Abstract
Aims
NKX3.1 is an androgen-regulated tumour suppressor gene that is downregulated in prostate carcinoma. Immunohistochemistry for NKX3.1 is primarily specific for prostatic-derived tumours and tissue but is reported in a small number of breast carcinomas. NKX3.1 is also shown to inhibit estrogen receptor (ER) signalling in breast carcinoma models. Here, we investigate labelling of NKX3.1 in invasive ductal (IDC) and lobular (ILC) carcinomas of the breast with full characterisation of ER, progesterone receptor (PR), androgen receptor (AR) and Her2 status.
Methods
Tissue microarrays of 86 primary IDC and 37 ILC were labelled for NKX3.1. The IDC consisted of 20 luminal A, 7 luminal B, 14 Her2, and 45 triple negative carcinomas. The ILC consisted of 34 luminal A and 3 luminal B cases. NKX3.1 expression was scored as percentage nuclear labelling and labelling intensity.
Results
Nuclear NKX3.1 labelling was seen in 2 IDC (2%) and 10 ILCs (27%). labelling intensity was weak in all cases (1–100% nuclear positivity). Positive NKX3.1 labelling was significantly associated with ILC (p<0.0001). NKX3.1 labelling was seen only in ER and AR-positive carcinomas, which showed a significant correlation (p=0.0003 and p=0.0079, respectively). Expression was not correlated with tumour stage, size, Her2 expression, presence of lymph node metastases or age.
Conclusions
This is the first study to evaluate NKX3.1 expression in breast carcinomas with known ER, PR, AR and Her2 status. Further studies are needed to evaluate what potential role NKX3.1 plays in ER and AR signalling and hormonal treatment response in breast carcinomas.
INTRODUCTION
NKX3.1 is an androgen-regulated, homeobox tumour suppressor gene that is downregulated in prostate carcinoma and associated with prostate carcinoma progression.1–4 Immunohistochemical (IHC) expression of NKX3.1 is largely specific for prostatic-derived tumours and tissue, however, it was also originally described in normal testis, in 9% of invasive ductal carcinomas (IDC) and in 26% of invasive lobular carcinomas (ILC).1 Although initial studies suggested that NKX3.1 expression by IHC was decreased in metastatic prostate carcinoma,1 newer antibodies show greater sensitivity for NKX3.1 in prostate metastases.5,6 This greater sensitivity did not appear to compromise specificity, as NKX3.1 labelling was examined in a wide range of tumour types and only seen in one non-prostatic case,6 which was a case of breast ILC.
The finding that NKX3.1 labelling is limited to prostatic and breast carcinomas is an interesting one, as both tissue types are hormonally regulated, namely through the androgen receptor (AR)7 and estrogen receptor (ER),8 respectively. In prostate carcinoma, NKX3.1 colocalises with AR across the cancer genome,9 and NKX3.1 correlates with AR expression.3 Interestingly, NKX3.1 was shown to inhibit ER signalling in murine models of breast cancer.10 Furthermore, AR signalling is increasingly understood to have a role in breast carcinoma progression and is a candidate for targeted therapies in breast carcinoma.11 AR expression in breast carcinomas is associated with better clinical outcomes independent of ER status12,13; decreased AR expression is seen in end-stage breast carcinoma metastases14; and loss of AR labelling predicts earlier recurrences in triple negative breast carcinomas.15
To date, the relationship between NKX3.1, AR and ER in human breast carcinoma has not been examined. Here, we investigate the expression of NKX3.1 in primary IDCs and ILCs of the breast with full characterisation of clinicopathologic features including AR, ER, progesterone receptor (PR), and Her2 status.
MATERIALS AND METHODS
Tissue microarray construction and case selection
This study was approved by the institutional review board of the Johns Hopkins Medical Institutions. Tissue microarrays (TMA) were created from archived, formalin-fixed paraffin-embedded tissues from 36 cases of primary ILC. Each TMA consisted of 99 cores measuring 1.4 mm in diameter, with five cores taken per tumour to minimise sampling error, including one core that contained benign lobules as an internal control. We also evaluated and expanded upon previously described TMAs16 containing 1 case of primary ILC and 86 cases of primary IDC, subdivided into the categories of luminal A (ER and/or PR+, Her2−), luminal B (ER and/or PR+, Her2+), Her2 (ER−/PR−/Her2+), and triple negative carcinomas (ER−/PR−/Her2−), using established IHC surrogate markers of gene expression profiles.17,18
Immunohistochemistry and expression scoring
Briefly, hormone expression for AR, ER, and PR was scored as labelling intensity (none, weak, moderate or strong) and percentage nuclear labelling (0–100%), with any labelling greater than 1% considered positive. Her2 IHC expression was scored from 0–3+ using established criteria using labelling intensity and proportion complete membranous labelling. To qualify as Her2-positive, a tumour had to demonstrate an IHC score of 3+ (defined as greater than 10% complete, strong membranous labelling) or a Her2 fluorescence in situ hybridisation ratio greater than 2.2. All ER-positive cases in this series showed greater than 70% ER labelling, such that they were unequivocally ER positive; conversely, all cases classified as ER and PR negative showed 0% nuclear labelling.
NKX3.1 labelling was scored as labelling intensity (absent, weak, moderate and strong) and labelling extent (0–100%), with any nuclear immunoreactivity considered positive. A core of benign prostate tissue was included on the TMAs and served as a positive control for NKX3.1 labelling. IHC of TMAs for NKX3.1 was performed using a prediluted rabbit polyclonal antibody (Biocare Medical, Concord, California, USA). Sections from formalin-fixed paraffin-embedded tissue were cut at 4 μm thickness on charged slides, and immunostaining was performed on automated Ultra Benchmark system (Ventana Medical Systems, Tucson, Arizona, USA) using a biotin-free Ultra-View detection (Ventana Medical Systems, Tucson, Arizona, USA). Staining followed routine basic steps of IHC including baking, deparaffinisation, antigen retrieval with high pH EDTA buffer (CC1) for 36 min and primary antibody incubation for 36 min. An amplification step was added that was followed by an ultra-wash step. For visualisation, reaction was developed with 3′3′ diaminobenzidine, and then counterstained with haematoxylin.
Statistical analysis
NKX3.1 nuclear labelling was correlated to carcinoma type (IDC vs ILC), hormone receptor status (AR, ER and PR), tumour stage, tumour size, Her2 expression, presence of lymph node metastases and mean patient age using a Fisher’s exact test calculated with GraphPad Software. A p value of <0.05 was considered to be statistically significant.
RESULTS
Clinicopathologic characteristics
The previously described 86 IDC cases consisted of 20 luminal A, 7 luminal B, 14 Her2 positive, and 45 triple negative carcinomas.16 The patients represented in these TMAs had an average age of 55 years (range, 30–95 years); three cases were Elston grade I, 15 cases were Elston grade II, and 68 cases were Elston grade III. The 37 ILC cases consisted of 34 luminal A and 3 luminal B. Lobular carcinoma in situ (LCIS) was present in 18 cases, 3 of which were pleomorphic LCIS. Clinicopathologic data was available for the 36 ILC on the newly created TMAs, which had an average patient age of 58 years (range, 34–87 years) and average primary tumour size of 2.9 cm (range, 0.7–10 cm). Fourteen cases were stage I, 13 cases were stage II, 8 cases were stage III, and one case presented as stage IV with metastatic disease to the bone. Three cases were Elston grade III, with the remaining 33 cases grades I–II. Androgen receptor labelling was able to be interpreted on 121 of the 122 carcinomas in this series. Of the 121 carcinomas, there were 58 ER +/AR+, 4 ER+/AR−, 20 ER−/AR+, and 39 ER−/AR−.
NKX3.1 labelling
The clinicopathologic and demographic details of cases with positive NKX3.1 labelling are described in table 1. Nuclear NKX3.1 labelling was seen in 2 IDC (2%) and 10 ILC (27%) (figure 1), as well as in the associated LCIS in three of the ILC. All but one case with NKX3.1 labelling were luminal A, with the remaining case being luminal B. One case of the positive ILC had pleomorphic features (case 6). The nuclear labelling intensity was weak in all cases, with variable percentages of cells demonstrating expression (1–100%). NKX3.1 positivity was seen only in ER-positive and AR-positive breast carcinomas, which showed a significant correlation (p=0.0003 and p=0.0079, respectively, Fisher’s exact test). The NKX3.1-positive cases representing 21% of all ER+/AR+ carcinomas in this series. Positive labelling was also significantly associated with the lobular histologic subtype (p<0.0001, Fisher’s exact test; table 2). NKX3.1 expression did not statistically correlate with tumour stage, presence of lymph node metastases, tumour size, Her2 expression, or patient age.
Table 1.
Clinicopathologic characteristics of breast carcinomas with positive NKX3.1 labelling
| Case | Age | Race | Type | Elston Grade | Pathologic Stage | Tumour Size (cm) | Phenotype | ER % | PR % | AR % | NKX3.1 (%) | Her2 Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 60 | W | IDC | II | T1N1 | 2 | Luminal A | 95 S | 70 S | 90 S | 5 | Negative |
| 2 | 73 | B | IDC | I | T2N1mi | 3 | Luminal A | 100 S | 100 S | 95 MS | 30 | Negative |
| 3 | 42 | W | ILC | II | T3N1mi | 5.7 | Luminal A | 90 S | 80 MS | 90 S | <5 | Negative |
| 4 | 41 | W | ILC | II | T2N1mi | 2.1 | Luminal B | 80 MS | 90 S | 60 M | 90 | Positive |
| 5 | 52 | W | ILC | II | T2N1 | 2.6 | Luminal A | 90 S | 70 S | 85 S | 5–10 | Negative |
| 6 | 47 | O | ILC | II | T2N2 | 2.4 | Luminal A | 95 S | 75 S | 95 S | <5 | Negative |
| 7 | 57 | U | ILC | I | T1N0 | 1.6 | Luminal A | 95 S | 5 M | 90 S | 10 | Negative |
| 8 | 68 | W | ILC | II | T2N0 | 2.5 | Luminal A | 90 MS | 50 MS | 75 M | 25 | Negative |
| 9 | 68 | W | ILC | II | T1N0 | 1.8 | Luminal A | 100 S | 100 S | 95 S | 50 | Negative |
| 10 | 41 | W | ILC | II | T2N0 | 3.3 | Luminal A | 75 MS | 65 MS | 80 MS | <5 | Negative |
| 11 | 75 | W | ILC | I | T3N1mi | 8 | Luminal A | 95 S | 80 M | 85 MS | <5 | Negative |
| 12 | 79 | W | ILC | I | T2N0 | 2.2 | Luminal A | 95 S | 90 S | 90 S | 100 | Negative |
AR, androgen receptor; B; black; cm, centimetres; ER, estrogen receptor; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; M, moderate; MS, moderate-strong; N, node; O, other; PR, progesterone receptor; S, strong; T, tumour; U, unknown; W, white.
Figure 1.
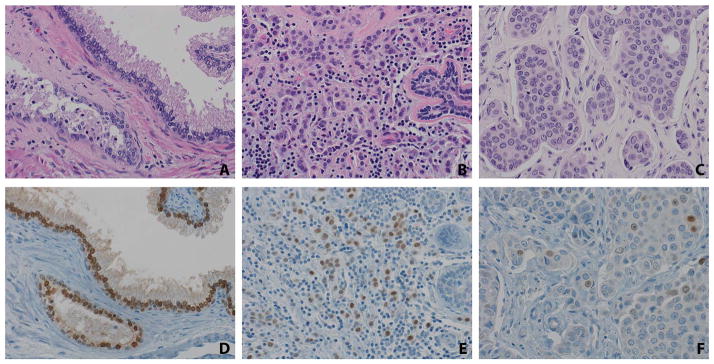
NKX3.1 labelling is seen in invasive lobular and ductal carcinomas of the breast. Benign prostatic glands (A) show strong nuclear NKX3.1 labelling (D), serving as a control on the tissue microarrays. Over 25% of invasive lobular carcinomas (B) showed weak-moderate nuclear labelling for NKX3.1 (E). Benign breast lobules did not label for NKX3.1 in any cases (E, right-hand side). Just 2% of invasive ductal carcinomas (C) showed weak nuclear NKX3.1 labelling (F). (×40)
Table 2.
NKX3.1 nuclear positivity was significantly associated with ER and PR positivity, AR positivity, and lobular histologic subtype
| ER+ | ER− | AR+ | AR− | ILC | IDC | |
|---|---|---|---|---|---|---|
| NKX3.1+ | 12 | 0 | 12 | 0 | 10 | 2 |
| NKX3.1− | 50 | 61 | 66 | 43 | 27 | 84 |
| p Value | 0.0003 | 0.0079 | <0.0001 |
AR, androgen receptor; ER, estrogen receptor; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; PR, progesterone receptor.
In control tissues included on the TMA, NKX3.1 labelling was positive in benign prostatic tissue and negative in gastric mucosa, small intestine, large intestine, skin, placenta, benign fibroadipose tissue, pancreas, endometrium, smooth muscle, lung, liver, kidney, spleen, lymph node, thymus, cerebellum and gallbladder. No NKX3.1 labelling was seen in any of the benign breast lobules.
DISCUSSION
Breast cancer is the most common malignancy and cause of cancer death in women in the USA.19 IDC represents the most common type of invasive breast carcinoma,20 with ILC comprising the second largest subtype. IDC and ILC have different biologic behaviours21–23; ILC tends to occur more often in older women, has a higher degree of hormone receptor positivity, a lower pathologic response to neoadjuvant therapy, and metastasises to unusual sites such as the gastrointestinal tract and ovary. Hormone receptor status is a critical component of breast carcinoma prognosis and predicts response to therapy. ER and PR-positive breast carcinomas have a better clinical outcome than ER or PR-negative carcinomas,8,24 and it is increasingly understood that androgens also have an important role in breast cancer progression. AR-positive carcinomas also have a better overall clinical outcome,12,13 and loss of AR expression is associated with breast cancer metastases14 and the development of early recurrence.15
NKX3.1 is a tumour suppressor gene that is regulated by androgens in the human prostate. It colocalises with AR across the prostate cancer genome9 and is associated with AR expression in human prostate carcinomas.3 NKX3.1 is implicated in human prostate development and progression of prostate carcinoma.1–4 Loss of NKX3.1 protein expression in prostate carcinoma has been associated with hormone-refractory disease2 and advanced tumour stage2,3 in limited studies. However, NKX3.1 expression by IHC has been primarily used to support the diagnosis of prostate carcinoma in primary1 and metastatic sites.5,6
Previous studies reported NKX3.1 expression in primary breast carcinomas,1,6 but not in benign breast tissue.1 Interestingly, NKX3.1 is shown to inhibit ER signalling in murine breast carcinoma.10 Given the reported positive labelling of NKX3.1 in a number of IDC and ILC, as well as the association of NKX3.1 with AR in the prostate and ER in mouse breast cancer lines, we hypothesised that NKX3.1 would have an association with ER and AR-positive breast carcinomas. However, no previous study had examined NKX3.1 and hormone receptor expression in breast carcinomas. Here, we present the first systematic study of NKX3.1 labelling in primary breast carcinomas with known AR, ER, PR and Her2 status.
Our study demonstrates nuclear NKX3.1 labelling in 2% of IDC and 27% of ILC, which is in keeping with previously reported rates of 0–9% labelling in IDC1,6 and 25–26% labelling in ILC.1,6 NKX3.1 labelling significantly correlated with the ILC histologic subtype and with ER/PR and AR-positive hormonal status. All but one case of NKX3.1 positivity were seen in luminal A carcinomas, which are an ER/PR-positive and Her2 negative subset of carcinomas with lower risk of local recurrence25 and overall better prognosis.26
It is possible that NKX3.1 is regulated by androgens in the breast in the same way as in the prostate; however, not all AR-positive breast carcinomas in our series demonstrated NKX3.1 labelling. Furthermore, it is unclear if NKX3.1 is aberrantly induced by ER, or if it is inhibiting the ER signalling pathway in these carcinomas. Further studies are needed to correlate NKX3.1 expression with clinical response to hormonal therapies, as well as disease progression and overall survival. Finally, the underlying mechanism for the increased NKX3.1 labelling seen in ILC than IDC is unclear. This may be due to the increased proportion of hormone receptor positivity in ILC than IDC, however, this cannot entirely explain the difference as still only 7% of ER-positive IDC labelled with NKX3.1. Additional studies are warranted.
Finally, in the work-up of a metastatic carcinoma of unknown primary, the presence of NKX3.1 labelling in prostate and breast carcinomas is unlikely to cause diagnostic confusion with exception of a rare circumstance of a male patient with lobular breast carcinoma and prostate carcinoma. The clinical history and other organ-specific markers, such as prostrate specific antigen (PSA) for prostate and gross cystic disease fluid protein (GCDFP) or mammaglobin for breast, would certainly help resolve this issue.
In summary, this is the first study to evaluate the expression of the androgen-regulated tumour suppressor NKX3.1 in breast carcinomas with known AR, ER, PR and Her2 status. NKX3.1 labelling is limited to ER, PR and AR-positive carcinomas, with greater expression seen in ILC than IDC. Future studies are needed to determine if NKX3.1 may serve as a predictive or prognostic factor in breast carcinomas, and to examine the expression in male breast carcinomas.
Take home messages.
This is the first systematic study of NKX3.1 labelling in primary breast carcinomas with known androgen receptor (AR), ER, PR and Her2 status.
NKX3.1 labelling significantly correlated with the ILC histologic subtype and with ER/PR and AR-positive hormonal status.
Future studies are needed to determine if NKX3.1 may serve as a predictive or prognostic factor in breast carcinomas and to examine the expression in male breast carcinomas.
Footnotes
Contributors The authors RJA-K, MAS, MTL, APS, RS, PBI, PA, AC-M all contributed to the article with concept and design of work, analysis and interpretation of data. All authors were available to critically revise and assist in drafting the work. All authors consented to prior submission for publication and agree to be accountable for all aspects of the work.
Competing interests None.
Ethics approval The Johns Hopkins Hospital IRB.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Gelmann EP, Bowen C, Bubendorf L. Expression of NKX3. 1 in normal and malignant tissues. Prostate. 2003;55:111–17. doi: 10.1002/pros.10210. [DOI] [PubMed] [Google Scholar]
- 2.Bowen C, Bubendorf L, Voeller HJ, et al. Loss of NKX3. 1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–15. [PubMed] [Google Scholar]
- 3.Xu LL, Srikantan V, Sesterhenn IA, et al. Expression profile of an androgen regulated prostate specific homeobox gene NKX3. 1 in primary prostate cancer. J Urol. 2000;163:972–9. [PubMed] [Google Scholar]
- 4.Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3. 1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–27. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang AY, DeMarzo AM, Veltri RW, et al. Immunohistochemical differentiation of high-grade prostate carcinoma from urothelial carcinoma. Am J Surg Pathol. 2007;31:1246–55. doi: 10.1097/PAS.0b013e31802f5d33. [DOI] [PubMed] [Google Scholar]
- 6.Gurel B, Ali TZ, Montgomery EA, et al. NKX3. 1 as a marker of prostatic origin in metastatic tumors. Am J Surg Pathol. 2010;34:1097–105. doi: 10.1097/PAS.0b013e3181e6cbf3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Grann VR, Troxel AB, Zojwalla NJ, et al. Hormone receptor status and survival in a population-based cohort of patients with beast carcinoma. Cancer. 2005;103:2241–51. doi: 10.1002/cncr.21030. [DOI] [PubMed] [Google Scholar]
- 9.Tan PY, Chang CW, Chng KR, et al. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32:399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes KA, Song JS, Liu XS, et al. Nkx3-1 and LEF-1 function as transcriptional inhibitors of estrogen receptor activity. Cancer Res. 2008;68:7380–5. doi: 10.1158/0008-5472.CAN-08-0133. [DOI] [PubMed] [Google Scholar]
- 11.Garay JP, Park BH. Androgen receptor as a targeted therapy for breast cancer. Am J Cancer Res. 2012;2:434–45. [PMC free article] [PubMed] [Google Scholar]
- 12.Vera-Badillo FE, Templeton AJ, de Gouveia P, et al. Androgen receptor expression and outcomes in early breast cancer: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:djt319. doi: 10.1093/jnci/djt319. [DOI] [PubMed] [Google Scholar]
- 13.Qu Q, Mao Y, Fei XC, et al. The impact of androgen receptor expression on breast cancer survival: a retrospective study and meta-analysis. PLoS ONE. 2013;8:e82650. doi: 10.1371/journal.pone.0082650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimino-Mathews A, Hicks JL, Illei PB, et al. Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Hum Pathol. 2012;43:1003–11. doi: 10.1016/j.humpath.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thike AA, Yong-Zheng Chong L, Cheok PY, et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Mod Pathol. 2014;27:352–60. doi: 10.1038/modpathol.2013.145. [DOI] [PubMed] [Google Scholar]
- 16.Cimino-Mathews A, Subhawong AP, Illei PB, et al. GATA-3 expression in breast carcinoma: utility in triple negative, sarcomatoid and metastatic carcinomas. Human Pathol. 2013;44:1341–9. doi: 10.1016/j.humpath.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 19.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 20.Ellis IO, Collins L, Ichihara S, et al. Invasive carcinoma of no special type. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. World Health Organization classification of tumours of the breast. Lyon, France: IARC Press; 2012. pp. 34–8. [Google Scholar]
- 21.Arpino G, Bardou VJ, Clark GM, et al. Infiltrating lobular carcinoma of the breast: tumor characteristics and clinical outcome. Breast Cancer Res. 2004;6:R149–56. doi: 10.1186/bcr767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cristofanilli M, Gonzalez-Angulo A, Sneige N, et al. Invasive lobular carcinoma classic type: response to primary chemotherapy and survival outcomes. J Clin Oncol. 2005;23:41–8. doi: 10.1200/JCO.2005.03.111. [DOI] [PubMed] [Google Scholar]
- 23.Ferlicot S, Vincent-Salomon A, Medioni J, et al. Wide metastatic spreading in infiltrating lobular carcinoma of the breast. Eur J Cancer. 2004;40:336–41. doi: 10.1016/j.ejca.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Dawood S, Hu R, Homes MD, et al. Defining breast cancer prognosis based on molecular phenotypes: results from a large cohort study. Breast Cancer Res Treat. 2011;126:185–92. doi: 10.1007/s10549-010-1113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voduc KD, Cheang MC, Tyldesley S, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28:1684–91. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Park I, Cho HJ, et al. Analysis of the potent prognostic factors in luminal-type breast cancer. J Breast Cancer. 2012;15:401–6. doi: 10.4048/jbc.2012.15.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]


