSB induction in YMV80 wild-type and mec1Δ tel1Δ sml1Δ cells. The relative PCR signal from the TailU2 primer pair decreased to 0.027 by 12 hr in the wild-type strain, but did not drop below 0.24 in the mec1Δ tel1Δ sml1Δ mutant (Fig 4D, lower panels). Thus, deletion of both Mec1 and Tel1 reduced the efficiency of 3′ NH tail removal.
4. Discussion
We previously showed that Slx4 is required for SSA and that mutation of a combination of six Ser/Thr residues phosphorylated by Mec1 and Tel1 reduces SSA efficiency. To understand the role of Slx4 phosphorylation in the context of HR-dependent DSB repair, we investigated the kinetics and genetic dependency of Slx4 phosphorylation during repair of HO-induced DSBs by SSA. Slx4 was strongly phosphorylated after DSB induction and this required Mec1 and Tel1 but not Rad1 or Saw1 (Fig. 1). It is interesting to note that in the YMV80 strain (Fig. 1A), pairing of the repeat sequences occurs at approximately 1 h after HO induction and one of the two ends likely form a 3′ NH tail at this early time point. However, Slx4 phosphorylation occurs at 6 h but not earlier suggesting that Slx4 phosphorylation may be involved in dissociating Slx4 from the flap. This will be interesting to investigate, especially since Slx4 dephosphorylation also coincided with completion of DSB repair.
To test the possibility that Slx4 promotes Rad1–Rad10 activity by targeting it to its 3′ tail substrates, we used chromatin immunoprecipitation to investigate the ability of Slx4 to localise to these structures in vivo. We were able to detect recruitment of Slx4 in the vicinity of persistent 3′ NH tails. Although we have not proven that it is NH tails that Slx4 binds to, the data in Fig. 2C shows that Slx4 is not recruited to a DSB that cannot be repaired and so it is unlikely that Slx4 recognises a DSB or ssDNA tails created by DSB resection. More work will be required to determine the structures recognised by Slx4 during DNA repair. Surprisingly, this recruitment of Slx4 in the vicinity of a DSB was independent of Saw1 that was previously shown to be required for recruitment of Rad1–Rad10 [36]. At present it is not clear how Slx4 is recruited to NH tails but it may be that it binds directly by virtue of the conserved SAP domain towards the C–terminus [25].
So what step of SSA does Slx4 regulate? We thought it likely that Slx4 regulates NH tail cleavage since it interacts with Rad1–Rad10. Indeed, we found that NH tails generated during SSA persist in the absence of Slx4 and that this defect is as severe as that seen in cells lacking Rad1, consistent with a recent report [23]. Thus, Slx4 is essential for 3′ NH tail cleavage by Rad1–Rad10 activity. It is not yet clear why Slx4 is so important for NH tail cleavage and in vitro reconstitution experiments will be important in addressing this question. It is possible that Slx4 directly affects the activity and/or substrate specificity of Rad1–Rad10 or that Slx4 could deform or distort DNA substrates to enable efficient cleavage. We favour the latter hypothesis. It was recently shown that the activity of XPF from the archaeon Sulfolobus solfataricus towards branched DNA substrates is stimulated by PCNA binding, which increases the catalytic rate constant 7000–fold in vitro. S. solfataricus XPF was inactive towards a 3′ flap substrate without PCNA [39]. It is possible that Slx4 regulates Rad1–Rad10 in a manner analogous to stimulation of crenarchaeal XPF by PCNA. The PCNA–interaction motif in archaeal XPF is missing from yeast Rad1 and it will be interesting to test if this region of Rad1 binds Slx4 instead. We also note that NH tail removal also requires both the strand-annealing protein Rad59 [19] and the Msh2-Msh3 [31] mismatch repair proteins, especially when the regions that can anneal and stabilize an annealed intermediate with a NH tail is only a few hundred base pairs. It is possible that Slx4 might also interact with these components.
An Slx4 mutant with all six in vitro/in vivo Mec1/Tel1 sites mutated to alanine (Slx4-MUT6A) was previously shown to be unable to support SSA [22]. We tested individual Ala point mutants found that 3′ NH tail removal was impaired in the Slx4-T113A mutant or in cells lacking Mec1 and Tel1 that are both required for Slx4 phosphorylation. Furthermore, an Slx4 mutant in which Thr 113 was mutated instead to Gln, mimicking phosphorylation of this residue, was proficient in 3′ NH tail removal and SSA. So how does Mec1/Tel1–mediated phosphorylation of Slx4 affect its function to stimulate Rad1–Rad10 cleavage of NH tails? It is clear that phosphorylation of Slx4 is not required for its interaction with Rad1–Rad10 [40] or for the recruitment of Slx4 to sites 3′ NH tails and answering this question will first require a basic understanding of how Slx4 functions at the molecular level. Why should Slx4 phosphorylation be necessary for Rad1–Rad10 activity towards 3′ tails? It may be that Slx4 phosphorylation ensures that Rad1–Rad10 is are active towards flaps only when DNA damage is present – since only then will Slx4 be juxtaposed with Mec1/Tel1. This would restrict Rad1–Rad10 flap cleavage activity, possibly guarding against inappropriate cleavage events that could compromise genome integrity.
It was previously reported that mec1Δ sml1Δ cells repair a DSB by SSA with similar kinetics to the wild–type, but eventually lose viability after HO induction. This defect was attributed to an inability to trigger a G2 arrest that would prevent loss of acentric fragments from the broken chromosome, and was rescued by holding cells in G2/M with nocodazole during DSB induction [41]. Mec1 has a well–known role in mediating cell cycle arrest in response to damage, by activating numerous checkpoint effectors, including the checkpoint kinase Rad53. Unlike Rad53, Slx4 phosphorylation, although reduced, is not completely lost in mec1Δ sml1Δ cells. Some phosphorylation, presumably by Tel1, is detectable at early time-points (Fig. 1D). This phosphorylation of Slx4 appears to be sufficient to support tail cleavage, since when repair is measured directly by Southern blotting, SSA product formation occurs in mec1Δ sml1Δ cells with wild-type kinetics [41]. Our data are thus consistent with an additional role for the Mec1 kinase, functioning redundantly with Tel1, in directly activating the Slx4–Rad1–Rad10 complex for DSB repair.
Acknowledgments
This work was supported by the Medical Research Council (UK) (JR), NIH grants GM20056 and GM61766 (JEH) and NIH grant GM71011 (SEL). We are grateful to Ivan Muñoz and Karolina Hain for their comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. Embo J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 6.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1999;63:349. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippo JS, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annual Review of Biochemistry. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 9.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 10.Bardwell L, Cooper AJ, Friedberg EC. Stable and Specific Association between the Yeast Recombination and DNA-Repair Protein-Rad1 and Protein-Rad10 Invitro. Molecular and Cellular Biology. 1992;12:3041–3049. doi: 10.1128/mcb.12.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annual Review of Biochemistry. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 12.Schiestl RH, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov EL, Haber JE. Rad1 and Rad10, but Not Other Excision-Repair Genes, Are Required for Double-Strand Break-Induced Recombination in Saccharomyces-Cerevisiae. Molecular and Cellular Biology. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colaiacovo MP, Paques F, Haber JE. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151:1409–1423. doi: 10.1093/genetics/151.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans E, Sugawara N, Haber JE, Alani E. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Molecular Cell. 2000;5:789–799. doi: 10.1016/s1097-2765(00)80319-6. [DOI] [PubMed] [Google Scholar]
- 19.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Molecular and Cellular Biology. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyndaker AM, Goldfarb T, Alani E. Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics. 2008;179:1807–1821. doi: 10.1534/genetics.108.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 22.Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouse J. Control of genome stability by SLX protein complexes. Biochem Soc Trans. 2009;37:495–510. doi: 10.1042/BST0370495. [DOI] [PubMed] [Google Scholar]
- 25.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, Russell P, Fuchs RP, McGowan CH, Gaillard Human PHL. SLX4 Is a Holliday Junction Resolvase Subunit that Binds Multiple DNA Repair/Recombination Endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 Assembles A Holliday Junction Resolvase and Is Required for DNA Repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flott S, Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldfarb T, Alani E. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics. 2005;169:563–574. doi: 10.1534/genetics.104.035204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 31.Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci U S A. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Holmes A, Haber JE. Physical monitoring of HO-induced homologous recombination. Methods Mol Biol. 1999;113:403–415. doi: 10.1385/1-59259-675-4:403. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies AA, Friedberg EC, Tomkinson AE, Wood RD, West SC. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J Biol Chem. 1995;270:24638–24641. doi: 10.1074/jbc.270.42.24638. [DOI] [PubMed] [Google Scholar]
- 36.Li FY, Dong JC, Pan XW, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Molecular Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haber JE. Uses and abuses of HO endonuclease. Guide to Yeast Genetics and Molecular and Cell Biology, Pt B. 2002;350:141–164. doi: 10.1016/s0076-6879(02)50961-7. [DOI] [PubMed] [Google Scholar]
- 38.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 39.Hutton RD, Roberts JA, Penedo JC, White MF. PCNA stimulates catalysis by structure-specific nucleases using two distinct mechanisms: substrate targeting and catalytic step. Nucleic Acids Res. 2008;36:6720–6727. doi: 10.1093/nar/gkn745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. Phosphorylation of Slx4 by mec1 and tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Molecular and Cellular Biology. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]