Abstract
Long-term in vivo expression of a broad and potent entry inhibitor could circumvent the need for a conventional vaccine for HIV-1. Adeno-associated virus (AAV) vectors can stably express HIV-1 broadly neutralizing antibodies (bNAbs)1,2. However even the best bNAbs neutralize 10–50% of HIV-1 isolates inefficiently (IC80 > 5 μg/ml), suggesting that high concentrations of these antibodies would be necessary to achieve general protection3–6. Here we show that eCD4-Ig, a fusion of CD4-Ig with a small CCR5-mimetic sulfopeptide, binds avidly and cooperatively to the HIV-1 envelope glycoprotein (Env) and is more potent than the best bNAbs (geometric mean IC50 < 0.05 μg/ml). Because eCD4-Ig binds only conserved regions of Env, it is also much broader than any bNAb. For example, eCD4-Ig efficiently neutralized 100% of a diverse panel of neutralization-resistant HIV-1, HIV-2, and SIV isolates, including a comprehensive set of isolates resistant to the CD4-binding site bNAbs VRC01, NIH45-46, and 3BNC117. Rhesus macaques inoculated with an AAV vector stably expressed 17 to 77 μg/ml of fully functional rhesus eCD4-Ig for 40 weeks, and these macaques were protected from multiple infectious challenges with SHIV-AD8. Rhesus eCD4-Ig was also markedly less immunogenic than rhesus forms of four well characterized bNAbs. Our data suggest that AAV-delivered eCD4-Ig can function like an effective HIV-1 vaccine.
Rhesus macaques inoculated with an AAV-based gene-therapy vector express antibody-like immunoadhesins for years, and these immunoadhesins afforded partial protection from a neutralization-sensitive simian immunodeficiency virus (SIV)2, suggesting that long-term sterilizing protection from HIV-1 might be achievable without a conventional vaccine. Full-length AAV-expressed bNAbs also protected humanized mice from an HIV-1 challenge1,7. However a large fraction of HIV-1 isolates remain partially or wholly resistant to even the best bNAbs, with IC80s greater than 5 μg/ml measured under optimal in vitro conditions (Extended Data Table 1)3–6. Higher concentrations will likely be necessary for broad-based protection in vivo, but primate studies suggest that these concentrations will be difficult to establish in humans2,8. An effective AAV-based vaccine may therefore require broader and more potent inhibitors of HIV-1 entry.
The breadth of an antibody depends on the conservation of its epitope. The two most conserved epitopes of HIV-1 Env are its CD4- and coreceptor-binding sites9–11. The immunoadhesin form of CD4, CD4-Ig, has been extensively studied as a therapeutic. It neutralizes most isolates, irreversibly inactivates Env, and is demonstrated safe for use in humans12–15. However, its affinity for Env is lower than those of bNAbs16, and its potency is further compromised by its parallel ability to promote infection17. Mimetics of the primary HIV-1 coreceptor CCR5, in particular peptides based on its tyrosine-sulfated amino-terminus, have also been characterized18,19. These sulfopeptides bind Env specifically but with low affinity in the absence of CD4, in part because they include hydrophobic residues and O-linked glycosylation that impede their association with Env18,20. CCR5mim1, a 15-amino acid sulfopeptide derived from the HIV-1 neutralizing antibody E5121, lacks these interfering elements (Fig. 1a) and binds Env with higher affinity than CCR5-based peptides20,22. Reflecting the conservation of the sulfotyrosine-binding pockets of Env9,10, CCR5mim1 binds both CCR5- and CXCR4-dependent Envs from all HIV-1 clades20,22.
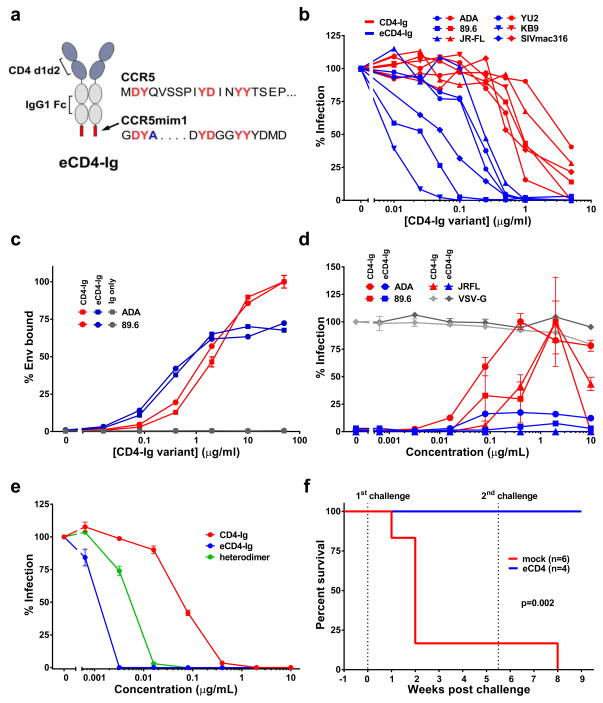
Figure 1. Functional characterization of eCD4-Ig.
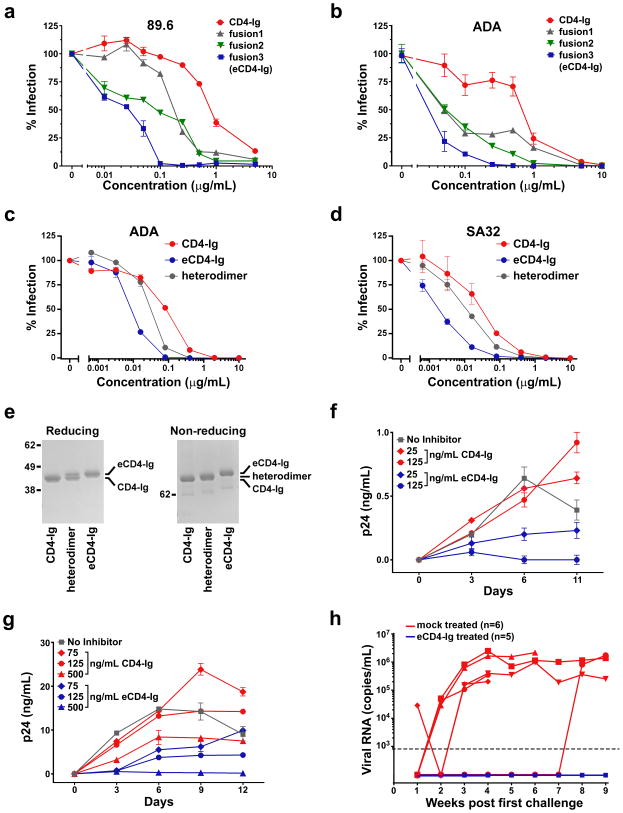
a, CD4-Ig is comprised of CD4 domains 1 and 2 (blue) fused to the human IgG1 Fc domain (grey). In eCD4-Ig, the sulfopeptide CCR5mim1 is fused to the carboxy-terminus of CD4-Ig. The sequence of the CCR5 amino terminus is provided for comparison. Common residues, including four CCR5 sulfotyrosines, are shown in red. CCR5mim1 alanine 4 (blue) is substituted with tyrosine in CCR5mim2, described below. b, HIV-1 pseudotyped with the Envs of the indicated HIV-1 or SIV isolates was incubated with GHOST-CCR5 cells and varying concentrations of CD4-Ig (red) or eCD4-Ig (blue). Infection was measured as GFP-expression by flow cytometry. Errors of replicates are less than 20% of indicated values but not indicated for clarity. c, 293T cells transfected to express 89.6 or ADA Envs were incubated with the indicated concentrations of CD4-Ig (red), eCD4-Ig (blue), or IgG (grey) and analyzed by flow cytometry. d, HIV-1 expressing luciferase and pseudotyped with the Envs of the indicated isolates was incubated with Cf2Th-CCR5 cells in the presence of varying concentrations of CD4-Ig (red) or eCD4-Ig (blue). Experiment was controlled with HIV-1 pseudotyped with the VSV-G protein (grey). Infection normalized to the maximum value observed for each pseudovirus. e, HIV-1 pseudotyped with the 89.6 Env was incubated with TZM-bl cells and varying concentration of CD4-Ig (red), eCD4-Ig (blue), or a CD4-Ig/eCD4-Ig heterodimer (grey). Similar experiments using additional Envs are presented in Extended Data Figs. 2c and d. f, Infection curves of humanized NSG mice with 2–4 mg/ml of serum eCD4-Ig at time of HIV-1NL4-3 challenges (blue line, n = 5), or mock treated (red line, n = 6) are shown. Three uninfected eCD4-Ig treated mice and the sole uninfected mock treated mouse were rechallenged 5 weeks post-first challenge. Significant protection (p=0.002; Mantel-Cox test) was observed in the eCD4-Ig treated group. Viral load measurements are shown in Extended Data Fig. 2h. Experiments shown in panels b-e were performed at least twice with each indicated isolate with similar results. Errors bars of duplicates denote one s.e.m.
We reasoned that a fusion of CD4-Ig and CCR5mim1 would bind Env cooperatively and with higher avidity than either molecule alone. Accordingly, three fusion proteins were generated (sequences in Extended Data Fig. 1). CCR5mim1 was inserted at either the CD4-Ig amino-terminus (fusion 1), between the CD4 and Fc domain (fusion 2), or at the CD4-Ig carboxy terminus (fusion 3, renamed eCD4-Ig). All three CD4-Ig variants neutralized CCR5- and CXCR4-dependent isolates more efficiently than did CD4-Ig, with eCD4-Ig consistently the most potent (Extended Data Fig. 2a and b). eCD4-Ig neutralized a wider panel of HIV-1 isolates and SIVmac316 with 10- to 100-fold lower IC50s than CD4-Ig (Fig. 1b). Improved neutralization of SIVmac316 is consistent with conservation of the Env’s sulfotyrosine-binding pockets 9,10, and a first indication of the exceptional breadth of eCD4-Ig.
To better understand the markedly greater potency of eCD4-Ig relative to CD4-Ig, we compared their abilities to bind cell-surface expressed Env trimers (Fig. 1c). At low concentrations, eCD4-Ig bound these trimers more efficiently than did CD4-Ig. Surprisingly, eCD4-Ig saturated trimer-expressing cells with approximately one-third less bound protein than CD4-Ig, suggesting that eCD4-Ig’s sulfopeptides made some CD4-binding sites inaccessible. eCD4-Ig also less efficiently promoted HIV-1 infection of CCR5-positive, CD4-negative cells than CD4-Ig (Fig. 1d), presumably because its sulfopeptides blocked virion access to cell-surface CCR5. CD4-Ig/eCD4-Ig heterodimers23 neutralized less potently than eCD4-Ig (Fig 1e; Extended Data Figs. 2c–e), indicating that both eCD4-Ig sulfopeptides engage the Env trimer, consistent with a model of eCD4-Ig bound to Env (Extended Data Fig. 3) and previous studies of CCR5mim124. Thus the markedly greater potency of eCD4-Ig relative to CD4-Ig is due in part to the higher avidity with which it binds Env and to its decreased ability to promote infection.
We next assessed eCD4-Ig under more physiological conditions. We observed that eCD4-Ig, but not CD4-Ig, halted replication of infectious viruses in human PBMC at concentrations as low as 125 ng/mL (Extended Data Figs. 1f and g). We administered sufficient eCD4-Ig to humanized NOD/SCID/γc (NSG) mice to maintain serum concentrations of 2–4 μg/ml at the time of challenge. Five eCD4-Ig treated mice and six control mice were challenged intravenously with 5×104 infectious units of HIV-1NL4-3. Five of six control mice, but no eCD4-Ig inoculated mice, were infected (Figs. 1h; Extended Data Fig. 2h). Five weeks later, three eCD4-Ig-treated mice and the uninfected control mouse were rechallenged. Again, no eCD4-Ig-treated mouse was infected, whereas the control mouse became infected.
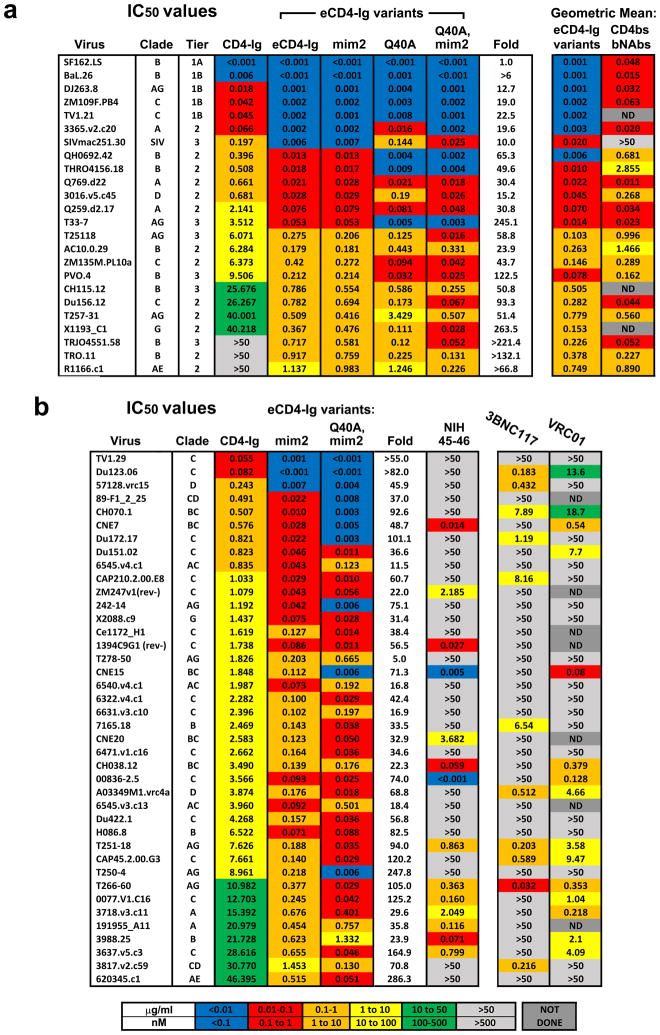
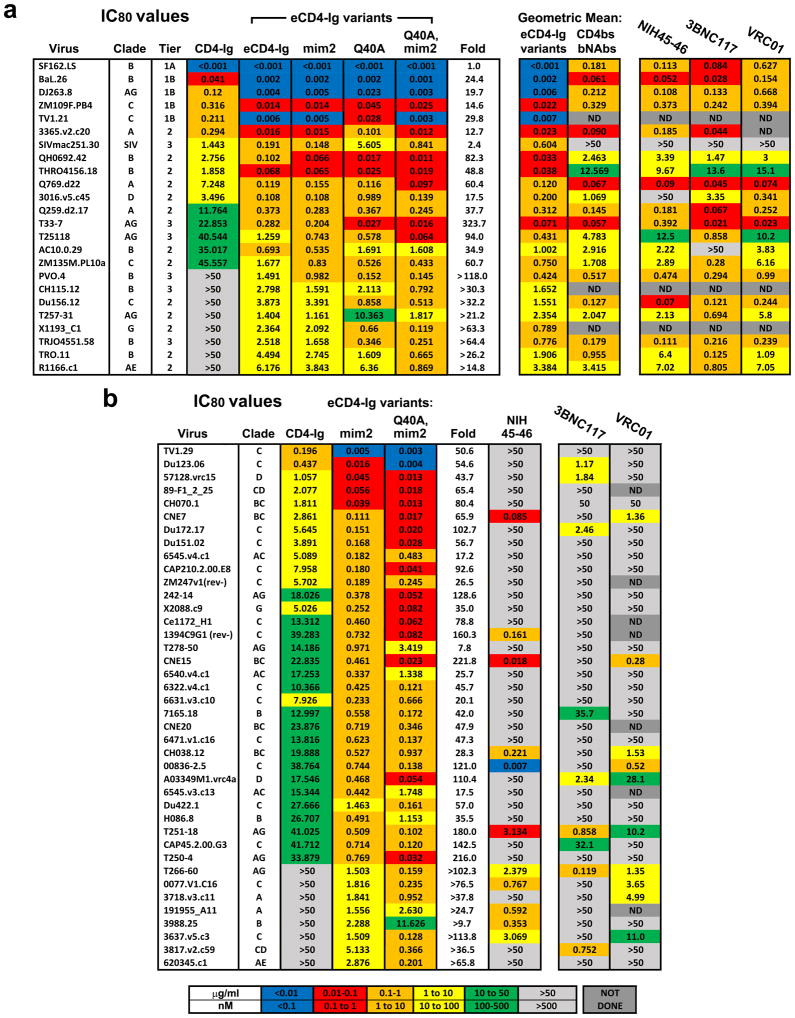
We then characterized the ability of eCD4-Ig to neutralize a diverse panel of neutralization resistant tier 2 and 3 viruses25 (Extended Data Figs. 4a and 5a). In parallel, we assayed three additional eCD4-Ig variants. In the first, eCD4-Igmim2, CCR5mim1 was replaced by CCR5mim2, which differs from CCR5mim1 by single alanine to tyrosine substitution22. We also introduced a previously characterized glutamine 40 to alanine mutation into CD4 domain 1 of eCD4-Ig (eCD4-IgQ40A)16. Both mutations were combined in a final variant (eCD4-IgQ40A,mim2). eCD4-Ig and these variants substantially outperformed CD4-Ig for every virus in the panel, typically improving neutralization potency by 20 to >200-fold. Underscoring its breadth, eCD4-Ig neutralized SIVmac251 33 times more efficiently than CD4-Ig. In general, the more neutralization resistant a virus, the better eCD4-Ig and its variants performed relative to CD4-Ig. In most cases, replacement of CCR5mim1 with CCR5mim2 modestly improved neutralization. Similarly, the Q40A mutation also improved neutralization of most HIV-1 isolates, but not of SIVmac251.
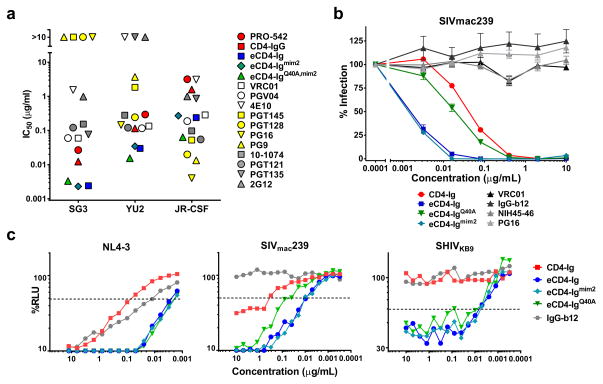
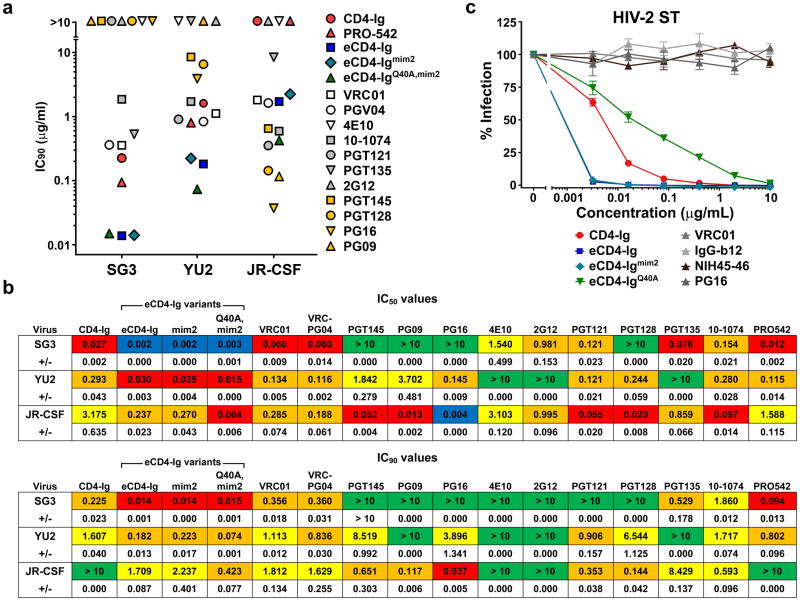
We compared eCD4-Ig, eCD4-Igmim2 and eCD4-IgQ40A,mim2 with a panel of 12 antibodies and inhibitors using three additional HIV-1 isolates (Fig. 2a; Extended Data Fig. 6a and b). eCD4-Ig and its variants neutralized the SG3 and YU2 isolates more efficiently than any of these inhibitors. Five bNAbs neutralized JR-CSF more efficiently than any eCD4-Ig variant, but four of these could not neutralize SG3. All eCD4-Ig variants neutralized these isolates with IC50s less than 0.3 μg/ml, more efficiently than CD4-Ig, the tetrameric CD4-Ig variant PRO-54212,14, or the antibodies 2G12, 4E10, and VRC01. eCD4-Ig and its variants, but not three CD4-binding site bNAbs, neutralized the neutralization-resistant SIVmac239 as well as HIV-2 ST (Fig. 2b; Extended Data Fig. 6c). As observed with SIVmac251, the Q40A variant was less efficient at neutralizing SIVmac239 and HIV-2. The potency of these eCD4-Ig variants was also reflected in their abilities to mediate antibody-dependent cell-mediated cytotoxicity (ADCC). eCD4-Ig, eCD4-Igmim2, and eCD4-IgQ40A,mim2 each facilitated 30–40 times more killing of infected cells by CD16+ natural killer cells26 than did CD4-Ig or the antibody IgGb12 (Fig. 2c). Thus the carboxy-terminal modification of eCD4-Ig did not interfere with the ADCC effector function of its Fc domain.
Figure 2. Comparison of eCD4-Ig variants and HIV-1 neutralizing antibodies.
a, HIV-1 pseudotyped with the Envs of the indicated isolates were incubated with TZM-bl cells and varying concentrations of the indicated entry inhibitors, and the resulting IC50 values are plotted. IC90 values and standard errors are presented in Extended Data Figs. 6a and 6b. b, Experiments similar to those in (a) except that HIV-1 pseudotyped with the SIVmac239 Env was incubated with varying concentrations of CD4-Ig, eCD4-Ig variants, or CD4bs antibodies. Extended Data Fig. 6c shows a similar study using the HIV-2 ST Env. Errors bars of triplicates denote one s.e.m. of triplicates. c, ADCC activity was assessed using CEM.NKR-CCR5 target cells incubated with infectious HIV-1 NL4-3, SHIVKB9 or SIVmac239 for four days. Cells were then incubated with KHYG-1 NK effector cells26 for 8 hours in the presence of the indicated inhibitors. ADCC activity was measured as loss of luciferase activity from the target cells. All experiments represented in this figure were performed at least twice with each isolate and inhibitor with similar results.
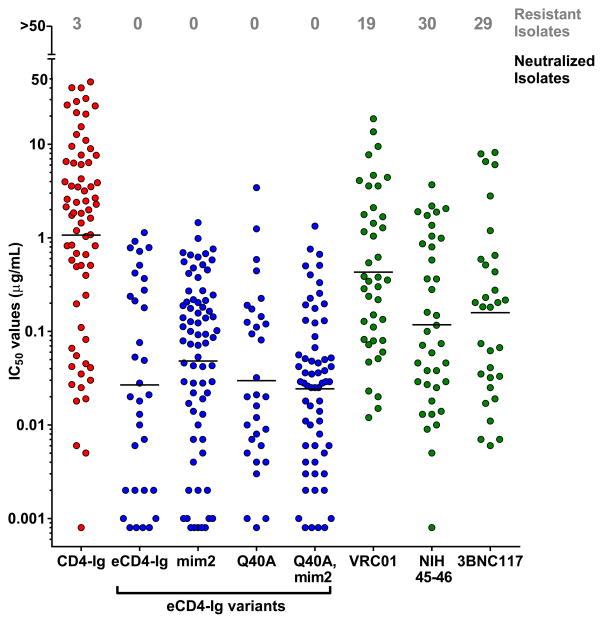
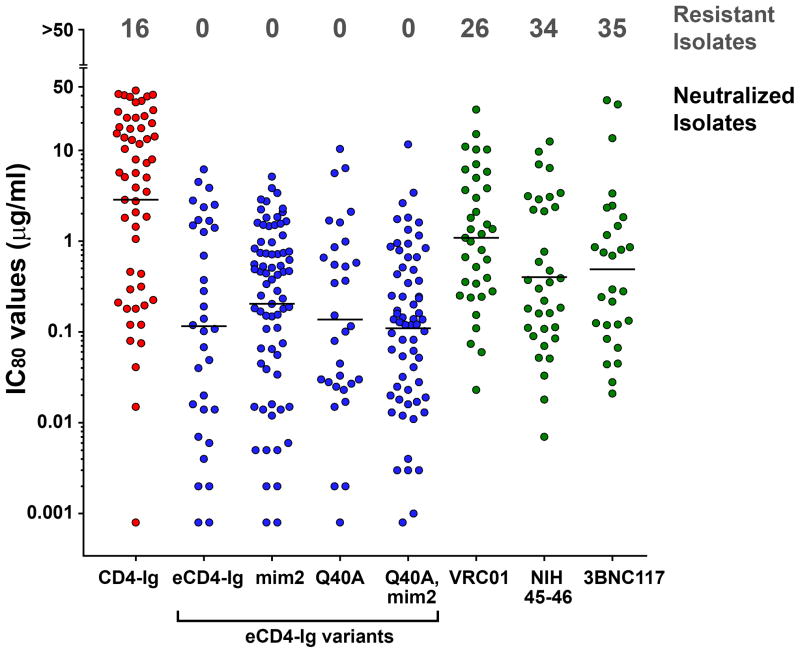
We further evaluated eCD4-Ig, eCD4-Igmim2, eCD4-IgQ40A,mim2 and the bNAb NIH45-46 using nearly every isolate reported to be resistant to either of the CD4bs antibodies NIH45-46 or 3BNC117 (Extended Data Figs. 4b and 5b). Both eCD4-Ig variants efficiently neutralized all 38 resistant isolates assayed with IC50s ranging from <0.001 μg/ml to 1.453 μg/ml. In contrast, 26 isolates in this panel were confirmed to be resistant to NIH45-46. 29 isolates and 18 isolates have been previously reported resistant to 3BCN117 and VRC01, respectively4,6. Fig. 3 and Extended Data Fig. 7 summarize the neutralization studies compiled from the experiments in Figs. 1 and 2, Extended Data Figs. 4–6, and from previous studies of VRC01 and 3BNC117 against the same isolates4. They show that the geometric mean IC50 and IC80 values of eCD4-Ig and its variants are less than 0.05 μg/ml (500 pM) and 0.2 μg/ml (2 nM), respectively, roughly 3–4 times lower than those of VRC01, NIH45-46, or 3BNC117. Importantly, our lead eCD4-Ig variant, eCD4-Igmim2, neutralized 100% of the isolates assayed at concentrations (IC50 < 1.5 μg/ml; IC80 < 5.2 μg/ml) likely sustainable in humans.
Figure 3. Summary of HIV-1, HIV-2 and SIV neutralization studies.
The IC50 values from studies of Figs. 1b, 2a, 2b, and Extended Data Figs. 4–6 are plotted. The number of isolates resistant to 50 μg/ml of the indicated inhibitors are indicated at top of figure. Geometric means are calculated for neutralized isolates and indicated with horizontal lines. Note that these data include 38 HIV-1 isolates selected for resistance to NIH45-46 or 3BNC117, so that isolates resistant to these antibodies are over-represented. Nonetheless, the geometric mean values of neutralized viruses are consistent with previous reports (Extended Data Table 1). Data for VRC01 and 3BNC117 were reported in Huang et al. and Scheid et al.4,6. IC80 values are presented in Extended Data Fig. 7.
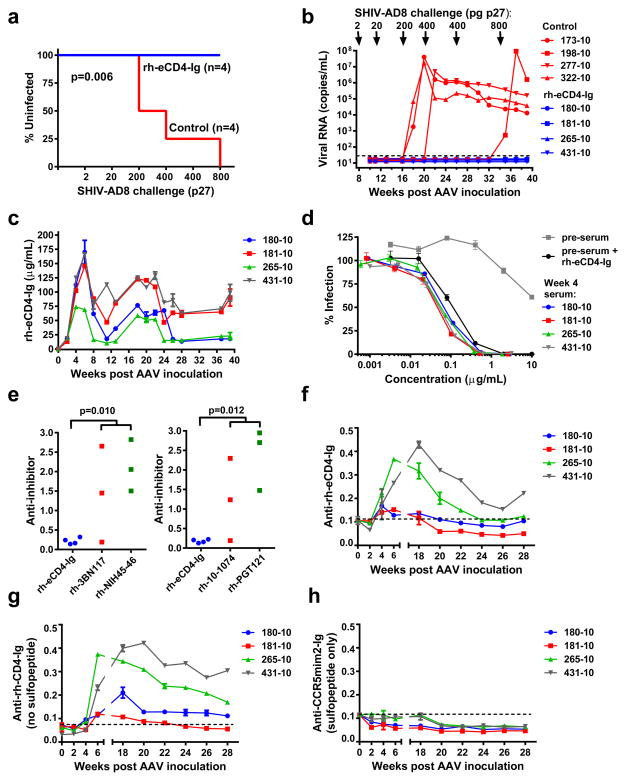
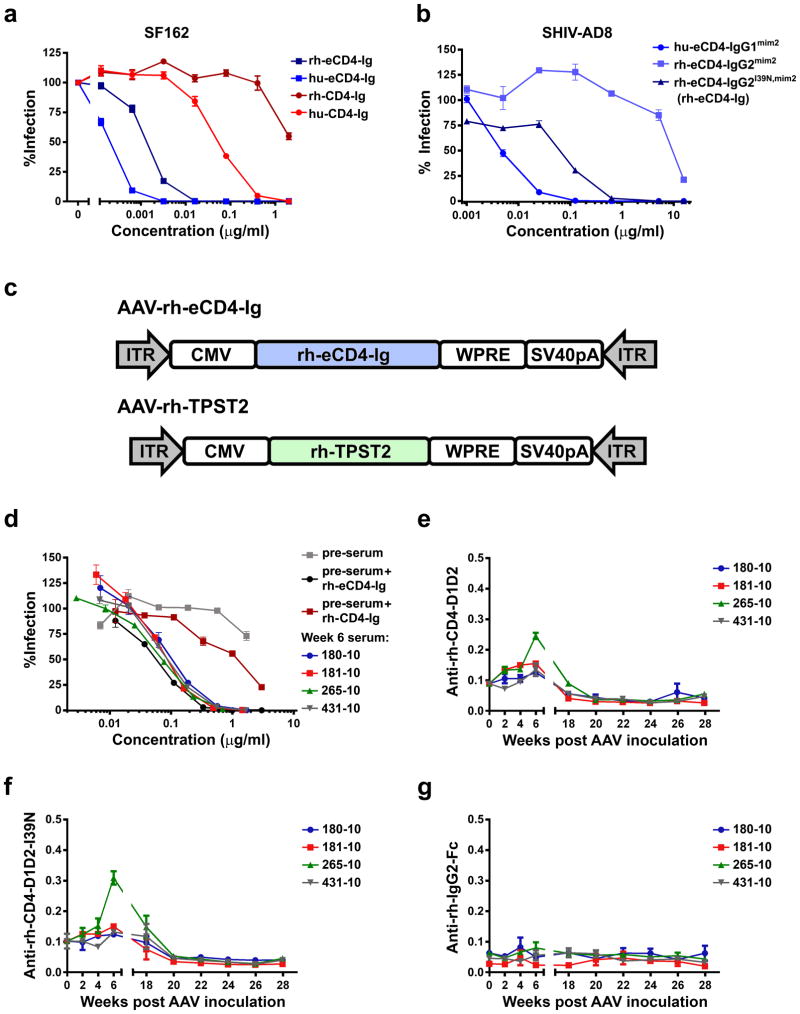
Finally, using a rhesus macaque form of eCD4-Igmim2, we investigated whether AAV-delivered eCD4-Ig could function like an HIV-1 vaccine. To minimize potential adverse reactions, the Fc domain of rhesus IgG2, which binds Fc receptors and complement less efficiently than IgG1, was used. We also introduced an I39N mutation into the CD4 domain27 to partially correct the lower affinity of rhesus CD4 for most HIV-1 isolates (Extended Data Figs. 8a and b). The gene for the resulting construct, rh-eCD4-IgG2I39N,mim2 (described hereafter as rh-eCD4-Ig) was cloned into a single-stranded AAV2 vector (AAV-rh-eCD4-Ig; Extended Data Fig. 8c). 2.5×1013 AAV1-encapsidated particles delivering this vector were administered into the quadriceps of four male 2-year old Indian-origin rhesus macaques. To promote rh-eCD4-Ig sulfation, a separate single-stranded AAV vector expressing rhesus tyrosine-protein sulfotransferase 2 (AAV-rh-TPST2; Extended Data Fig. 8c) was co-administered with AAV-rh-eCD4-Ig at a 1:4 ratio. No adverse reactions were observed in any of the AAV-rh-eCD4-Ig inoculated macaques. These macaques and four age- and gender-matched controls were challenged intravenously with increasing doses of SHIV-AD8 (Figs. 4a and b). 16 weeks post-AAV inoculation, two control macaques became infected following challenge with 200 pg p27. A subsequent 400 pg challenge infected a third control animal, and, after resisting an additional 400 pg challenge, the final control was infected with 800 pg, 34 weeks from the date of AAV inoculation. None of these challenges infected AAV-rh-eCD4-Ig inoculated macaques, indicating that eCD4-Ig protected them from four doses capable of infecting control animals.
Figure 4. AAV-rh-eCD4-Ig protects rhesus macaques from SHIV-AD8.
a, Infection analysis comparing four male Indian-origin rhesus macaques inoculated intramuscularly with 2×1013 AAV particles delivering rh-eCD4-Ig (blue) and four age- and gender-matched controls (red). At 8, 11, 16, 20, 26, and 34 weeks post-inoculation, macaques were challenged with the indicated p27 titers of SHIV-AD8. Significant protection (p=0.006; Mantel-Cox test) was observed in the AAV-rh-eCD4-Ig treated group. b, Viral loads of inoculated (blue) and control macaques (red) are shown, with the time and titer of challenge indicated above the graph. c, Concentrations of rh-eCD4-Ig in the sera of inoculated macaques were measured by ELISA through week 40 post-inoculation. d, The neutralizing potency of macaque sera obtained 4 weeks post-AAV-inoculation was compared to pre-inoculation sera (pre-sera), and pre-sera mixed with laboratory produced rh-eCD4-Ig, as in Fig. 2b. e, Anti-transgene antibody responses in AAV-rh-eCD4-Ig inoculated macaques were compared to those in macaques inoculated with AAV expressing the indicated bNAbs bearing constant regions of rhesus IgG2. Sera from four weeks post-inoculation were analyzed. Plates were coated with equivalent amounts of rh-eCD4-Ig or rhesus forms of bNAbs and incubated with sera and anti-rhesus lambda chain (left panel) or -kappa chain (right panel) antibody conjugated to horseradish peroxidase. Note that 3BNC117 and NIH45-46 bear a kappa light chain, whereas PGT121 and 10-1074 bear a lambda light chain, so that only host antibody responses were detected. Values indicate absorbance at 450 nM. P-values (Student’s 2-tailed t test) are indicated above the figures. f, The sensitivity of the assay in (e) was increased to measure longitudinally the anti-rh-eCD4-Ig activity in the sera of inoculated macaques. Both anti-kappa and anti-lamda secondary antibodies were used. Values are scaled for comparison to values in (e). g, h, The same assay as in (f) except that responses to rh-CD4-Ig, lacking CCR5mim2 (g) or to CCR5mim2 fused to a human IgG1 Fc domain (h) were measured. Experiments of panels c-h were performed at least twice with similar results. Errors bars denote one s.e.m. of duplicates.
Measured rh-eCD4-Ig titers in the serum stabilized to between 17 and 77 μg/ml over the last ten weeks of the 40-week study period (Fig. 4c). Two macaques expressed less than 20 μg/ml at the time of the final 800 pg challenge, suggesting that this concentration could prevent many otherwise infectious exposures in humans. Sera from inoculated macaques neutralized HIV-1 as efficiently as laboratory-prepared rh-eCD4-Ig mixed with pre-inoculation sera (Fig. 4d; Extended Data Fig. 8d), indicating that the eCD4-Ig was efficiently sulfated and fully active in vivo. We also compared macaque humoral responses to expressed rh-eCD4-Ig and to four AAV-expressed bNAbs inoculated for a separate study. 3BNC117, NIH45-45, 10-1074, and PGT121, each bearing rhesus IgG2 and light-chain constant domains, elicited markedly higher endogenous antibody responses than did rh-eCD4-Ig, consistent with their high levels of somatic hypermutation (Fig. 4e). To investigate the target of the anti-rh-eCD4-Ig responses, we increased the sensitivity of our assay and compared longitudinally the reactivity of inoculated rhesus sera to a series of antigens. rh-eCD4-Ig (Fig. 4f) and rh-CD4-Ig (without the CCR5mim2 sulfopeptide; Fig. 4g) were recognized by rhesus sera with nearly the same reactivity, whereas CCR5mim2 fused to a human IgG1 Fc domain was not (Fig. 4h), indicating that the sulfopeptide was not immunogenic. Rhesus CD4 domains 1 and 2 fused to a human IgG1 Fc was much less reactive than the same CD4 domains fused to the rhesus IgG2 Fc, without or with the I39N mutation (Extended Data Figs 8e and f), whereas an unrelated construct bearing the rhesus IgG2 Fc domain showed no reactivity (Extended Data Figs. 8g), suggesting that a neo-epitope formed by the rhesus CD4 and Fc domains was recognized by most anti-rh-eCD4-Ig antibodies. Thus eCD4-Ig is less immunogenic than bNAbs, and can be expressed for at least 40 weeks at concentrations that are well tolerated and protective against several robust SHIV-AD8 challenges.
A key question is whether eCD4-Ig or a similar construct could be used to prevent new HIV-1 infections in a population, and whether it might do so more effectively than a bNAb. We show that AAV-delivered rhesus eCD4-Ig protected all inoculated macaques from multiple infectious doses that are likely higher than those present in most human transmission events, although we have not yet tested protection from mucosal challenges. Protection lasted at least 34 weeks after inoculation (Fig 4b), and other studies indicate that these protective titers can be sustained for several years2. Previous studies of CD4-Ig indicate that it is safe when passively administered12,14, and in particular it does not engage MHC II or otherwise interfere with immune function13, although further safety studies of eCD4-Ig are warranted. eCD4-Ig has fewer non-self B- and T-cell epitopes than heavily hypermutated bNAbs, and thus elicits fewer endogenous antibodies that can impair its expression and activity (Fig. 4e). Its most prominent non-self element is its sulfopeptide, which did not elicit any measurable antibody responses (Figs. 4f–h). However the clearest advantage of eCD4-Ig over bNAbs is its potency and its unmatched breadth (Fig. 3; Extended Data Figs. 4–7). The breadth of eCD4-Ig arises from the necessary conservation of its binding sites on Env, suggesting that emergence of eCD4-Ig escape variants in a population is less likely than with bNAbs. Moreover, any virus that does bypass prophylaxis is likely to bind CD4 and CCR5 less efficiently in the continued presence of eCD4-Ig, and may therefore be less efficiently retransmitted. Its potency suggests that relatively lower concentrations of eCD4-Ig will be sufficient to protect against most circulating viruses, a feature that may be critical to its use with AAV in humans. Although there are remaining challenges, these observations suggest that AAV-expressed eCD4-Ig could provide effective, long-term, and near universal protection from HIV-1.
METHODS
Plasmids and cells
Plasmid expressing CD4-Ig was previously described20. Fusion constructs were created by adding sequences encoding CCR5mim1 and tetra-glycine linker to N-terminus (fusion1) or between domain 2 and human Fc (fusion2) of CD4-Ig by inverse PCR. eCD4-Ig (fusion3) and eCD4-Igmim2 were created by adding sequence encoding a tetra-glycine linker and CCR5mim1 or CCR5mim2, respectively, to the C-terminus of CD4-Ig by inverse PCR. The glutamine 40 to alanine mutation was introduced in eCD4-Ig and eCD4-Igmim2 by Quickchange PCR. The eCD4-Ig/CD4-Ig heterodimer was generated as previously described 23 and analyzed by SDS-PAGE under reducing and non-reducing conditions. rh-eCD4-Ig, consisting of rhesus CD4 domains 1 and 2 bearing an I39N mutation, rhesus IgG1 Fc, and CCR5mim2, was synthesized and cloned into a previously described single-stranded AAV plasmid 2. AAV expression plasmids for HIV-1 antibodies were created by synthesizing the variable heavy and light chains of 3BNC117, NIH45-46, PGT121, and 10-1074 with the rhesus heavy and light constant regions, and cloning these genes into a previously described ssAAV plasmid that has been previously described 2. The following reagent was obtained through the NIH AIDS Reagent Program (Division of AIDS, NIAID, NIH): CMVR-VRC01-H, CMVR-VRC01-L, from Dr. John Mascola28,29, pNL4-3.Luc.R-.E- from Dr. Nathaniel Landeau30,31, TZM-bl cells from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc32–36, SF162 gp160 from Dr. Leonidas Stamatatos and Dr. Cecilia Cheng-Mayer37, and GHOST-CCR5 and -CXCR4-cells from V. KewalRamani and D. Littman. Human embryonic kidney HEK293T cells were obtained from ATCC. Cf2Th-CD4+.CCR5+ and CfTh-CCR5+ cells were a generous gift from Dr. Hyeryun Choe. No testing for mycoplasma contamination was performed in any cell line after their receipt from these contributors. The variable heavy and light chains of IgG-b12, NIH45-46, 3BNC117, 10-1074, and PGT121 were cloned into the CMVR-VRC01-H and -L plasmids. Plasmids encoding TPST-2 or the envelope glycoproteins pNL4-3Δenv, 89.6, ADA, SG3, SA32, YU2, JRFL, KB9, VSV-G, HIV-2 ST, SIVmac239, SIVmac316, and replicative 89.6 or SG3 viruses were previously described20,21,38–40.
Purification of antibodies, CD4–Ig, and eCD4-Ig variants
Production of CD4-Ig, eCD4-Ig variants and antibodies was performed as previously described 41. Briefly, HEK293T cells in 140 mm plates were transfected with 25 ug/plate at 50% confluency by the calcium phosphate transfection method. Plasmids encoding sulfated proteins were cotransfected with a plasmid encoding human tyrosine protein sulfotranserase 2 (TPST2). At 12 hrs post-transfection, 10% FBS-DMEM media was replaced with serum-free 293 Freestyle media (Invitrogen). Media was collected after 48 hrs, debris was cleared by centrifugation for 10 min at 1,500 g and filtered using 0.45 μm filter flasks (Millipore). Complete protease inhibitor cocktail (Roche) was added to the filtered supernatants. 500 μl bed volume of Protein A sepharose beads (GE Healthcare) were added and were agitated 4°C overnight. The bead/media mixture was collected by gravity flow column (Biorad) and was washed with 30 mL phosphate buffered saline (PBS; Lonza) + 0.5M NaCl (0.65M NaCl final) followed by 10 mL PBS. Protein was eluted with 3M MgCl2 in PBS. Buffer was exchanged for PBS and protein was concentrated to 1 mg/ml by Ultrafiltration (Amicon Ultra) at 4,000 g.
Flow cytometry analysis of CD4-Ig and eCD4-Ig binding to cell-expressed envelope glycoprotein
HEK293T cells were transfected with plasmids expressing envelope glycoprotein lacking cytoplasmic residues 732 to 876 (HXBc2 numbering) together with plasmid encoding the tat protein. Transfection medium was replaced after an overnight incubation and cells were harvested 48 hours post transfection. Harvested cells were washed twice in flow cytometry buffer (PBS with 2% goat serum, 0.01% sodium azide). Cells were incubated with CD4-Ig or eCD4-Ig on ice for 1 hour and then washed twice with flow cytometry buffer. A secondary antibody recognizing human Fc (Jackson Immuno Research) was added to the cells for 30 minutes. Cells were washed twice with flow cytometry buffer, twice with PBS, and resuspended in 1% paraformaldehyde solution. Binding was analyzed with an Accuri C6 Flow Cytometer (BD Biosciences) and data analyzed with the C6 Software (BD Biosciences).
Viral entry enhancement assay
HIV-1 pseudovirus expressing firefly luciferase was pre-incubated with titrated amounts of CD4-Ig or eCD4-Ig variants in DMEM (10% FBS) for 1 hour at 37°C. CD4-negative Cf2Th-CCR5 cells were harvested and diluted in DMEM (10% FBS) to 100,000 cells/mL and added to the pseudovirus/inhibitor mixture. Cells were then incubated for 48 hours at 37°C. Viral entry was analyzed using Britelite Plus (Perkin Elmer) and luciferase activity of cell lysates was read using a Victor X3 plate reader (Perkin Elmer).
HIV-1 neutralization assays
GHOST-CCR5 or –CXCR4 cells were plated into 12-well plates at 50,000 cells per well. HIV-1 pseudovirus was diluted in RPMI and titrated amounts of CD4-Ig, fusion1, fusion2, or eCD4-Ig were added. Virus and inhibitor were incubated at room temperature for 20 minutes and added to the cells for 2 hours at 37°C. Cells were then washed with serum free medium and then incubated in 1 mL of DMEM (10% FBS) for 48 hours at 37°C. Cells were harvested by trypsinization, fixed in 1% paraformaldehyde in PBS, and viral entry was determined by flow cytometry based on GFP expression.
For studies of infectious virus, unstimulated PBMCs were harvested and resuspended in RPMI medium (15% FBS, 20 U/mL IL-2). Cells were plated in a 12-well plate at 106 cells per well. HIV-1 was diluted in RPMI and varying amounts of inhibitor were added. The virus and inhibitor was incubated at room temperature for 20 minutes and added to the cells for 3 hours at 37°C. Cells were then washed with serum-free medium and resuspended in fresh RPMI medium (15% FBS, 20 U/mL IL-2). At 3-day intervals post infection, supernatants were collected and fresh RPMI medium (15% FBS, 20 U/mL IL-2) was added to the cells. Supernatants were analyzed for viral infection by ELISA with Alliance HIV-1 p24 antigen ELISA kit (Perkin Elmer).
TZM-bl neutralization assays were performed as previously described42. Briefly, HIV-1 pseudoviruses were pre-incubated with titrated amounts of CD4-Ig or eCD4-Ig variants in DMEM (10% FBS) for 1 hour at 37°C. TZM-bl cells were harvested and diluted in DMEM (10% FBS) to 100,000 cells/mL and added to the pseudovirus/inhibitor mixture. Cells were then incubated for 48 hours at 37°C. Viral entry was analyzed using Britelite Plus (Perkin Elmer) and luciferase activity was read using a Victor X3 plate reader (Perkin Elmer). All neutralization and enhancement studies of Figs 1–4 were performed at least twice in triplicate. All error bars represent S.E.M.
Antibody-dependent cell-mediated cytotoxicity assays
ADCC activity was performed as previously described 43. Briefly, CEM.NKR CCR5 CD4+ T cells were infected 4 days with infectious HIV-1 NL4.3, SHIV-KB9, or SIVmac239. After 4 days, KHYG-1 effector cells were co-incubated with infected cells in the presence of titrated CD4-Ig, eCD4-Ig variants, or the b12 antibody for 8 hours. ADCC activity was measured by luciferase activity as above.
Production of HIV-1NL4-3 stocks and SHIV-AD8-EO stocks for in vivo studies
A molecular clone of HIV-1NL4-3 was obtained from the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH from material deposited by Suzanne Gartner, Mikulas Popovic, Robert Gallo and Malcolm Martin. Virus stocks were produced in 293T cells by transient transfection using TurboFect (Thermo Scientific) and 12 μg of proviral plasmid. Supernatants were harvested at 40 hours, filtered through 0.45 μm filters, and dispensed into single use doses and frozen at −80°C. Viruses were quantified by p24 ELISA (Zeptometrix, Buffalo, NY) and by GHOST cell titer44 to determine infectious units per mL (IU/mL). Titering was performed per the GHOST cell line protocol obtained through ARRRP. The molecular clone of SHIV-AD8-EO was a generous gift from Dr. Malcom Martin45. 293T cells were plated in 140 mm flasks and transfected with 80 μg DNA/plate by calcium phosphate technique. At 12 hour post transfection, flasks were replaced with fresh DMEM (10% FBS). Medium was harvested at 48 hours post transfection, frozen at −80C, and tittered using an SIV p27 ELISA kit (ABL).
Hematopoietic stem cell isolation and NSG mouse transplantation
Human CD34+ hematopoietic stem cells (HSC) were isolated from fetal livers obtained from Advanced Bioscience Resources, INC (ABR, Alameda, CA). Tissue was disrupted and incubated with 1mg/mL Collagenase/Dispase (Roche Applied Sciences) for 15 min at 37°C. Cells were isolated by passing the disrupted tissue through a 70 μm filter. Red blood cells were lysed in BD Pharm Lyse (BD Biosciences, San Jose, CA), with CD34+ cells being isolated using CD34 MACS microbeads (Miltenyi) according to manufacturer’s instructions with an additional purification step using a second column. NOD.Cg-Prkdc scid Il2rγ tm1Wj/Szj (NOD/SCID/IL2rγnull, NSG) mice were obtained from Jackson Laboratories (Bar Harbor, ME). Neonatal mice received 150 cGy radiation, and 2–4 hours later 1×106 CD34+ HSCs in 1% heparin (Celgene, Summit, NJ) via intrahepatic injection. Mice were monitored for engraftment levels of human CD45+ cells and development of T cells and B cells at 8, 10, and 12 weeks post engraftment.
Mouse infections, treatment, and analysis
Humanized mice with evidence of human CD4+ T cell development in blood were infected with 5×104 IU of HIV-1NL4.3 by intraperitoneal injection. Mice were administered with 65 μg of eCD4-Ig once weekly for the first 2 weeks, starting at 8 day prior to the HIV-1 challenge, and then twice weekly starting week 3 by retro-orbital injection while under anesthetization by 2.5% isofluoane. Mock treated mice received a retro-orbital injection of PBS one and eight days preceding HIV-1 challenge, and were anesthetized in parallel with eCD4-Ig mice throughout. Every week post-infection the mice were anesthetized by inhalation of 2.5% isoflourane and blood was collected retro-orbitally for analysis. At week 6, three eCD4-Ig treated mice and one mock treated mouse (who had not become infected) were challenged a second time with 5×104 IU HIV-1NL4-3. Mouse blood was blocked for 20 minutes at room temperature in FBS (Denville) and stained with appropriate antibodies for 15 minutes at room temperature. Red blood cells were removed by incubation in BD FACS Fix/Lysing Solution (BD Biosciences), which was removed by dilution with PBS prior to analysis by flow cytometry. HIV-1 levels in peripheral blood were determined by extracting viral RNA from mouse plasma at each blood draw using a viral RNA isolation kit (Qiagen, Germantown, MD) followed by Taqman One-Step RT-PCR (Life Technologies, Carlsbad, CA) using a primer and probe set targeting the HIV-1 LTR region, as previously described46,47. Reactions were performed and analyzed using a 7500 Fast Realtime PCR System (Life Technologies). To analyze engrafted T cells by flow cytometry, stained cells were acquired on a FACS Canto II (BD Biosciences) and analyzed using FlowJo software v7.6.5 (Tree Star Inc., Ashland, OR). Blood samples were stained using human-specific antibodies at a 1:20 dilution for CD4-V450 (RPA-T4), CD8-APC (RPAT8), CD3-PE (UCHT1), and CD45-PerCP (TUI16) (BD Bioscience). Up to 10,000 events were recorded for viable cell populations and gated based on fluorescence minus one controls as previously described46. All mouse studies were performed in accordance with the Scripps Research Institute Institutional Review Board, Protocol number 14-018.
AAV inoculation of rhesus macaques
Eight four-year old AAV1-negative male Indian-origin rhesus macaques were housed at the New England Primate Research Center in accordance with standards set forth by the American Association for Accreditation of Laboratory Animal Care. Their weights at the time of AAV inoculation ranged from 5.2 to 8.2 kg. Macaques were separated into age- and weight-matched control groups, but blinding and randomization were not performed. Four macaques were inoculated with 1 mL saline containing 2.5×1013 AAV1 particles delivering 80% of a single-strand rh-eCD4-Ig transgene (IgG2 isotype) and 20% of a single-strand rhesus TPST-2 transgene into both quadriceps muscle (two 0.5 mL per injections per quadriceps muscle). 1 mL of sera was obtained every one to two weeks post-AAV inoculation beginning at Week 4. Animals were challenged at Week 8 post-inoculation with 2 pg p27 of SHIV-AD8-EO. SHIV-negative animals were repeatedly challenged with escalating doses of SHIV-AD8-EO up to 800 pg p27. Plasma viral loads were quantified as previously described45.
For AAV studies of bNAbs, six two-year old AAV1-negative Indian-origin rhesus macaques (two males and four females) were housed at the New England Primate Research Center in accordance with standards set forth by the American Association for Accreditation of Laboratory Animal Care. Three macaques were inoculated with 1 mL saline containing 1×1013 AAV1 particles delivering single-strand rh-3BNC117-IgG2 transgene into one quadriceps (two 0.5 mL injections) and 1 mL saline containing 1×1013 AAV1 particles delivering single-strand rh-10-1074-IgG2 transgene into the second quadriceps (two 0.5 mL injections). The other three macaques were incoluated with 1 mL saline containing 1×1013 AAV1 particles delivering single-strand rh-NIH45-46-IgG2 transgene into one quadriceps (two 0.5 mL injections) and 1 mL saline containing 1×1013 AAV1 particles delivering single-strand rh-PGT121-IgG2 transgene into the second quadriceps (two 0.5 mL injections). 1 mL of sera was obtained every two weeks beginning at Week 2 and analyzed by ELISA. All primate studies were performed in accordance with the Harvard Medical School Standing Committee on Animals protocol number 04888.
eCD4-Ig, rh-eCD4-Ig and anti-transgene antibody concentrations in NSG mice and rhesus macaque sera
In vivo concentrations of eCD4-Ig, rh-eCD4-Ig were measured by ELISA as previously described2. Briefly, to measure NSG mouse and macaque serum concentrations, ELISA plates (Costar) were coated with 5μg/mL SIV gp120 overnight at 4°C. Plates were washed with PBS-T (PBS + 0.05% Tween-20) twice and blocked with 5% milk in PBS for 1 hour at 37°C. Sera serially diluted in 5% milk in PBS were added to the plate and incubate for 1 hour at 37°C. Samples were washed five times with PBS-T and a horseradish peroxidase secondary antibody (Jackson Immuno Research) recognizing human IgG1. Plates were incubated for 1 hour at 37°C and then washed ten times with PBS-T. TMB solution (Fisher) was added for 10 minutes at room temperature and then stopped with TMB Stop Solution (Southern Biotech). Absorbance was read at 450 nm by a Victor X3 plate reader (Perkin Elmer) and compared with a standard curve generated using a rh-eCD4-Ig mixed with pre-inoculation sera. Anti-rh-eCD4-Ig antibodies and anti-bNAb antibodies were measured in the same way except that ELISA plates were coated with 5 μg/mL of various constructs. Constructs so assayed included rh-eCD4-Ig, rh-CD4-IgI39N, rh-CD4-Ig domains 1 and 2 (with or without I39N) bearing a human IgG1 Fc and hinge domain, C-terminal CCR5mim2-Ig (human IgG1 Fc and hinge, no CD4 domains), NIH45-46 bearing the rhesus IgG2 Fc domain and hinge, or HIV-1 bNAbs. Serum samples were diluted 10- or 20-fold and blocked in 5% milk in PBS. Anti-transgene antibodies were measured using secondary antibodies detecting either the kappa or lambda light chain (Southern Biotech) that was opposite of the antibody being assayed when comparing the anti-bNAb response to that to rh-eCD4-Ig. Both anti-kappa and anti-lambda secondary antibodies were used when measuring anti-rh-eCD4-Ig responses alone. TMB solution was added for 10–15 minutes at room temperature and measured as described above.
Extended Data
Extended Data Figure 1. Sequences of CD4-Ig and eCD4-Ig variants.
The amino-acid sequences of CD4-Ig, eCD4-Ig, fusion1, fusion2, eCD4-Igmim2, eCD4-IgQ40A, eCD4-IgQ40A,mim2 and rh-eCD4-Ig (rh-eCD4-IgG2I39N,mim2) are shown. Leader peptides are underlined, CD4 domains 1 and 2 are indicated in red, Fc domains are indicated in cyan, CCR5-mimetics peptides are indicated in green, and linker sequences are shown in black.
Extended Data Figure 2. Extended Data for Figure 1.
a, b, Experiments similar to those of Fig. 1b except that CD4-Ig (red), fusion1 (grey), fusion2 (green), and fusion3 (eCD4-Ig; blue) are compared using HIV-1 pseudotyped with the envelope glycoproteins of the 89.6 (a) or ADA (b) isolates. c, d, Experiments similar to those in Fig. 1e except that CD4-Ig (red), eCD4-Ig (blue), or heterodimers thereof (grey) are compared. e, CD4-Ig, eCD4-Ig, and the CD4-Ig/eCD4-Ig heterodimer assayed in Fig. 1e and (c) and (d) were analyzed by SDS-PAGE and stained with Coomassie blue under reducing (left) and non-reducing (right) conditions. f, g, Infectious 89.6 (f) or SG3 (g) HIV-1 was incubated with human PBMC in the presence of the indicated concentrations of CD4-Ig (red) or eCD4-Ig (blue), or without either inhibitor (grey). Culture supernatants were collected on the indicated day and viral p24 levels were measured by ELISA. h, Viral loads in RNA copies/mL are shown for each humanized mouse of Fig. 1f. Mice treated with eCD4-Ig are indicated with blue lines and mice treated with PBS are indicated with red lines. The 800 copies/mL limit of detection of this assay is indicated by a dashed line. Experiments shown in panels a–g were performed at least twice with similar results. Error bars denote s.e.m. of triplicates.
Extended Data Figure 3. A model of eCD4-Ig bound to the HIV-1 Env trimer.
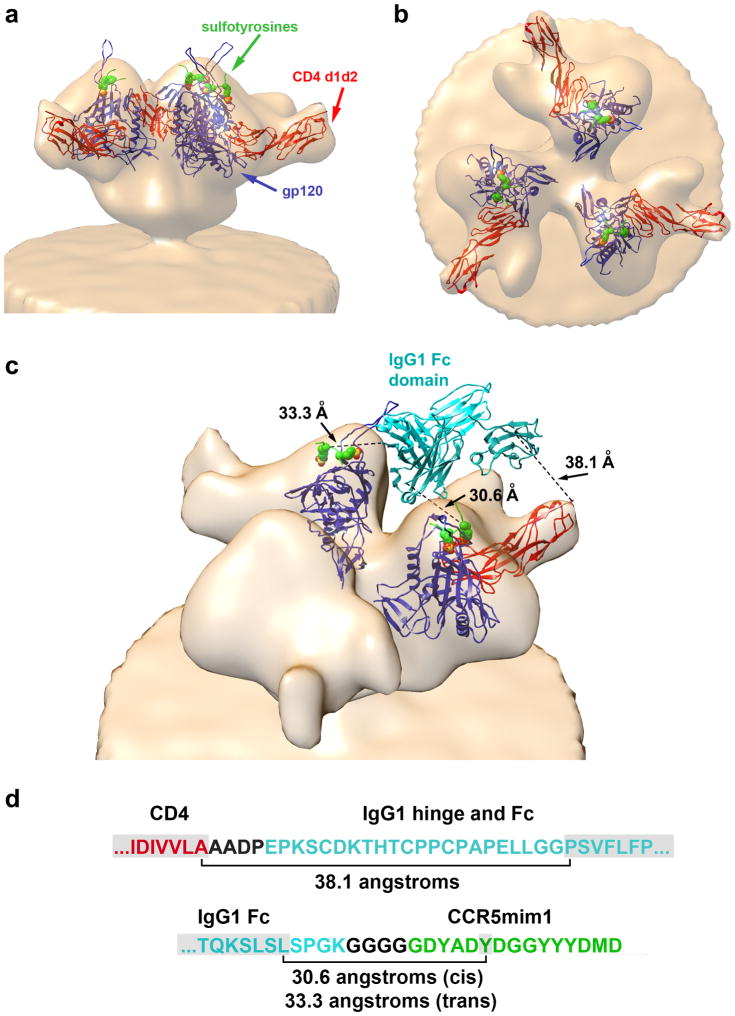
a, The structure (2QAD) of gp120 (YU2 isolate) bound to the tyrosine-sulfated CD4i antibody 412d and CD4 domains 1 and 210, was fitted into a cryoelectron micrograph of the HIV-1 envelope glycoprotein trimer (Env; Bal isolate) bound to CD448. gp120 and CD4 domains 1 and 2 are shown in blue and red, respectively. 412d sulfotyrosines are represented as green (carbon), red (oxygen), and yellow (sulfur) spheres. The remainder of 412d was excluded for clarity. b, The same structure shown in (a) rotated 90 degrees about the horizontal axis. Note that the sulfotyrosine-binding pockets are proximal to the trimer axis, whereas the carboxy-terminus of CD4 domain 2 is distal from the trimer axis, preventing both CD4 domains of CD4-Ig from simultaneously binding the same Env trimer. c, A model of how eCD4-Ig may associate with Env is presented. The Fc domain of human IgG1 (1FCC, cyan)49 was positioned to be proximal to the gp120 sulfopeptide-binding pocket occupied by sulfotyrosine 100 (Tys 100) of the 412d heavy chain while avoiding steric interaction with Env. Tys 100 occupies a pocket in gp120 thought to bind CCR5 sulfotyrosine 1050. This pocket is also critical for binding of CCR5mim1 and CCR5mim220,22. In this model, the Fc domain is oriented to allow each eCD4-Ig sulfopeptide to engage a different gp120 protomer24. A single CD4 domain also binds one of the sulfopeptide-bound protomers. Distances between the carboxy-terminus of CD4 and the amino-terminus of one Fc domain monomer (38.1 angstroms), between the carboxy-terminus of the Fc domain and Tys 100 pocket of the CD4-bound gp120 protomer (30.6 angstroms), and between the carboxy-terminus of the Fc domain and Tys 100 pocket of an adjacent gp120 protomer (33.3 angstroms), are indicated. d, Residues not visible in the crystal structures used to construct this model are shown between brackets. In the model shown in (c), these residues span the distances indicated. Note that these distances are well under the extension of a typical beta strand. CD4-, IgG1- and CCR5mim1-derived residues are shown in red, cyan, and green, respectively, with linker regions shown in black. Residues visible in the crystal structures, including the CCR5mim1 sulfotyrosine presumed to fill the Tys 100 pocket, are highlighted in grey. Modeling was performed using UCSF Chimera version 1.8.
Extended Data Figure 4. IC50 values of eCD4-Ig variants against neutralization-resistant isolates.
a, The IC50 values (μg/mL) of CD4-Ig, eCD4-Ig, eCD4-Igmim2 (mim2), eCD4-IgQ40A (Q40A), and eCD4-IgQ40A,mim2 (Q40A,mim2) against 24 HIV-1 and SIV isolates selected for their neutralization resistance are shown. The clade and tier of each isolate is listed. HIV-1 pseudotyped with the indicted envelope glycoprotein was incubated in triplicate with TZM-bl cells and varying concentrations of CD4-Ig or eCD4-Ig variant. Luciferase activity was determined two days post-infection. ‘Fold’ indicates the ratio of the IC50 value of CD4-Ig to the geometric mean of the IC50 values of the assayed eCD4-Ig variants. The geometric mean of eCD4-Ig variants and the CD4bs antibodies 3BCN117, NIH45-46, and VRC01 calculated from values reported in Huang et al. and Sheid et al.4,6 are shown in the two rightmost columns. b, Neutralization studies similar to those in (a) except that the IC50 values of CD4-Ig, eCD4-Igmim2 (mim2), eCD4-IgQ40A,mim2 (Q40A,mim2) and NIH45-46 were determined for a panel of 40 viral isolates selected for their resistance to the CD4bs bNAbs 3BNC117 and NIH45-46. IC50 values of the CD4bs antibodies VRC01 and 3BNC117 listed in the two rightmost columns were reported in Huang et al. and Scheid et al.4,6
Extended Data Figure 5. IC80 values of eCD4-Ig variants against neutralization-resistant isolates.
a, b, The IC80 values (μg/mL) of the experiments described in Extended Data Fig. 5a (a) and 5b (b) are shown.
Extended Data Figure 6. Extended Data for Figure 2.
a, IC90 values for the same experiments shown in Fig. 2a, presented in the same format. b, Numeric IC50 and IC90 values of the experiment shown in (a) and Fig. 2a are shown, using the same color coding of Extended Data Figs. 4 and 5. S.e.m of triplicates are indicated below their IC50 and IC90 values. c, Experiments similar to those in Fig. 2b except that HIV-1 pseudotyped with the Env of the HIV-2 isolate ST were incubated with the indicated concentrations of CD4-Ig, eCD4-Ig variants, or the CD4bs antibodies IgG-b12, VRC01, or NIH45-46. Error bars denote s.e.m. of triplicates.
Extended Data Figure 7. Extended Data for Figure 3 (IC80 values).
The IC80 values from studies of Figs. 1b, 2a, 2b, and Extended Data Figs. 4–6 are plotted. The number of isolates resistant to 50 μg/ml of the indicated inhibitors are indicated on top. Geometric means are calculated for neutralized isolates and indicated with horizontal lines.
Extended Data Figure 8. Extended Data for Figure 4.
a, An experiment similar to that in Fig. 2b, except that rhesus and human CD4-Ig and eCD4-Ig are compared for their ability to neutralize HIV-1 pseudotyped with the SF162 envelope glycoprotein. All variants have wild-type rhesus or human CD4 domains. Note that variants bearing rhesus CD4 are markedly less potent at neutralizing HIV-1. b, Experiment similar to Fig. 2b and to (a) except that human eCD4-Igmim2 and its rhesus analog bearing or not the I39N mutation are compared using SHIV-AD8. Note that the I39N mutation largely restores the neutralization activity of rhesus eCD4-Igmim2. c, A representation of the AAV vectors used in the non-human primate studies of Fig. 4. Rh-eCD4-Ig (rh-eCD4-IgG2I39N,mim2; blue) and rhesus tyrosine protein sulfotransferase 2 (TPST2; green) were introduced into a single-stranded AAV vector downstream of a CMV promoter. A woodchuck response element (WPRE), used to promote expression, and the SV40 polyadenylation signal (SV40pA) were also included. AAV inverted terminal repeats (ITR) are indicated in grey arrows. d, An experiment similar to that in Fig. 4d except that sera from week 6 were analyzed. e, f, g, Experiments similar to those in Figs.4f–h except that the reactivity of rhesus sera was examined for a construct bearing wild-type rhesus CD4 domains 1 and 2 fused to the human IgG1 Fc domain (e), one bearing rhesus CD4 domains 1 and 2 with the I39N mutation, again fused to the human IgG1 Fc domain (f), or the antibody NIH45-46 fused to the rhesus IgG2 constant regions, used here to present the rhesus IgG2 Fc domain (g). Experiments shown in a,b, and d–g represent at least two with similar results. Error bars denote s.e.m. of triplicates.
Extended Data Table 1.
Potencies and breadth of well characterized broadly neutralizing antibodies.
A summary of antibody neutralization potencies compiled using the Los Alamos National Laboratory Database CATNAP tool (www.hiv.lanl.gov/components/sequence/HIV/neutralization/main.comp). The geometric mean IC50 and IC80 values are listed for the indicated broadly neutralizing antibodies (bNAbs) against all reported isolates, excluding those with values greater than 50 μg/ml. The percentage of isolates neutralized with IC50 values less than 50 μg/ml, or with IC80 values less than 5 μg/ml are shown. bNAbs are ranked by their geometric mean IC50 values. See Fig. 3 and Extended Data Figure 7 for comparisons of eCD4-Ig variants with the bNAbs NIH45-46, 3BNC117, and VRC01.
| Antibody | IC50 | IC80 | IC50 < 50 μg/ml | IC80 < 5 μg/ml |
|---|---|---|---|---|
| 10-1074 | 0.053 | 0.217 | 57.8% | 51.2% |
| 35022 | 0.057 | n.a. | 61.9% | n.a. (<61.9%) |
| PGT121 | 0.060 | 0.274 | 63.0% | 47.9% |
| PGT128 | 0.069 | n.a. | 62.9% | n.a. (<62.9%) |
| PG16 | 0.092 | 0.178 | 55.6% | 43.6% |
| 3BNC117 | 0.111 | 0.345 | 82.2% | 61.0% |
| VRC07 | 0.114 | 0.187 | 83.2% | 86.0% |
| NIH45-46 | 0.139 | 0.540 | 83.7% | 57.4% |
| 12A12 | 0.171 | 1.101 | 93.2% | 68.9% |
| PG9 | 0.176 | 0.427 | 77.3% | 62.0% |
| 10E8 | 0.262 | 1.536 | 98.3% | 75.5% |
| VRC01 | 0.306 | 0.913 | 88.0% | 69.7% |
Acknowledgments
This project was supported by National Institutes of Health grants R01 AI091476 and R01 AI080324 (M.F.), P01 AI100263 (G.G., R.C.D., M.F.), RR000168 (M.R.G., L.M.K, D.T.E, R.C.D., M.F), R01 AI058715 (B.H.H), by the Intramural Research program of the Vaccine Research Center, NIAID, NIH (J.G, B.Z., P.D.K), and by federal funds from the National Cancer Institute, NIH under Contract No. HHSN261200800001E. The authors would like to thank Drs. Hyeryun Choe and Malcolm Martin for critical advice.
Footnotes
AUTHOR CONTRIBUTIONS
M.R.G and L.M.K contributed equally to this work. M.R.G, L.M.K., H.R.K, M.V.S, T.D., J.J.C, M.D.A., M.P., J.D.L., R.C.D., D.T.E, B.H.H., P.M.C., M.S.S., A.P. and M.F. designed experiments. M.R.G, L.M.K., H.R.K, M.V.S, T.D., J.J.C, K.G.H., J. M. D., M.D.A, C.C.B., C.H.F, V. R. J., B.D.Q., and A.Y.Y. performed experiments. L.M.K. conducted all non-human primate studies. J.G. and P.D.K. assisted with modeling. J.M.N., H.M., B.Z., P.P., M.S., J. R. M., M.C.N., and D. R. B. contributed advice and critical reagents. M.F. conceived the study and, with important assistance from M.R.G. and L.M.K., wrote the manuscript.
The authors declare they have no competing financial interests.
References
- 1.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson PR, et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis AD, Chen R, Montefiori DC, Johnson PR, Clark KR. Generation of neutralizing activity against human immunodeficiency virus type 1 in serum by antibody gene transfer. J Virol. 2002;76:8769–8775. doi: 10.1128/JVI.76.17.8769-8775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greig JA, et al. Intramuscular Injection of AAV8 in Mice and Macaques Is Associated with Substantial Hepatic Targeting and Transgene Expression. PloS one. 2014;9:e112268. doi: 10.1371/journal.pone.0112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 10.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagenaur LA, Villarroel VA, Bundoc V, Dey B, Berger EA. sCD4-17b bifunctional protein: extremely broad and potent neutralization of HIV-1 Env pseudotyped viruses from genetically diverse primary isolates. Retrovirology. 2010;7:11. doi: 10.1186/1742-4690-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher CV, et al. Nonlinear pharmacokinetics of high-dose recombinant fusion protein CD4-IgG2 (PRO 542) observed in HIV-1-infected children. The Journal of allergy and clinical immunology. 2007;119:747–750. doi: 10.1016/j.jaci.2006.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussey RE, et al. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331:78–81. doi: 10.1038/331078a0. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson JM, et al. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. The Journal of infectious diseases. 2000;182:326–329. doi: 10.1086/315698. [DOI] [PubMed] [Google Scholar]
- 15.Haim H, et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moebius U, Clayton LK, Abraham S, Harrison SC, Reinherz EL. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan N, et al. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 19.Farzan M, et al. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- 20.Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J Biol Chem. 2006;281:28529–28535. doi: 10.1074/jbc.M602732200. [DOI] [PubMed] [Google Scholar]
- 21.Choe H, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 22.Chiang JJ, et al. Enhanced recognition and neutralization of HIV-1 by antibody-derived CCR5-mimetic peptide variants. J Virol. 2012;86:12417–12421. doi: 10.1128/JVI.00967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein engineering. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 24.Kwong JA, et al. A tyrosine-sulfated CCR5-mimetic peptide promotes conformational transitions in the HIV-1 envelope glycoprotein. J Virol. 2011;85:7563–7571. doi: 10.1128/JVI.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alpert MD, et al. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol. 2012;86:12039–12052. doi: 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humes D, Emery S, Laws E, Overbaugh J. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol. 2012;86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barouch DH, et al. A Human T-Cell Leukemia Virus Type 1 Regulatory Element Enhances the Immunogenicity of Human Immunodeficiency Virus Type 1 DNA Vaccines in Mice and Nonhuman Primates. Journal of Virology. 2005;79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, et al. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. Journal of Virology. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connor RI, Chen BK, Choe S, Landau NR. Vpr Is Required for Efficient Replication of Human Immunodeficiency Virus Type-1 in Mononuclear Phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 32.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that Ecotropic Murine Leukemia Virus Contamination in TZM-bl Cells Does Not Affect the Outcome of Neutralizing Antibody Assays with Human Immunodeficiency Virus Type 1. Journal of Virology. 2009;83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi Y, McClure MO, Pizzato M. Identification of Gammaretroviruses Constitutively Released from Cell Lines Used for Human Immunodeficiency Virus Research. Journal of Virology. 2008;82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X, et al. Emergence of Resistant Human Immunodeficiency Virus Type 1 in Patients Receiving Fusion Inhibitor (T-20) Monotherapy. Antimicrobial Agents and Chemotherapy. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derdeyn CA, et al. Sensitivity of Human Immunodeficiency Virus Type 1 to the Fusion Inhibitor T-20 Is Modulated by Coreceptor Specificity Defined by the V3 Loop of gp120. Journal of Virology. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 Cell Surface Concentrations on Infections by Macrophagetropic Isolates of Human Immunodeficiency Virus Type 1. Journal of Virology. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harouse JM, et al. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3) J Virol. 2001;75:1990–1995. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choe H, et al. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choe H, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 40.Farzan M, et al. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlan BD, Gardner MR, Joshi VR, Chiang JJ, Farzan M. Direct expression and validation of phage-selected peptide variants in mammalian cells. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M113.452839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, et al. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. Journal of Virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alpert MD, et al. ADCC Develops Over Time during Persistent Infection with Live-Attenuated SIV and Is Associated with Complete Protection against SIVmac251 Challenge. PLoS Pathog. 2012;8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morner A, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shingai M, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013 doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouet F, et al. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J Clin Microbiol. 2005;43:2709–2717. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 50.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. Epub 2004 Feb 2723. [DOI] [PMC free article] [PubMed] [Google Scholar]