Abstract
We investigated the expression of gross cystic disease fluid protein 15 (GCDFP) and mammaglobin (MGB) by immunohistochemical analysis in 71 invasive breast carcinomas (IBCs) subtyped into luminal (A and B), HER2, basal-like carcinoma (BLC), and unclassified triple-negative carcinoma (UTNC) by established surrogate immunohistochemical profiles. GCDFP and MGB were less likely to be expressed in BLC than in HER2 cancers (P = .000021 and P = .013, respectively) or luminal cancers (P = .00002 and P = .00008, respectively). However, the difference in GCDFP or MGB expression between HER2 and luminal cancers was not significant (P = 1.0 and P = .671, respectively). Our results suggest that luminal cancers demonstrate similar degrees of apocrine differentiation as HER2 cancers. Most BLCs and UTNCs are negative for MGB and GCDFP. Correlation with clinical findings may be needed to exclude the possibility of a metastasis to the breast when BLCs or UTNCs are encountered in a limited sample such as a core biopsy sample.
Keywords: Breast, Invasive carcinoma, Gross cystic disease fluid protein, Mammaglobin, HER2
In 2000, Perou et al1 found initial evidence for molecular subtypes of breast cancer from a complementary DNA–microarray study of gene expression among breast tumor samples and several benign control samples. Based on their findings, a novel molecular classification of breast cancer into basal-like carcinoma (BLC), luminal, HER2, and normal was proposed. In 2001, the same group subdivided the luminal subgroup into A and B.2 Although the “gold standard” for determining molecular invasive breast carcinoma (IBC) subtypes is gene expression array analysis, this method requires frozen tissue. Surrogate immunohistochemical profiles correlating to the molecular subtypes have been developed.3 By surrogate immunohistochemical profiles, IBCs can be subtyped into luminal A (estrogen receptor [ER]+ and/or progesterone receptor [PR]+ and HER2−), luminal B (ER+ and/or PR+ and HER2+), HER2 (ER−, PR−, and HER2+), BLC (ER−, PR−, HER2−, cytokeratin [CK] 5/6+, and/or epidermal growth factor receptor [EGFR]+), and unclassified triple-negative carcinoma (UTNC; ER−, PR−, HER2−, CK5/6−, and EGFR−).
Gross cystic disease fluid protein 15 (GCDFP) was identified by Haagensen et al4 in 1977 as 1 of 4 glycoproteins present in breast gross cystic disease fluid. GCDFP, a 15-kDa monomer mapped to chromosome 7, is uniformly expressed in cells of apocrine differentiation and functions as an aspartyl protease with fibronectin as its substrate.5 It also inhibits T-cell apoptosis induced by CD4 cross-linking and subsequent T-cell receptor activation.6 Normal breast ducts and lobules do not express GCDFP; however, apocrine metaplastic epithelium expresses GCDFP.7 GCDFP expression by immunohistochemical methods has been reported in approximately 25% of breast carcinomas.8
Recent studies have suggested that the HER2 subtype of IBC is better classified as a molecular “apocrine” subtype, which preferentially demonstrates apocrine morphologic features and androgen receptor (AR) expression.9-12 However, GCDFP expression in the different molecular subtypes of breast cancer has not been previously studied.
Gene sequence fragments of mammaglobin (MGB) were first isolated from breast carcinoma tissue in 1994 by Watson and Fleming13 using a modified differential display polymerase chain reaction technique. In 1996, the same authors isolated the full-length complementary DNA clone of MGB.14 In addition to breast carcinoma, MGB is also expressed in benign breast epithelium. MGB gene architecture is similar to that of a family of related genes that includes uteroglobin and rat prostatein subunits C1, C2, and C3. The MGB gene was mapped by fluorescence in situ hybridization to chromosome 11q13, a genomic region frequently amplified in breast neoplasia.15 MGB expression by immunohistochemical analysis has been reported to be found in 55.4% of breast carcinomas,8 although its expression in the different molecular subtypes of breast cancer has not been well studied.
In the present study, we investigated GCDFP and MGB expression by immunohistochemical analysis in IBCs subtyped into luminal (A and B), HER2, BLC, and UTNC.
Materials and Methods
Case Selection and Tissue Microarray Construction
This study was approved by the institutional review board of the Johns Hopkins Medical Institutions, Baltimore, MD. We reviewed cases of IBC resected at our institution between the years 2001 and 2007. All of these cases were sectioned in a fresh state and fixed overnight in 10% neutral buffered formalin before processing to ensure 24 to 33 hours of formalin fixation. IBCs showing processing artifacts that were small (<1 cm) or from cases treated with neoadjuvant chemotherapy were excluded from consideration.
Tissue microarrays (TMAs) were constructed as previously described.16,17 Each TMA contained 99 tissue cores, each 1.4 mm in diameter. These were arranged as 9 rows and 11 columns. Column 6 consisted of unrelated control tissue, leaving 90 cores on the array for carcinoma samples. For each carcinoma case, 5 areas were identified on the H&E-stained slides, punched from the corresponding donor blocks, and placed on the array. Therefore, each array contained 18 different IBCs. Among the 5 samples of each case, we attempted to include normal tissue in 1 sample, if possible, leaving 4 to 5 cores of IBC per case.
Immunohistochemical Staining and Scoring
Immunohistochemical studies for ER, PR, HER2, CK5/6, and EGFR were performed as previously described.17 Immunohistochemical studies for ER, PR, and HER2 were previously performed on all cases as part of a routine panel applied on all cases of IBC at The Johns Hopkins Hospital, Baltimore. The slides were reviewed by 2 of us (A.P.S. and P.A.) to confirm the reported interpretation. IBCs demonstrating weak, moderate, or strong nuclear labeling for ER or PR in greater than 1% of cells were considered ER+ or PR+, respectively. To qualify as HER2+ for this study, a case had to demonstrate a 3+ (strong positive) immunohistochemical score or an HER2 fluorescence in situ hybridization amplification ratio of greater than 4. Cases with equivocal ratios (1.8-2.2) or low-level amplification (ratios, 2.2-4.0) were excluded from this study owing to their uncertain clinical significance.
CK5/6 and EGFR immunohistochemical studies were performed on IBCs that were negative for ER, PR, and HER2 to identify BLC cases as previously described.17 For CK5/6, cases were scored on the basis of percentage of positive cells into 1+ (1%-25%), 2+ (26%-50%), 3+ (51%-75%), and 4+ (76%-100%) categories. Cases demonstrated convincing membranous or cytoplasmic labeling in more than 25% of neoplastic cells were considered positive. Cases with equivocal or less extensive labeling, which was difficult to distinguish from biotin artifact, were excluded from the study, so as to include only unequivocal cases in the BLC category. For EGFR, cases were scored on the basis of percentage of positive cells into 1+ (1%-25%), 2+ (26%-50%), 3+ (51%-75%), and 4+ (76%-100%) categories. Any strong membranous labeling for EGFR was considered a positive result. In general, positive cases demonstrated labeling in 10% to 50% of neoplastic cells.
Immunohistochemical studies for GCDFP and MGB were performed on TMAs. Immunohistochemical labeling was performed using standard methods. Immunohistochemical labeling for GCDFP and MGB was performed on the BenchMark XT autostainer (Ventana, Tucson, AZ). The antibodies, dilutions, and sources were as follows: GCDFP, monoclonal antibody, catalog No. 611-01, dilution 1:200, Signet, Dedham, MA; MGB, monoclonal antibody, catalog No. M7401, dilution 1:200, DAKO, Carpinteria, CA. GCDFP and MGB labeling was scored by 2 of us (G.H.L and P.A.) as the percentage of tumor cells showing positive labeling multiplied by the intensity (weak, 1+; moderate, 2+; or strong, 3+), yielding possible H scores of 0 to 300. Any labeling with these markers was considered a positive result.
Statistical Analysis
Data were analyzed by using the Student t test and the Fisher exact test.
Results
Patient demographics, tumor grade, and tumor stage for this cohort have previously been described.17 Details of age, grade, and lymph node status of the cases used in this study are summarized in Table 1
Table 1. Demographics of Cases.
| Cancer Subtype | Mean Age (y) | Elston Grade | Nodal Status | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| 1 | 2 | 3 | Involved | Uninvolved | ||
| Luminal A (n = 17) | 64.1 | 4 | 8 | 5 | 8 | 1 |
| Luminal B (n = 7) | 50.3 | 0 | 3 | 4 | 4 | 2 |
| HER2 (n = 14) | 56.1 | 0 | 0 | 14 | 7 | 7 |
| BLC (n = 21) | 51.5 | 0 | 1 | 20 | 11 | 9 |
| UTNC (n = 12) | 53.0 | 0 | 0 | 12 | 2 | 10 |
BLC, basal-like carcinoma; UTNC, unclassified triple-negative carcinoma.
GCDFP labeling was present in 37% (26/71) of total cases. Among the different subtypes, 5% (1/21) of BLCs, 65% (11/17) of luminal A tumors, 71% (5/7) of luminal B tumors, and 64% (9/14) of HER2 tumors expressed GCDFP. No GCDFP labeling was identified in UTNCs. The mean GCDFP H score (percentage of tumor cells showing positive labeling multiplied by intensity) among positive cases was 6 in BLCs, 5.18 in luminal A tumors, 1.2 in luminal B tumors, and 4.22 in HER2 tumors. Averaged over total cases (positive for GCDFP labeling and negative), GCDFP H scores were 0.286 in BLCs, 3.35 in luminal A tumors, 0.857 in luminal B tumors, and 2.71 in HER2 tumors. Because there was no GCDFP labeling in the UTNCs, the GCDFP H score was 0.
MGB labeling was present in 54% (38/71) of total cases. Among the different subtypes, 24% (5/21) of BLCs, 17% (2/12) of UTNC, 82% (14/17) of luminal A tumors, 86% (6/7) of luminal B tumors, and 79% (11/14) of HER2 tumors expressed MGB. The mean MGB H score among positive cases was 8.4 in BLCs, 2.5 in UTNCs, 58.4 in luminal A tumors, 88.8 in luminal B tumors, and 95.2 in HER2 tumors. Averaged over total cases, the MGB H score was 2 in BLCs, 0.417 in UTNCs, 48.1 in luminal A tumors, 76.1 in luminal B tumors, and 74.8 in HER2 tumors. These data are summarized in Table 2. Representative labeling is shown in Image 1.
Table 2. GCDFP and MGB Immunolabeling*.
| GCDFP | MGB | |||
|---|---|---|---|---|
|
|
|
|||
| Cancer Subtype | Positive | Mean H Score | Positive | Mean H Score |
| Luminal A (n = 17) | 11 (65) | 3.35 | 14 (82) | 48.1 |
| Luminal B (n = 7) | 5 (71) | 0.857 | 6 (86) | 76.1 |
| HER2 (n = 14) | 9 (64) | 2.71 | 11 (79) | 74.8 |
| BLC (n = 21) | 1 (5) | 0.286 | 5 (24) | 2 |
| UTNC (n = 12) | 0 (0) | 0 | 2 (17) | 0.417 |
BLC, basal-like carcinoma; GCDFP, gross cystic disease fluid protein; MGB, mammaglobin; UTNC, unclassified triple-negative carcinoma.
Data in the positive columns are given as number positive (percentage).
Image 1.
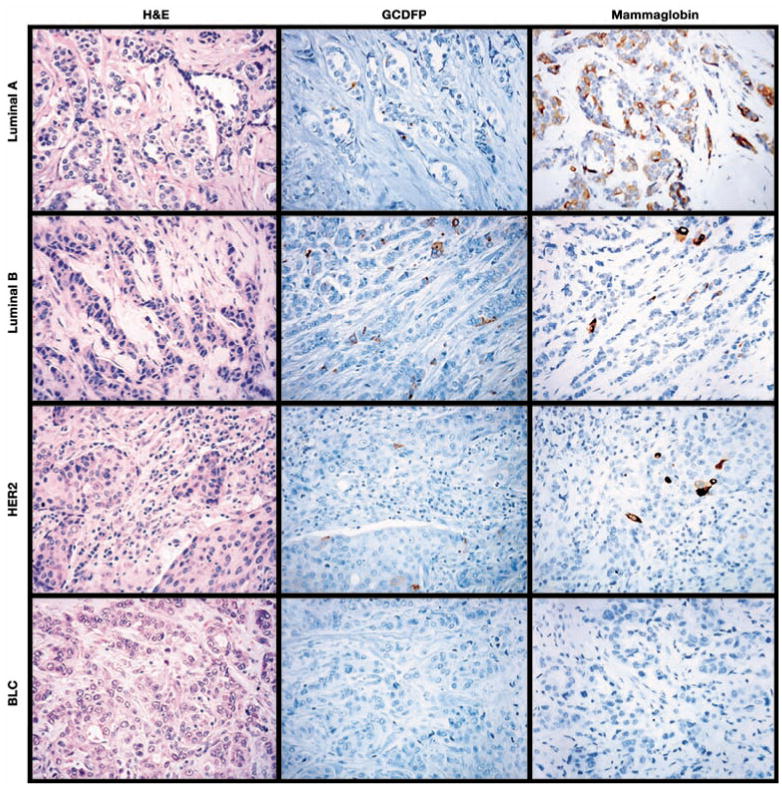
Representative cases of luminal A, luminal B, HER2, and basal-like carcinomas (BLC) immunolabeled for gross cystic disease fluid protein (GCDFP) and mammaglobin. While the luminal A and B cancers and the HER2 cancer are focally positive for GCDFP and mammaglobin, the basal-like carcinoma is negative for both markers (all parts, ×400).
Comparing the scoring between the different groups revealed that GCDFP and MGB labeling are least likely in UTNCs. GCDFP and MGB labeling is less likely in BLCs than in HER2 (P = .000021 and P = .013, respectively) and luminal groups (P = .00002 and P = .00008, respectively). However, differences in GCDFP and MGB labeling between HER2 and the luminal subgroups were not significant (P = 1.0 and P = .671, respectively).
Discussion
IBCs have been classified into molecular subtypes based on gene expression profiling. Immunohistochemical expression can be used as a surrogate for molecular profiling. In this study, IBCs were subtyped by surrogate immunohistochemical profiles into luminal A (ER+ and/or PR+ and HER2−), luminal B (ER+ and/or PR+ and HER2+), HER2 (ER−, PR−, and HER2+), BLC (ER−, PR−, HER2−, CK5/6+, and/or EGFR+), and UTNC (ER−, PR−, HER2−, CK5/6−, and EGFR−). GCDFP and MGB have been reported to be expressed in 23.1% and 55.4% of IBCs, respectively.8 In the present study, we investigated the expression of GCDFP and MGB by immunohistochemical analysis in these molecular subtypes of IBC.
In this study, GCDFP labeling was identified in 37% of the cases and MGB expression in 54% of cases. These results are very similar to the percentages reported by Bhargava et al,8 who found MGB and GCDFP labeling in 55.4% and 23.1% of breast carcinomas, respectively. These results indicate that MGB is a more sensitive marker of breast cancer than is GCDFP; however, because MGB labels more nonbreast cancers such as endometrial carcinoma and melanomas, MGB is a less specific marker of breast cancer than GCDFP.8
Recent studies have suggested that a molecular apocrine subtype (which has apocrine morphologic features and AR expression) overlaps with the HER2 subtype.9-12 Farmer et al9 analyzed 49 breast cancers using Affymetrix U133A gene expression microarrays (Affymetrix, Santa Clara, CA) and subdivided them by principal components analysis and hierarchical clustering into 3 groups: basal, luminal, and molecular apocrine. They showed that the molecular apocrine group is AR+ and that HER2 amplification is most common in this subgroup. These authors postulated that the molecular apocrine subtype corresponds to the HER2 group defined by previous authors. These authors also showed that tumors in the molecular apocrine group showed more apocrine morphologic features (such as abundant eosinophilic cytoplasm and pronounced nucleoli) than the basal or luminal tumors. Similarly, Doane et al10 showed that an ER−, AR+ subgroup of breast cancers tends to be HER2+, proliferates in response to androgen, and also shows strong apocrine morphologic features (abundant eosinophilic and granular cytoplasm, vesicular nuclei with prominent nucleoli, and apocrine snouts).
However, most pathologists would accept that the determination of “apocrine morphologic features” is somewhat subjective. In contrast, immunoreactivity for GCDFP is a more objective correlate of apocrine differentiation. To our knowledge, GCDFP expression has not previously been correlated with different molecular subtypes of breast cancer. Our results show that luminal cancers demonstrate a similar degree of GCDFP immunolabeling as HER2 cancers and that BLCs and UTNCs show less frequent immunoreactivity. Moreover, we have found similar degrees of AR expression in luminal cancers as in HER2 cancers (unpublished data, A. Cimino-Matthews and P.A., 2011). Hence, reclassification of HER2 cancers as preferentially apocrine may be premature at this time.
To our knowledge, only 1 other study11 has evaluated MGB expression in the different molecular subgroups of breast cancer. These authors found MGB expression to be more common in luminal (50%) and HER2 cancers (53%) than in BLCs (20%), findings that are very similar to our results.
Our study also showed that most BLCs and UTNCs are negative for GCDFP and MGB. BLCs and UTNCs often grow with a pushing border and frequently lack a prominent ductal carcinoma in situ component, both of which raise the possible differential diagnosis of a metastatic carcinoma to the breast when BLCs or UTNCs are encountered in a needle core biopsy specimen. Given the frequent absence of specific markers of breast origin (ER, PR, GCDFP, and MGB) in BLCs and UTNCs, our results emphasize that correlation with clinical findings may be needed to exclude the possibility of a metastasis to the breast when BLCs or UTNCs are encountered on a limited sample such as from a needle core biopsy. The value of open communication with the radiologist in such circumstances cannot be overstated.
Upon completion of this activity you will be able to.
define the major subtypes of breast carcinoma as defined by gene expression profiling.
correlate the expression of mammaglobin and gross cystic disease fluid protein 15 (GCDFP) with the major subtypes of breast carcinoma as defined by gene expression profiling.
Acknowledgments
Supported by grant P50 CA88843 from the National Institutes of Health, Bethesda, MD.
Footnotes
The ASCP is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians. The ASCP designates this educational activity for a maximum of 1 AMA PRA Category 1 Credit™ per article. This activity qualifies as an American Board of Pathology Maintenance of Certification Part II Self-Assessment Module.
The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Questions appear on p 644. Exam is located at www.ascp.org/ajcpcme.
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 4.Haagensen DE, Mazoujian G, Holder WD, et al. Evaluation of a breast cystic fluid protein detectable in the plasma of breast carcinoma patients. Ann Surg. 1977;185:279–285. doi: 10.1097/00000658-197703000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputo E, Camarca A, Moharram R, et al. Structural study of GCDFP-15/gp17 in disease versus physiological conditions using a proteomic approach. Biochemistry. 2003;42:6169–6178. doi: 10.1021/bi034038a. [DOI] [PubMed] [Google Scholar]
- 6.Gaubin M, Autiero M, Basmaciogullari S, et al. Potent inhibition of CD4/TCR-mediated T cell apoptosis by a CD4-binding glycoprotein secreted from breast tumor and seminal vesicle cells. J Immunol. 1999;162:2631–2638. [PubMed] [Google Scholar]
- 7.Mazoujian G, Pinkus GS, Davis S, et al. Immunohistochemistry of a breast gross cystic disease fluid protein (GCDFP-15): a marker of apocrine epithelium and breast carcinomas with apocrine features. Am J Pathol. 1983;110:105–111. [PMC free article] [PubMed] [Google Scholar]
- 8.Bhargava R, Beriwal S, Dabbs DJ. Mammaglobin vs GCDFP-15. Am J Clin Pathol. 2007;127:103–113. doi: 10.1309/TDP92PQLDE2HLEET. [DOI] [PubMed] [Google Scholar]
- 9.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumors by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 10.Doane AS, Danso M, Lal P, et al. An estrogen-receptor–negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki E, Tsunoda N, Hatanaka Y, et al. Breast-specific expression of MGB1/mammaglobin: an examination of 480 tumors from various organs and clinicopathological analysis of MGB1-positive breast cancers. Mod Pathol. 2007;20:208–214. doi: 10.1038/modpathol.3800731. [DOI] [PubMed] [Google Scholar]
- 12.Bhargava R, Sushil B, Striebel JM, et al. Breast cancer molecular class ERBB2: preponderance of tumors with apocrine differentiation and expression of basal phenotypic markers CK5, CK5/6 and EGFR. Appl Immunohistochem Mol Morphol. 2010;18:113–118. doi: 10.1097/PAI.0b013e3181b94ff1. [DOI] [PubMed] [Google Scholar]
- 13.Watson MA, Fleming TP. Isolation of differentially expressed sequence tags from human breast cancer. Cancer Res. 1994;54:4598–4602. [PubMed] [Google Scholar]
- 14.Watson MA, Fleming TP. Mammaglobin, a mammary-specific member of the uteroglobin gene family, is overexpressed in human breast cancer. Cancer Res. 1996;56:860–865. [PubMed] [Google Scholar]
- 15.Watson MA, Darrow C, Zimonjic DB, et al. Structure and transcriptional regulation of the human mammaglobin gene, a breast cancer associated member of the uteroglobin gene family localized to chromosome 11q13. Oncogene. 1998;16:817–824. doi: 10.1038/sj.onc.1201597. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Fackler MJ, Halushka MK, et al. Heterogeneity of breast cancer metastasis: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subhawong AP, Subhawong T, Nassar H, et al. Most basal-like breast carcinomas demonstrate the same Rb−/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]


