Abstract
Objective
The diagnostic boundaries of sleep disorders are under considerable debate. The main sleep disorders are partly heritable; therefore, defining heritable pathophysiologic mechanisms could delineate diagnoses and suggest treatment. We collected clinical data and DNA from consenting patients scheduled to undergo clinical polysomnograms, to expand our understanding of the polymorphisms associated with the phenotypes of particular sleep disorders.
Methods
Patients at least 21 years of age were recruited to contribute research questionnaires, and to provide access to their medical records, saliva for deoxyribonucleic acid (DNA), and polysomnographic data. From these complex data, 38 partly overlapping phenotypes were derived indicating complaints, subjective and objective sleep timing, and polysomnographic disturbances. A custom chip was used to genotype 768 single-nucleotide polymorphisms (SNPs). Additional assays derived ancestry-informative markers (eg, 751 participants of European ancestry). Linear regressions controlling for age, gender, and ancestry were used to assess the associations of each phenotype with each of the SNPs, highlighting those with Bonferroni-corrected significance.
Results
In peroxisome proliferator-activated receptor gamma, coactivator 1 beta (PPARGC1B), rs6888451 was associated with several markers of obstructive sleep apnea. In aryl hydrocarbon receptor nuclear translocator-like (ARNTL), rs10766071 was associated with decreased polysomnographic sleep duration. The association of rs3923809 in BTBD9 with periodic limb movements in sleep was confirmed. SNPs in casein kinase 1 delta (CSNK1D rs11552085), cryptochrome 1 (CRY1 rs4964515), and retinoic acid receptor-related orphan receptor A (RORA rs11071547) were less persuasively associated with sleep latency and time of falling asleep.
Conclusions
SNPs associated with several sleep phenotypes were suggested, but due to risks of false discovery, independent replications are needed before the importance of these associations can be assessed, followed by investigation of molecular mechanisms.
Keywords: Sleep apnea, Sleep-related periodic leg movements, DIMS, rs10766071, rs3923809, PPARGC1B
1. Introduction
Sleep disorders are an expanding arena of medical research. For decades, insomnia has been the primary focus and it may remain the most frequently treated sleep disorder with the greatest total medical cost. Insomnia is strongly associated with depression and other emotional disorders, which seemingly contribute to each other. However, expanding data indicate that sleep-disordered breathing may be more common than insomnia [1,2]. Further, sleep-disordered breathing might be associated with higher morbidity and mortality. With or without comorbid insomnia complaints, reported sleep durations either several hours longer or shorter than the population median are associated with elevated mortality risks and numerous morbidities [3–5]. Contrary to popular belief, reported sleep durations longer than the epidemiologic optimum appear more common than short sleep [4]. Reported long sleep may be the best-documented mortality risk factor among the sleep disorders, and long sleep is associated with more serious morbidities [3,6]. Willis–Ekbom disease (restless legs syndrome or RLS) might have a prevalence of 3–15% [7,8], but its associations with morbidity and mortality have not yet been fully clarified. Among the circadian rhythm sleep disorders, delayed sleep-phase disorders (DSPDs) are an increasing concern among adolescents and young adults [9,10]. Delayed sleep phase is associated with broad health impairments and possibly, when the disturbance is persistent, a DSPD leads to excess mortality [11]. Narcolepsy was one of the first sleep disorders characterized. Although narcolepsy can be quite disabling, the prevalence appears to be less than one per thousand [12].
The diagnostic definitions of these sleep disorders have been somewhat controversial and have often varied among successive presentations of standard criteria, suggesting uncertainties in the diagnostic formulations. The population prevalence of these sleep disorders is poorly defined due to frequent changes in the diagnostic formulations. Validations of the most recent criteria by predicted prognoses and responses to treatment are scanty. Various sleep disorders are frequently comorbid and may have common symptoms such as trouble falling asleep, excess arousals during sleep, daytime sleepiness, and fatigue. It may be difficult for the clinician to isolate the various factors causing the symptoms in order to focus on the most useful targets for intervention.
All of the sleep disorders mentioned above are somewhat or strongly heritable, with genetic components of causation [12–16]. Clarification of the genetic predispositions might lead to better understanding of pathophysiologic mechanisms, more useful diagnostic formulations, and ultimately better approaches to treatment. Up to now, with the exception of polymorphisms associated with narcolepsy [12,17,18], the identified polymorphisms associated with sleep disorders explain only small parts of their heritability and prevalence.
To expand understanding of genetic variants associated with sleep disorders, we systematically collected research questionnaires and DNA from patients scheduled to undergo clinical polysomnograms at the Scripps Clinic Viterbi Family Sleep Center.
2. Methods
2.1. Recruitment and procedure
From June 2006 to May 2010, whenever practical, patients scheduled for polysomnography (PSG) were invited to participate in genetic research. Patients 21 years and older were included, provided they competently signed informed written consent, under supervision of the Scripps Human Research Participant Protection Program and Institutional Review Board and in compliance with the Helsinki Declaration. Patients were not paid for research participation. Saliva was collected in Oragene saliva kits (DNA Genotek Inc., Kanata, Ontario, Canada). Then DNA was purified according to the Oragene protocol and the samples were frozen.
An extensive symptom questionnaire was completed by patients for clinical purposes with the help of family or staff as needed, and the same questionnaire was used for the research. The questionnaire included queries about sleep habits and schedules; sleep onsets and awakening times on weekdays and weekends; napping; and numerous items concerning symptoms of disturbed sleep, daytime sleepiness, narcoleptic symptoms, restless legs, and sleep-disordered breathing. Limited demographic items were included. The questionnaire included four key questions about restless legs used to recognize Willis–Ekbom disease [19], the Epworth Sleepiness Scale (ESS) [20], the Basic Language Morningness scale (BALM, a circadian morningness–eveningness trait measure) [21], and the Quick Inventory of Depressive Symptomatology-Self Report (QIDS-SR) scale evaluating current depressive symptoms [22]. Both International Classification of Sleep Disorders (ICSD) [23] research diagnoses recorded by the sleep specialist administering the intake questionnaire and the International Classification of Diseases (ICD)-9 diagnoses recorded over recent months in our electronic medical information system were retrieved and coded.
2.2. Polysomnography
After the polysomnogram was recorded and reviewed, the research database was coded with statistics such as the polysomnographic total sleep time, sleep latency, rapid eye movement (REM) latency, sleep efficiency, REM and slow-wave sleep percentages, total number and number per hour of obstructive and central apneas, apnea–hypopnea index (AHI), pulse-oximetry measurement of the 3% oxygen desaturation index (ODI3), percent of sleep time when oxygen saturation was <80%, and the periodic leg movement index. Logarithmic transforms were also tested for highly skewed parameters. Polysomnography scoring used contemporary American Academy of Sleep Medicine (AASM) criteria, which underwent several modifications during the study. When a patient was observed to display substantial sleep-disordered breathing in the first part of the polysomnogram, often a split-night procedure was implemented to save the clinical expense of extra recording nights. On 416 split nights, the first portion of the night was devoted to uninterrupted recording to assess the degree of sleep-disordered breathing. Then the remainder of the night was devoted to sometimes-disturbing technician interventions to initiate continuous positive airway pressure (CPAP) treatment; adjust masks; titrate pressures to find optimal responses; change CPAP to bilevel, auto-adjusting, and paced ventilatory support protocols; provide supplemental oxygen, etc. For analyses of polysomnographic total sleep time (tstpsg), only recordings with uninterrupted time in bed (tib) > 300 min were used (N = 517), which included 99.6% of the full-night recordings but excluded 99% of the split-night recordings wherein sleep was artificially curtailed. If the patient wore CPAP or used supplemental oxygen during an entire undisturbed recording (without technician interventions and titrations), those data were also used. Patients received their usual medications on recording nights.
Strengths and limitations of the polysomnographic data collection strategy should be noted. The patient sample was recruited from a busy academic sleep practice, first established in 1983. The polysomnographic recordings followed approved AASM procedures in this accredited sleep center, overseen by clinicians most of whom were diplomats of the American Board of Sleep Medicine. As such, the clinical sample was generally representative of patients referred for sleep disorders specialist consultation, except that there was a bias against participants for whom polysomnography might not be indicated (eg, a bias against patients referred primarily for insomnia, restless legs complaints, or circadian rhythm phase disorders). Indeed, almost all patients for whom polysomnography was ordered carried a before-polysomnography diagnosis of some form of sleep apnea (whether the clinician thought the pretest probability high or low), perhaps influenced by clinician knowledge that other diagnoses were less likely to earn insurance preapproval for laboratory polysomnographic testing. As our sleep center is located at a department of chest medicine and staffed mainly by pulmonologists (not atypical), when the clinical focus was on sleep apnea, comorbid conditions such as periodic limb movement disorders, insomnia, or circadian rhythm disorders might not be explored with equal interest, and when comorbid with sleep apnea, these diagnoses might not be coded. The severe limitation of these procedures for research was the variability of polysomnographic recording conditions, especially recording duration, for measurement of polysomnographic parameters such as total sleep time or REM sleep percentage. The advantage of the strategy was that assessments of sleep-disordered breathing are often thought to have adequate clinical reliability on split nights [24,25], whereas the cost of performing a research recording for each participant would have added $1,000,000–$2,000,000 to research expenses and would have limited the sample as well. It is doubtful that as high a percentage of our patients would have agreed to recordings primarily for research purposes, and some already using CPAP would not have agreed to recordings without CPAP.
Even when performed for a research purpose, recordings using standard methods to monitor sleep-disordered breathing and periodic limb movements are known to produce discomforts and substantial sleep disturbances not usually experienced by patients in their home environments, although the home may produce disturbances as well. Single-night recordings do not provide highly repeatable measurements of total sleep time, sleep apnea, and periodic limb movements, whether performed on a clinical or research basis [26–28].
2.3. Phenotype assembly
From an enormous quantity and complexity of questionnaire, diagnostic, and polysomnographic data, the clinical data were prospectively distilled to 38 phenotype measures, focusing on complaints of insomnia and excessive daytime sleep, symptoms of depression, duration and time of day spent in bed, total subjective and polysomnographic sleep time, symptoms of restless legs [29], polysomnographic periodic leg movements, and several interrelated parameters of sleep apnea. Definitions of the phenotype variables are presented in the “Phenotype Definitions” worksheet of Supplement S1, which explains how each phenotype was coded and lists the numerical distributions of each phenotype. The text of the research questionnaire is attached as Supplement S2.
2.4. Genotyping
DNA was assayed primarily with a custom Illumina Golden Gate assay for 768 single nucleotide polymorphisms, designed as part of a multi-center collaboration [30]. Most targeted polymorphisms were selected for relevance to circadian rhythm regulation, but a smaller number were selected based on previous reports of association with sleep disorders [31]. Previous aspects of our research including detailed assay methods for this genotyping have been presented in prior reports, especially in two reports of associations of these same genotypes with circadian rhythm sleep disorders (delayed sleep phase and non-24-h sleep–wake cycles) and with mood symptoms [32–37]. A few single-nucleotide polymorphisms (SNPs) of special interest were assayed using additional Sequenom, SNPlex, and Taqman assays, as well as some of 27 ancestry-informative markers (AIMS) used to develop multidimensional scale (MDS) dimensions within participants of European ancestry [38]. The MDS components were used as regression covariates to control for ancestry stratification within the European ancestry portion of the sample (751 of European ancestry) and were usually useful in reducing genomic inflation of the regression results. Additional MDS components for all ancestries were derived to examine participants of all ancestries together or to examine those 86 of non-European ancestry separately. The latter included those of Asian, African, Native American, and mixed ancestries.
2.5. Statistical analyses
The PLINK whole genome analysis tool set [39] was used to compute linear or logistic regression analyses of the association between each phenotype and the available SNPs. The regression models were selected with a goal of maximizing validity. A primary goal was curtailing genomic inflation as much as possible, by using age, sex, and multidimensional scale factors as covariates to control stratification and by excluding violations of Hardy–Weinberg equilibrium or low minor allele frequencies. In some cases, eliminating one or more covariates reduced the genomic inflation. It appeared advisable at times to allow minor alleles with <1% frequency, two of which relatively rare variants appeared of considerable interest as reported below. In most cases, we only tested additive models a priori to avoid further inflation of the multiple-testing burden, but prior data on heritability suggested dominant models for the BALM and DSPS phenotypes [40,41]. Regressions for up to 626 SNPs were computed, requiring Bonferroni correction for multiple comparisons to obtain the family-wise error rates, in order to minimize false positives. We emphasize those associations with a phenotype which met a Bonferroni-corrected criterion of P < 0.05, but we also flag nominally significant P < 0.05 values, which should not be considered significant by the Bonferroni criteria. To obtain an overall perspective on the statistical outcomes considering the linkage of many of the SNPs and the correlations among many of the dependent phenotypes, we computed the q-values (false discovery rates) of all of the P values harvested from the 38 regressions combined, using the method and R statistical program with the default parameters of Storey and Tibshirani [42].
Because most of the participants had European ancestries, we focused on those participants to minimize stratification, as we generally did not find the MDS factors sufficient to control genomic inflation in samples of mixed ancestry, nor did adding non-Europeans to the European sample yield more powerful outcomes. The 86 samples of the non-European ancestry group were too few in themselves to expect adequate statistical power after Bonferroni correction.
3. Results
There were 13 Bonferroni-significant associations of SNPS with 11 phenotypes, which are presented in Table 1. This included three SNPs of the fragile X mental retardation 1 (FMR1) gene associated with a reduced QIDS-SR depression self-rating, which have been discussed in more detail elsewhere [36], but a somewhat different statistical model is presented here to provide perspective and comparison. Additional Bonferroni-significant results were as follows. Rs6888451 in peroxisome proliferator-activated receptor gamma, coactivator 1 beta (PPARGC1B) was associated with the number of obstructive apneas, with the obstructive apnea index, with the percentage of the sleep time that oxygen saturation was below 80%, and with the ODI3, which are all related phenotypes representing slightly different perspectives on obstructive sleep-disordered breathing. Rs3923809 of BTBD9 was inversely associated with the log transform of the leg movement index, loglmindexp1. Rs10766071 of aryl hydrocarbon receptor nuclear translocator-like (ARNTL) was associated with reduced polysomnographic total sleep time, tstpsg, in the recordings with tib >300 min. Rs11552085 in casein kinase 1 delta (CSNK1D) was associated with polysomnographic sleep latency, sleeplatpsg. The distribution of central apneas was highly skewed, with most patients scored as having none, so the linear regression of central apneas with retinoic acid receptor-related orphan receptor A (RORA) rs8025324 shown as Bonferroni significant was not evidence of reliable association (a Kruskal–Wallis nonparametric comparison of the three genotypes showed no significant association with central apneas). Rs11071547 in RORA was associated with questionnaire-reported sleep latency. Finally, rs4964515 in cryptochrome 1 (CRY1) was associated with the questionnaire-reported weekday time of falling asleep, fallasleep. The main results are presented in more detail in Supplement S1 along with additional results not achieving Bonferroni criteria of significance.
Table 1.
Linear regression P values for key phenotypes.
 |
Note: For definitions of the phenotypes, details of the regression models, regression results for additional phenotypes, and more detailed regression results, see Supplement 1. Beta (B) is the unstandardized regression coefficient (the same for all three Bonferroni-significant linked FMR1 SNPs). Blanks represent SNP associations not estimated by the models due to the low minor allele frequencies of rs4964515 and rs11552085 among Europeans and the impact of gender covariates on PLINK chromosome X analyses. Positive beta (B) values, flagged for nominal P < 0.05 associations by ↑ symbols, indicate that the minor allele was associated with increased phenotype quantity. Negative B values, flagged by ↓ symbols, indicate a minor allele associated with decreased phenotype quantity. The Bonferroni P is the probability of the association, conservatively adjusted for the number of SNPs tested within each regression.
The reliability of 13 Bonferroni-significant associations among regressions with a total of 38 phenotypes is difficult to appraise, considering that various SNPs were genetically linked and various phenotypes were intercorrelated. An overall perspective was offered by q false-discovery statistics, which estimate the chances of a false-positive discovery among multiple comparisons [43]. The overall q statistics were computed by compiling the P values for all of the SNP regressions for all phenotypes in combination, a total of 23,294 P estimates. The false-discovery estimates were as follows: for rs4964515 in CRY1, P = 1.99E-07 and q = 0.0045. For P values of 8.28E-06 to 1.09E-05, the risk of false discovery was estimated as q = 0.062. For P values of 2.62E-05 to 7.82E-05, q was estimated as 0.107–0.125. In other words, the Bonferroni-significant P values ranging from 8.28E-06 to 7.82E-05 had chances of roughly 6–13% of representing false discoveries. For P values of 0.000107–0.000895, q was estimated as 0.161–0.495. These estimates indicate that P values exceeding 0.000934 were >50% likely to be false discoveries, for example, a nominal P > 0.0208 was about 90% likely to be a false discovery. A graphic perspective is presented by Fig. 1, a quantile–quantile plot of the same 23,294 P values, which shows the observed P values below the Bonferroni criteria to be rather close to the random expectation. The Bonferroni-significant values were somewhat above expectation, but only the observed P value near 10−7 was impressively above the random expectation illustrated in the Q–Q plot.
Fig. 1.
Quantile–quantile plot of P values. The ranked P values of SNP associations with sleep phenotypes are plotted with x symbols for 23,294 P values. The values above the Bonferroni-corrected criteria for P < 0.05 (thick dashed line) were approximately one log unit above the random expectations (i.e., P values ~0.10 times random expectations). Only the single association for rs4964515 in CRY1 with the questionnaire-reported weekday time of falling asleep was substantially above the random trend (P = 1.99E10–7).
We examined the genotypes from the 86 subjects of non-European ancestry to investigate if any of the Bonferroni criteria associations shown in Table 1 were replicated among the non-Europeans, but none of the 13 specific associations met even a nominal P < 0.05 criterion for replication significance.
4. Discussion
A genetic survey of a heterogeneous group of sleep disorders patients yielded a substantial collection of findings, largely novel and surprising. Recruiting unselected patients undergoing polysomnograms for genetic studies appears to be a fruitful strategy. From the false-discovery statistical estimates, we would anticipate that the Bonferroni-significant SNPs in Table 1 include some false positives, but most of the Bonferroni-significant results might be true discoveries. Of course, most SNPs merely nominally significant in reality were likely to be “false discoveries.” Even Bonferroni-significant associations should be considered tentative until replicated by independent research groups, particularly since none of the associations could be replicated in our small sample of non-European ancestry.
Perhaps the most important result was the association of PPARGC1B rs6888451 with several measures reflecting obstructive sleep apnea. Of the 601 patients (83%) who had <10 obstructive apneas per hour, only 19% had one or two copies of the minor allele of rs6888451, but of the 17 (2%) who had >50 obstructive apneas per hour, 65% had one minor allele. As shown in Fig. 2, those with at least one minor G allele averaged almost twice as many total obstructive apneas as the homozygotes for the common allele. Likewise, those with one minor allele had almost twice the obstructive apneas per hour, 61% more oxygen desaturations per hour, and six times as many central apneas (NS). The less significant associations of rs6888451 with the number of central apneas and the AHI (the apnea–hypopnea index includes central apneas) might suggest that an obstructing apnea mechanism was mainly involved, such as airway anatomic compromise or collapsibility or inadequate airway dilator responses [44,45], or it might simply reflect that most patients had no central apneas scored. It was confirmatory that, besides the direct indices of obstructive apnea, the ODI3 (≥3% oxygen desaturations per hour) and the under 80% desaturation index (phenotype indices reflecting sleep-disordered oxygen desaturations) were also strongly associated with rs6888451. That the apnea score was associated with only weak nominal significance might suggest either a closer association of rs6888451 with objective measures of apnea than with clinician diagnostic formulations, or it might suggest that the logarithmic transform of ODI3 and AHI weaken the correlation by de-emphasizing the most severe cases of respiratory disturbances with which the rs6888451 minor allele was predominantly associated.
Fig. 2.
Association of rs6888451 with the number of obstructive apneas. The numbers of obstructive apneas per night (columns labeled with the numeric mean within each column) are plotted versus the genotypes of rs6888451. The numbers of participants having each genotype are in parentheses. The error bars show the 95% confidence intervals for the means: the confidence limits for the GG genotype were too large for contrasts versus either CG or CC genotypes to be reliable.
We had no a priori reason to expect that PPARGC1B would influence sleep apnea. The gene had been included in the SNP panel because of previous indications relating PPARGC1B to depression [36,46] and because PPARGC1B is a transcription factor binding at RRE sites of circadian genes, for example, weakly associated with delayed sleep phase and eveningness in a different sample [37]. An article reporting PPARGC1B rs7732671 associated (P = 0.004) with reduced obesity [47] was cited by NCBI OMIM, but we could not find replication of any obesity association with PPARGC1B in more recent very large genome-wide association studies (GWASs) [48]. We did not observe an association of rs6888451 either with rs7732671 or with weight and body mass index (BMI), and using BMI as a covariate in the regressions had no substantial effect on the association of rs6888451 with apnea variables. However, rs32574, an SNP 6643 nucleotides 3′ of rs7732671, would have met the Bonferroni criteria for association with body weight, had the P value not been corrected for genomic inflation. According to the Scripps Genome Adviser [49], rs6888451 is part of a SMAD3 transcription factor binding site. The University of Washington “Seattle SNPs” Genome Variation Server 138 and the SNAP Annotation and Proxy Search website [50] suggested nine additional common intronic SNPs with R2 = 1.0 linkage to rs6888451 and at least 13 other intronic SNPs with R2 >0.5, at least one of which is part of muscle initiator sequence 19 [49]. If the association of rs6888451 with sleep-disordered breathing can be replicated, it might thus require considerable molecular biology to identify which among the linked variants might be causal. Among the participants of non-European ancestry, although there were no significant associations with PPARGC1B, the four homozygotes for the rs6888451 minor allele tended to have the most respiratory disturbance, with the heterozygote intermediate.
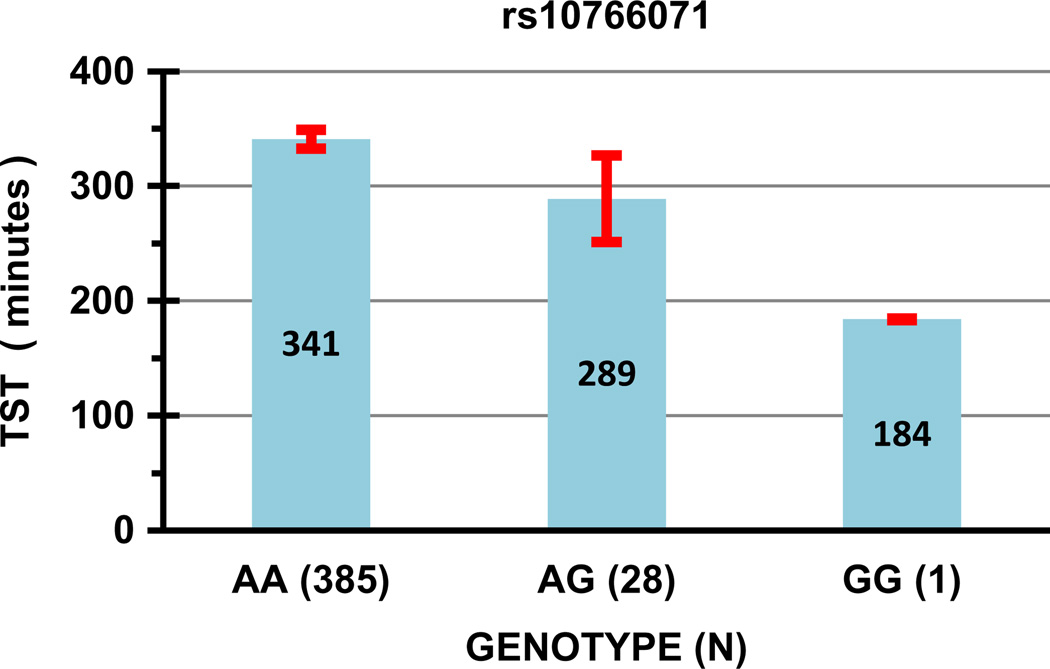
Another important group of findings was SNPs associated with short and disturbed sleep. The minor allele of ARNTL rs10766071 was significantly associated with shorter polysomnographic total sleep (Bonferroni P < 0.05) and nominally associated with increased sleep latency (both questionnaire and PSG), with delayed sleep-phase classification, with poor sleep efficiency, and with reduced REM sleep. See Fig. 3. The Scripps Genome Adviser [49] identified no specific functional role for this SNP, but it is perfectly linked (R2 = 1.0) with a nearby intronic linkage block, and with one promoter SNP, rs7126225. In a large GWAS [14], rs10766071 was significantly associated with questionnaire-reported home total sleep time (P = 0.043), but a number of other ARNTL SNPs including rs7126225 (P = 0.012) were more significantly associated, and the strongest association with home total sleep was with a potassium channel on chromosome 12, which happens to be rather near a homologue, ARNTL2. Our study might be thought a partial replication of the GWAS ARNTL results [14], except that our own similar questionnaire-reported home sleep time data were not significantly associated with ARNTL SNPs. When our polysomnographic data were limited to tib >300 min., tib was also nominally associated with rs10766071 (P = 0.0003), so it is uncertain whether there might be a mutual interaction of tib and tstpsg in this data set limited to tib >300. Possibly delayed sleep onset, as indicated by long sleep latencies and DSPS symptoms, caused both curtailed tib and tstpsg in interaction with laboratory scheduling of polysomnography. Interestingly, some RORA SNPs were nominally associated with questionnaire sleep duration in the GWAS [14] as were rs8025324, rs9806633, and rs12907550 in our study, but the SNPs highlighted in the GWAS supplement were not among those we assayed, and the RORA gene is so large that nominally significant false discoveries are to be expected. The association of RORA rs11071547 we observed with the subjective sleep latency met the Bonferroni criteria in our study, with nominally significant associations with leg movement indices and a restless legs complaint, hinting that rs11071547 may mediate prolonged sleep latency partly through restless legs and periodic limb movements. Nominally significant associations of different RORA SNPs with sleep duration were also reported by Utge et al. [51] along with Bonferroni-significant associations of subjective sleep duration (among females only) with an SNP in GRIA3. Suggestive association of SNPs with questionnaire sleep duration was also reported by Ollila et al. [52]. Since short and long sleep are associated with excess mortality, it is possible that some of these SNPs might be found to be mortality predictors. If so, more complex analyses based on Mendelian randomization of the SNP variables considered as instrumental variables influencing sleep duration might allow exploration of whether sleep duration variation in itself mediates the mortality with which it is associated.
Fig. 3.
Association of rs10766071 with polysomnographic total sleep time. Columns with the numeric mean within each column show the sleep time (TST) associated with each rs10766071 genotype. The ordinate is the minutes of polysomnographic sleep time, using only recordings with time in bed (tib) > 300 min. Error bars are the 95% confidence limits of the means. The horizontal axis specifies genotypes of rs10766071, with the number of DNA samples having each genotype in parenthesis.
As previously reported [53], BTBD9 rs3923809 was associated with the leg movement indices with Bonferroni significance. The less common allele is associated with fewer leg movements (Fig. 4). BTBD9 rs3923809 was also nominally associated with polysomnographic total sleep time, sleep efficiency, and REM latency, all of which associations might be mediated by periodic limb movements causing sleep disturbance. Our findings partially replicate the previous reports of Stefansson et al. [53] and Winkelmann, et al. [54]. However, we did not replicate any association of rs3923809 with restless legs, nor could we replicate an association of any of five MAP2K5 SNPs with restless legs [54], an outcome rather similar to that in the Wisconsin cohort [55]. One limitation of our method was that we did not isolate an RLS subsample focused on those with familial RLS [56]. But our strategy was directed towards SNPs that would impact RLS susceptibility in a sleep clinic sample in which familiality was not a criterion. An association of MEIS1 rs6710341 with restless legs [57] was confirmed by the replication criterion of P < 0.05, but the association of rs2300478 [13] was not replicated in the questionnaire data. In a supplementary logistic regression based on physician recorded research diagnoses of restless legs syndrome (not shown), of the nine SNPs of BTBD9, MEIS1, and MAP2K examined for replication, only MEIS1 rs2300478 reached nominal significance (P < 0.05).
Fig. 4.
Association of rs3923809 with the log10 periodic leg movement index. The association of rs3923809 genotypes with the leg movement index (log scale) is shown. The ordinate is the loglmindexp1, that is, LOG10[lmindex + 1], where lmindex is the leg movements per hour of sleep (plus 1). The error bars show the 95% confidence limits of the mean loglmindexp1. The horizontal axis specifies genotypes of rs3923809, with the number of DNA samples having each genotype in parenthesis.
Two other SNPs related to phenotypes of disturbed sleep were CSNK1D rs11552085 and CRY1 rs4964515. Both were nominally associated with reduced subjectively reported total sleep time, with subjective insomnia, and with later reported clock times to fall asleep both on weekdays and on weekends. In CSNK1D, heterozygotes (N = 8) with the minor allele of rs11552085 were associated with 3.37 times the polysomnographic sleep latency (Bonferroni P < 0.02), but the five participants of non-European ancestry with one or two minor alleles did not have longer sleep latencies. For CRY1 rs4964515, the Bonferroni P = 1.99E-07 for reported time falling asleep was the smallest P value obtained in all the analyses, and as mentioned above, this was the only association with a false-discovery probability of q = 0.0045, suggesting statistical reliability. As shown in Fig. 5, this highly significant contrast was derived largely from one outlying participant’s very late reported time of falling asleep on weekdays (among only 11 with the minor allele), as the reported time of falling asleep on weekends was several orders of magnitude less significant, and the polysomnographic sleep latency was not associated with CRY1 SNPs at all. The Mann–Whitney rank-order contrast of the two groups was significant only at the P = 0.0001 level. Moreover, our sample of non-European ancestry had included 23 participants with an rs4964515 minor allele, and the minor allele had no significant association with any phenotype. No indication of a similar association of rs4964515 with delayed falling asleep was found in our companion sample of DSPS cases and controls [37]. For these reasons, despite the very small estimated q value, we must be quite skeptical that this rs4964515 association was a true positive until replicated, and the same is true of the rs11552085 findings. It is notable that neither rs11552085 nor rs4964515 showed evidence of DSPS on the BALM scale, so the meaning of a delayed sleep onset would be uncertain.
Fig. 5.
CRY1 rs4964515 versus subjective time when falling asleep. Questionnaire-reported time to fall asleep (ordinate) is plotted against the rs4964515 AG genotype (group at left) and the GG genotype (group at right), both groups spread out for better visualization. Although the median time of reported falling asleep was certainly later for the AG genotype with one rare allele, the linear regression P = 2.0E-07 value was strongly influenced by one outlying point. The Mann–Whitney rank-order contrast of the two groups was significant only at the P = 0.0001 level, which would not meet Bonferroni criteria.
The significant protective association of FMR1 SNPs with the QIDS-SR depression scale has been more extensively discussed in a previous paper [36], but here we note the interesting nominally significant protective associations of the same SNPs with excessive sleepiness as indicated by decreased ESS. From a genetic viewpoint, it might appear that depression might be the main mechanism of reported daytime sleepiness in this Sleep Center sample. Genotypes aside, we have similarly found the ESS to be more closely correlated with the QIDS-SR than with any apnea, RLS, or leg movement index (data not presented), a finding resembling previous reports of Bardwell et al. [58]. Similarly, the insomnia scale (which was derived from the first three items of the QIDS-SR) was also associated with the three FMR1 SNPs.
To highlight the variability of clinical samples and the risks of false discovery, it is worth reviewing some pertinent negative results of this study. For example, Gottlieb et al. confirmed the heritability of sleep disorder phenotypes in Framingham data; however, using FBAT statistics, which are not subject to genomic inflation on Framingham GWAS data, they located no SNP associated with a sleep phenotype with Bonferroni significance after correcting for multiple-testing inflation [31]. The one SNP association meeting Bonferroni significance with population-based statistics, rs1823068, did not replicate at all in our data (see Supplement S1.). Some tentative associations (inconsistent in two races and of questionable statistical reliability) have been reported for obstructive sleep apnea [59,60], but a recent review found apnea-association reports that could be meta-analyzed only for angiotensin I converting enzyme (ACE), apolipoprotein E (APOE4) [33,61], and tumor necrosis factor alpha (TNFalpha) [62–65], and only TNFalpha was supported by meta-analysis (possibly mediated by associations with heart failure). We could not confirm associations of APOE or TNFalpha in our data. The dominant polymorphism in period circadian clock 2 (PER2) associated with advanced sleep phase disorder (ASPD) seems quite rare [66,67], and we did not find it. A much-studied polymorphism reported to be associated with DSPD, the C3111T SNP in CLOCK (rs1801260) [68], was not a significant correlate of DSPD or morningness–eveningness in the sleep center data [37]. We did confirm some previous associations of periodic leg movements and RLS with rs3923809 and rs6710341 while failing to confirm other reported associations. In one report, a larger number of SNPs from three genes appeared to explain more than half of the total RLS risk [57]. In an expansion of that study examining the association of RLS with six genetic risk loci, the explained genetic variance was estimated at 6.8% [13,69].
5. Conclusions
In summary, this report introduces valuable clues to sleep disorders, which might be pursued by greatly expanding genetic research in patients studied in the sleep center clinical setting. Previous experience emphasizes the importance of independent replications of our results before their value can be confirmed in clinical application.
Supplementary Material
Acknowledgments
This study was supported by Scripps Clinic Academic Funds and by U.S. National Institutes of Health grants 5 UL1 RR025774 and R01AG030474. Katharine M. Rex contributed to human subject administration. Tatyana Shekhtman designed and performed the SNPlex and Taqman assays. Kep Wadiak, RPSGT, scored most of the polysomnograms.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.sleep.2014.11.003.
Footnotes
Conflict of interest
The ICMJE Uniform Disclosure Form for Potential Conflicts of Interest associated with this article can be viewed by clicking on the following link: http://dx.doi.org/10.1016/j.sleep.2014.11.003.
The authors declare that they have no competing financial interests.
References
- 1.Kripke DF, Ancoli-Israel S, Klauber MR, Wingard DL, Mason WJ, Mullaney DJ. Prevalence of sleep disordered breathing in ages 40–64 years: a population-based survey. Sleep. 1997;20:65–76. doi: 10.1093/sleep/20.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ancoli-Israel S, Kripke DF, Klauber MR, Mason WJ, Fell R, Kaplan O. Sleep disordered breathing in community-dwelling elderly. Sleep. 1991;14(6):486–495. doi: 10.1093/sleep/14.6.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurina LM, McClintock MK, Chen JH, Waite LJ, Thisted RA, Lauderdale DS. Sleep duration and all-cause mortality: a critical review of measurement and associations. Ann Epidemiol. 2013;23:361–370. doi: 10.1016/j.annepidem.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kripke DF, Garfinkel L, Wingard DL, Klauber MR, Marler MR. Mortality associated with sleep duration and insomnia. Arch Gen Psychiatry. 2002;59:131–136. doi: 10.1001/archpsyc.59.2.131. [DOI] [PubMed] [Google Scholar]
- 5.Kripke DF, Brunner R, Freeman R, Hendrix S, Jackson RD, Masaki K, et al. Sleep complaints of postmenopausal women. Clin J Womens Health. 2001;1:244–252. doi: 10.1053/cjwh.2001.30491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohayon MM, Reynolds CF, III, Dauvilliers Y. Excessive sleep duration and quality of life. Ann Neurol. 2013;73:785–794. doi: 10.1002/ana.23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wang W, Winkelman JW, Malhotra A, Ma J, Gao X. Prospective study of restless legs syndrome and mortality among men. Neurology. 2013;81:52–59. doi: 10.1212/WNL.0b013e318297eee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rye DB, Trotti LM. Restless legs syndrome and periodic leg movements of sleep. Neurol Clin. 2012;30:1137–1166. doi: 10.1016/j.ncl.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Asarnow LD, McGlinchey E, Harvey AG. The effects of bedtime and sleep duration on academic and emotional outcomes in a nationally representative sample of adolescents. J Adolesc Health. 2014;54:350–356. doi: 10.1016/j.jadohealth.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxvig IW, Pallesen S, Wilhelmsen-Langeland A, Molde H, Bjorvatn B. Prevalence and correlates of delayed sleep phase in high school students. Sleep Med. 2012;13:193–199. doi: 10.1016/j.sleep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Gale C, Martyn C. Larks and owls and health, wealth, and wisdom. BMJ. 1998;317:1675–1677. doi: 10.1136/bmj.317.7174.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tafti M, Hor H, Dauvilliers Y, Lammers GJ, Overeem S, Mayer G, et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulte EC, Kousi M, Tan PL, Tilch E, Knauf F, Lichtner P, et al. Targeted resequencing and systematic in vivo functional testing identifies rare variants in MEIS1 as significant contributors to restless legs syndrome. Am J Hum Genet. 2014;95:85–95. doi: 10.1016/j.ajhg.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RVDM, et al. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. 2013;18:122–132. doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- 15.Barclay NL, Gregory AM. Quantitative genetic research on sleep: a review of normal sleep, sleep disturbances and associated emotional, behavioural, and health-related difficulties. Sleep Med Rev. 2013;17:29–40. doi: 10.1016/j.smrv.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Byrne EM, Gehrman PR, Medland SE, Nyholt DR, Heath AC, Madden PA, et al. A genome-wide association study of sleep habits and insomnia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:439–451. doi: 10.1002/ajmg.b.32168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faraco J, Lin L, Kornum BR, Kenny EE, Trynka G, Einen M, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9:e1003270. doi: 10.1371/journal.pgen.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw P, Tafti M, Thorpy M. Genetic basis of sleep and sleep disorders. West Nyack, NY, USA: Cambridge University Press; 2013. [Google Scholar]
- 19.Allen RP, Picchietti DL, Garcia-Borreguero D, Ondo WG, Walters AS, Winkelman JW, et al. Restless legs syndrome/Willis-Ekbom disease diagnostic criteria: updated International Restless Legs Syndrome Study Group (IRLSSG) consensus criteria – history, rationale, description, and significance. Sleep Med. 2014;15:860–873. doi: 10.1016/j.sleep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 20.Johns MW. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 1991;14:540–545. doi: 10.1093/sleep/14.6.540. [DOI] [PubMed] [Google Scholar]
- 21.Brown FM. Psychometric equivalence of an improved Basic Language Morningness (BALM) Scale using industrial population within comparisons. Ergonomics. 1993;36:191–197. doi: 10.1080/00140139308967872. [DOI] [PubMed] [Google Scholar]
- 22.Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN, et al. The 16-item Quick Inventory of Depressive Symptomatology (QIDS), clinical rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry. 2003;54:573–583. doi: 10.1016/s0006-3223(02)01866-8. [DOI] [PubMed] [Google Scholar]
- 23.Winkelman J, Kotagal S, Olson E, Scammel T, Schenck C, Spielman A. The international classification of sleep disorders, second edition, pocket version: diagnostic and coding manual. Westchester, IL: American Academy of Sleep Disorders; 2006. [Google Scholar]
- 24.Khawaja IS, Olson EJ, van derWalt C, Bukartyk J, Somers V, Dierkhising R, et al. Diagnostic accuracy of split-night polysomnograms. J Clin Sleep Med. 2010;6:357–362. [PMC free article] [PubMed] [Google Scholar]
- 25.Patel NP, Ahmed M, Rosen I. Split-night polysomnography. Chest. 2007;132:1664–1671. doi: 10.1378/chest.06-1801. [DOI] [PubMed] [Google Scholar]
- 26.Moses JM, Lubin A, Naitoh P, Johnson LC. Reliability of sleep measures. Psychophysiology. 1972;9:78–82. doi: 10.1111/j.1469-8986.1972.tb00745.x. [DOI] [PubMed] [Google Scholar]
- 27.Mosko SS, Dickel MJ, Ashurst J. Night-to-Night variability in sleep apnea and sleep in sleep-related periodic leg movements in the elderly. Sleep. 1988;11:340–348. [PubMed] [Google Scholar]
- 28.Iber C, Redline S, Kaplan Gilpin AM, Quan SF, Zhang L, Gottlieb DJ, et al. Polysomnography performed in the unattended home versus the attended laboratory setting – Sleep Heart Healthy Study methodology. Sleep. 2004;27:536–540. doi: 10.1093/sleep/27.3.536. [DOI] [PubMed] [Google Scholar]
- 29.Gehrman PR, Pfeiffenberger C, Byrne E. The role of genes in the insomnia phenotype. Sleep Med Clin. 2013;8:323–331. doi: 10.1016/j.jsmc.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans DS, Parimi N, Nievergelt CM, Blackwell T, Redline S, Ancoli-Israel S, et al. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. Sleep. 2013;36:431–446. doi: 10.5665/sleep.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl. 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kripke DF, Rex KM, Ancoli-Israel S, Nievergelt CM, Klimecki W, Kelsoe JR. Delayed sleep phase cases and controls. J Circadian Rhythms. 2008;6:1. doi: 10.1186/1740-3391-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kripke DF, Shadan FF, Dawson A, Cronin JW, Jamil SM, Grizas AP, et al. Genotyping sleep disorders patients. Psychiatry Investig. 2010;7:36–42. doi: 10.4306/pi.2010.7.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kripke DF, Nievergelt CM, Tranah GJ, Murray SS, McCarthy MJ, Rex KM, et al. Polymorphisms in melatonin synthesis pathways: possible influences on depression. J Circadian Rhythms. 2011;9:8. doi: 10.1186/1740-3391-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee MK, Lee HJ, Rex KM, Kripke DF. Evaluation of two circadian rhythm questionnaires for screening for the delayed sleep phase disorder. Psychiatry Investig. 2012;9:236–244. doi: 10.4306/pi.2012.9.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kripke DF, Nievergelt CM, Tranah GJ, Murray SS, Rex KM, Grizas AP, et al. FMR1, circadian genes and depression: suggestive associations or false discovery? J Circadian Rhythms. 2013;11:3. doi: 10.1186/1740-3391-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kripke DF, Klimecki WT, Nievergelt CM, Rex KM, Murray SS, Shekhtman T, et al. Circadian polymorphisms in night owls, in bipolars, and in non-24-hour sleep cycles. Psychiatry Investig. 2014;11:345–362. doi: 10.4306/pi.2014.11.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nievergelt CM, Maihofer AX, Shekhtman T, Libiger O, Wang X, Kidd KK, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig Genet. 2013;4:13. doi: 10.1186/2041-2223-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barclay NL, Eley TC, Buysse DJ, Archer SN, Gregory AM. Diurnal preference and sleep quality: same genes? A study of young adult twins. Chronobiol Int. 2010;27:278–296. doi: 10.3109/07420521003663801. [DOI] [PubMed] [Google Scholar]
- 41.Koskenvuo M, Hublin C, Partinen M, Heikkila K, Kaprio J. Heritability of diurnal type: a nationwide study of 8753 adult twin pairs. J Sleep Res. 2007;16:156–162. doi: 10.1111/j.1365-2869.2007.00580.x. [DOI] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 44.Wellman A, Eckert DJ, Jordan AS, Edwards BA, Passaglia CL, Jackson AC, et al. A method for measuring and modeling the physiological traits causing obstructive sleep apnea. J Appl Physiol. 2011;110:1627–1637. doi: 10.1152/japplphysiol.00972.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckert DJ, White DP, Jordan AS, Malhotra A, Wellman A. Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets. Am J Respir Crit Care Med. 2013;188:996–1004. doi: 10.1164/rccm.201303-0448OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kripke DF, Nievergelt CM, Joo EJ, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms. 2009;7:2. doi: 10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen G, Wegner L, Yanagisawa K, Rose CS, Lin J, Glumer C, et al. Evidence of an association between genetic variation of the coactivator PGC-1beta and obesity. J Med Genet. 2005;42:402–407. doi: 10.1136/jmg.2004.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scripps Translational Science Institute; [accessed 2013]. Torkamani A Scripps genome adviser: annotation and distributed variant interpretation server. < http://genomics.scripps.edu/adviser/>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Utge S, Kronholm E, Partonen T, Soronen P, Ollila HM, Loukola A, et al. Shared genetic background for regulation of mood and sleep: association of GRIA3 with sleep duration in healthy Finnish women. Sleep. 2011;34:1309–1316. doi: 10.5665/SLEEP.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ollila HM, Kettunen J, Pietilainen O, Aho V, Silander K, Kronholm E, et al. Genome-wide association study of sleep duration in the Finnish population. J Sleep Res. 2014 doi: 10.1111/jsr.12175. [DOI] [PubMed] [Google Scholar]
- 53.Stefansson H, Rye DB, Hicks A, Petursson H, Ingason A, Thorgeirsson TE, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 54.Winkelman JW. Periodic limb movements in sleep–endophenotype for restless legs syndrome? N Engl J Med. 2007;357:703–705. doi: 10.1056/NEJMe078129. [DOI] [PubMed] [Google Scholar]
- 55.Moore H, Winkelmann J, Lin L, Finn L, Peppard P, Mignot E. Periodic leg movements during sleep are associated with polymorphisms in BTBD9, TOX3/BC034767, MEIS1, MAP2K5/SKOR1, and PTPRD. Sleep. 2014;37:1535–1542. doi: 10.5665/sleep.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kemlink D, Polo O, Frauscher B, Gschliesser V, Hogl B, Poewe W, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. 2009;46:315–318. doi: 10.1136/jmg.2008.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 58.Bardwell WA, Moore P, Ancoli-Israel S, Dimsdale JE. Fatigue in obstructive sleep apnea: driven by depressive symptoms instead of apnea severity? Am J Psychiatry. 2003;160:350–355. doi: 10.1176/appi.ajp.160.2.350. [DOI] [PubMed] [Google Scholar]
- 59.Larkin EK, Patel SR, Goodloe RJ, Li Y, Zhu X, Gray-McGuire C, et al. A candidate gene study of obstructive sleep apnea in European-Americans and African-Americans. Am J Respir Crit Care Med. 2010;182:947–953. doi: 10.1164/rccm.201002-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel SR, Goodloe R, De G, Kowgier M, Weng J, Buxbaum SG, et al. Association of genetic loci with sleep apnea in European Americans and African-Americans: the Candidate Gene Association Resource (CARe) PLoS ONE. 2012;7:e48836. doi: 10.1371/journal.pone.0048836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thakre TP, Mamtani MR, Kulkarni H. Lack of association of the APOE epsilon4 allele with the risk of obstructive sleep apnea: meta-analysis and meta-regression. Sleep. 2009;32:1507–1511. doi: 10.1093/sleep/32.11.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waters KA, Mast BT, Vella S, De La Eva R, O’Brien LM, Bailey S, et al. Structural equation modeling of sleep apnea, inflammation, and metabolic dysfunction in children. J Sleep Res. 2007;16:388–395. doi: 10.1111/j.1365-2869.2007.00614.x. [DOI] [PubMed] [Google Scholar]
- 63.Riha RL, Brander P, Vennelle M, McArdle N, Kerr SM, Anderson NH, et al. Tumour necrosis factor-{alpha} (−308) gene polymorphism in obstructive sleep apnoea-hypopnoea syndrome. Eur Respir J. 2005;26:673–678. doi: 10.1183/09031936.05.00130804. [DOI] [PubMed] [Google Scholar]
- 64.Imagawa S, Yamaguchi Y, Ogawa K, Obara N, Suzuki N, Yamamoto M, et al. Interleukin-6 and tumor necrosis factor-alpha in patients with obstructive sleep apnea-hypopnea syndrome. Respiration. 2004;71:24–29. doi: 10.1159/000075645. [DOI] [PubMed] [Google Scholar]
- 65.Vgontzas AN, Papanicolaou DA, Bixler EO, Kales A, Tyson K, Chrousos GP. Elevation of plasma cytokines in disorders of excessive daytime sleepiness: role of sleep disturbance and obesity. J Clin Endocrinol Metab. 1997;82:1313–1316. doi: 10.1210/jcem.82.5.3950. [DOI] [PubMed] [Google Scholar]
- 66.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, et al. An hPer2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 68.Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, et al. A clock polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 69.Winkelmann J, Czamara D, Schormair B, Knauf F, Schulte EC, Trenkwalder C, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility Loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.