Abstract
Objectives
Slower rates of aging distinguish humans from our nearest living cousins. Chimpanzees rarely survive their forties while large fractions of women are postmenopausal even in high-mortality hunter–gatherer populations. Cellular and molecular mechanisms for these somatic aging differences remain to be identified, though telomeres might play a role. To find out, we compared telomere lengths across age-matched samples of female chimpanzees and women.
Methods
We used a monochrome multiplex quantitative polymerase chain reaction to assay canonical telomere repeats in blood cells from captive female chimpanzees (65 individuals; age: 6.2–56.7 years) and compared them to the same measure in human females (43 individuals; age: 7.4–57.3 years).
Results
Our samples showed little difference in attrition rates between the species (~0.022 T/S per year for chimpanzees and ~0.012 T/S per year for humans with overlapping 95% confidence intervals), but telomeres were twice as long in chimpanzees as in humans (T/S ratios = 2.70 and 1.26, respectively).
Conclusions
Based on the longevity differences, we initially hypothesized that telomere shortening rates would be faster in chimpanzees than in humans. Instead, it is shorter telomere length that appears to be the derived state in humans. This comparison indicates that better characterization of physiological aging in our closest living relatives will be indispensable for understanding the evolution of distinctive human longevity.
Reconstructions of human life-history evolution rely heavily on nonhuman primates to help distinguish derived versus ancestral traits of growth, age-specific fertility, morbidity, and mortality that define our species. Chimpanzees (genus Pan) are especially useful for such comparisons. In addition to being our closest living relatives (Perelman et al., 2011), they are similar in brain and body size to the extinct genus Australopithecus from which our own genus Homo likely evolved (Wood, 2010 but see Plavcan, 2012 for complications.). Human–chimpanzee comparisons informed by the wider variation among living great apes can help identify the changes in the evolution of our own lineage.
Apart from larger brains, longer adult lifespan stands as a key derived feature of humans that distinguishes us from chimpanzees and the other living great apes (with whom we share the family Hominidae; Perelman et al., 2011). Our exceptional longevity is often obscured by mistaken assumptions that elders were rare in the past, assumptions fostered by the erroneous use of life expectancy at birth as an estimate of adult lifespan. Although life expectancy at birth has doubled in some human populations since the mid 19th Century, the increase through the 1950s is largely owing to declines in infant and juvenile mortality (Oeppen and Vaupel, 2002). In historical and traditional human populations with life expectancies of <40 years, a girl who survives to adulthood has about a 70% chance of surviving beyond the fertile years; among the living adults, one-third of the women are past the childbearing ages (Gurven and Kaplan, 2007). Yet, ages at last birth and rates of ovarian follicle depletion with age are similar in chimpanzees and humans (Jones et al., 2007; Robson et al., 2006). If female fertility decline remains similar to our sister species, yet overall somatic aging is slower in humans (Hawkes et al., 2009), then what physiological processes account for our slower somatic aging rates and our distinctive postmenopausal life spans (Alberts et al., 2013; Levitis et al., 2013)?
Aging is commonly viewed as a decline in functional performance and increased risk of morbidity and mortality across adulthood due to the accumulation of cellular damage (Kirkwood, 2005). Evolutionary life-history theory draws on assumptions about tradeoffs to explain it (Hamilton, 1966; Kirkwood and Rose, 1991). Although organisms have evolved mechanisms that prevent, reduce, or repair damage, allocation to those mechanisms comes at the cost of decreased investment in current reproduction. Whether selection favors increased allocation to maintenance depends on both the chance of being around at older ages and also the fitness gains likely to be earned then (Hamilton, 1966; Kirkwood and Rose, 1991). Selection favors those phenotypes more successful at leaving descendants; descendant numbers depend on the survival and reproduction of children and grandchildren (Hamilton, 1966). Many hypotheses exist to explain the reason why postmenopausal life spans evolved in our line-age, some with a focus on grandmothering (Hawkes, 2003; Hawkes et al., 1998; Kim et al., 2012; Lee, 2003, 2008), others employing alternative hypotheses (Cant and Johnstone, 2008; Ellison, 2010; Kaplan et al., 2010; Morton et al., 2013; Tuljapurkar et al., 2007).
Within this evolutionary framework, mutations that affect physiological maintenance are expected to spread when they net fitness benefits. Similarly, maintenance mechanisms depend on likely sources of damage. One hypothesis about accumulating somatic damage with age is the Oxidative Stress Theory of Aging in which free radical production as a result of metabolism causes increases in cellular damage over time that eventually lead to the various phenotypes of aging (reviewed in Selman et al., 2012). According to this hypothesis, differences in rates of damage, rates of repair, or both account for species differences in rates of aging. Although the case for oxidative damage shaping life histories is mixed (Selman et al., 2012), the search for molecular pathways controlling cellular aging has pointed to a potential marker: telomeres. Telomere shortening could be either a driver of aging (a source of increasing damage), a consequence of it, or some combination of both (Hornsby, 2006).
Telomeres are repetitive, noncoding DNA sequences at the ends of chromosomes that share a conserved sequence of six bases (5’-TTAGGG-3’)n across the vertebrates and a conserved primary function in all linear eukaryotic genomes studied. With a unique secondary structure and associated proteins, telomeres maintain chromosomal integrity during cell replication (Blackburn, 2005; reviewed in Haussmann and Marchetto, 2010). Telomeres are restored by the ribonucleoprotein telomerase. In the primate radiation, telomerase expression occurs mainly in proliferative cells and is repressed/absent in nonproliferative adult somatic tissue (Gomes et al., 2011). The high guanine content of telomere sequence along with suppression of DNA repair enzymes exposes telomeres to higher oxidative damage and subsequent unrepaired double-strand DNA breaks (Shay and Wright, 2007). In the absence of sufficient telomerase activity, each replication event leads to progressive telomere shortening, making telomeres detectors of both the number of cellular replications and the degree of DNA damage for each cell.
This fact has implicated telomeres in the Replicative Aging Theory of Cellular Senescence (Harley et al., 1992; Shay and Wright, 2007). This theory proposes that cellular senescence begins when a subset of telomeres shortens to a critical level, arresting the cell cycle. This could serve as a strong anticancer mechanism as it blocks further growth of a malignant cell (Caulin and Maley, 2011; Shay and Wright, 2007). However, cells with shortening telomeres might decrease an organism's ability to regenerate old or damaged tissues and, if cell-cycle arrest can be bypassed by further mutations, shortened telomeres might play a causal role in some carcinogensis (see discussions of this topic in Eisenberg, 2011; Hou et al., 2012). As a consequence, energy must be allocated to oxidative protection of these shorter telomeres. These trade-offs implicate telomeres in an Axis of Aging along with DNA damage response and mitochondrial function (Sahin and DePinho, 2012). Accumulation of senescent cells in an aging tissue also alters normal tissue homeostasis, likely undermining function through chronic secretion of proinflammatory molecules (Campisi, 2013; Wiemann et al., 2005).
Recently, Gomes and colleagues’ (2011) phylogenetic analysis determined that the ancestral mammalian phenotype incorporated replicative aging. Replicative aging may have served as a key response to the higher mutational load of endothermic organisms and might play a stronger role in managing cellular damage and cancer risk in those mammals that evolved greater longevity. This phylogenetic perspective highlights important variation among vertebrates and across mammalian lineages, underlining the special relevance of comparisons among closely related species. Circumstances that favored lifespan increases in the great apes versus other primates and in humans versus other great apes might have selected for changes in telomere length (TL) and/or attrition rates, as we investigate here.
Multiple studies of variation within species have correlated increasing age with decreasing TL. In vivo, this is particularly evident when sampled from replicating somatic cells (though Daniali et al. (2013) report that attrition is not unique to highly proliferative tissue, particularly by adulthood) and the link is clear across many taxa (reviewed in Haussmann and Marchetto, 2010). Individuals of similar age with shortened telomeres suffer more from age-related disease (Cawthon et al., 2003; Kimura et al., 2008), and TL might be a good proxy for an organism's “biological age,” if not necessarily its “chronological age” (Monaghan, 2010). Short telomeres themselves, even in the presence of normally functioning telomerase, are degenerative and pathogenic in a mouse model (Armanios et al., 2009). The magnitude of the telo-mere rate of change (TROC) or attrition rate is another potential marker of (as well as potential contributor to) aging; intraspecies studies suggest that shortening occurs rapidly early in an individual's life, reaches a stable plateau before adulthood, and might increase again late in life (Baerlocher et al., 2007; Nakagawa et al., 2004).
Most TL and attrition research has focused on birds, rodents, and humans though phylogenetic diversity in telo-mere biology (Gomes et al., 2011) limits the relevance of birds and rodents as model systems for humans. Research on phylogenetically closer species has been sparse though comparative nonhuman primate data have accumulated in the past decade (Table 1). For the most part, these studies use the “gold standard” of terminal restriction fragment (TRF) analysis to determine TL (Horn et al., 2010), but variation in TRF methodologies, particularly in chosen restriction enzymes, makes direct comparison of results difficult. These studies differ in the tissue type chosen and in the age range of individuals included, sometimes using cultured cell lines to represent species. In cross-sectional studies with live-donors, ages represented are often limited by the availability of subjects.
TABLE 1.
Published primate telomere length (TL in Kilobases) and telomere attrition (TROC in nt/yr)
| Organism | Tissue | Maximum life span (yrs) | Sex | Age range (yrs) | TL: qPCRa | TL: TRF | TL: flow-FISHa | TROC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Baboon (Papio hamadryas cynocephalus ) | Lymphoctyes | 37.5 | Male | 6.5–26.5 | 7–18 (C); see text | 280 (C); see text | Baerlocher et al 2003 | ||
| Baboon (Papio hamadryas cynocephalus) (L) | Lymphoctyes | 37.5 | unreported | Newborn - ~4 | 25–28, 13–15; see text | Baerlocher et al 2007 | |||
| Bonobo (Pan paniscus) | Cultured skin fibroblasts | 55 | unreported | unreported | 9.3–10.1 | 200 (bp/PD) | Steinert et al 2002 | ||
| Chimpanzee (Pan troglodytes) | PBMCs | 59.4 | unreported | 16–26 | 11.6 | Feng et al 1998 | |||
| Chimpanzee (Pan troglodytes) | PBMCs | 59.4 | unreported | unreported | 20.6, 14.5 | Bhatnagar et al 1995 | |||
| Black-handed spider monkey (Ateles geoffroyi) | Cultured skin fibroblasts | 47.1 | unreported | unreported | 4.4–6.4 | 49 (bp/PD) | Steinert et al 2002 | ||
| Cynomolgus monkey (Macacafascicularis) | Skin Tissue | 39 | Both | 3.6–23 | 13 - 18 (C) (X) | 15.4 | Gardner et al 2007 | ||
| Cynomolgus monkey (Macaca fascicularis) | PBMCs | 39 | unreported | Newborn - 34 | 12–16 | 62.7 | Lee et al 2002 | ||
| Cynomolgus monkey (Macaca fascicularis) | PBMCs | 39 | unreported | 3.9–8.2 | 14.1 | 140 | Shibata et al 1999 | ||
| Gorilla (Gorilla gorilla) | PBMCs | 55.4 | unreported | unreported | 15.4 | Bhatnagar et al 1995 | |||
| Human | Leukocytes | 122.5 | Male | 18–68 | 4.9-9.2 | 19 | Aston et al 2012 | ||
| Human | Leukocytes | 122.5 | Both | 41–70 | 5.4–7.5 (C) | 4.8-7.2 (C) | 30 | Aviv et al 2011 | |
| Human | Lymphoctyes | 122.5 | Both | 4–90 | 4–11 (C) | 52 | Rufer et al 1999 | ||
| Human | PBMCs | 122.5 | unreported | unreported | 8 | Bhatnagar et al 1995 | |||
| Human | Cultured fibroblasts | 122.5 | Both (female biased) | Fetal - 91 | 6.2 - 8.6 (earliest PD) | 48 (bp/PD) (C) | Harley et al 1990 | ||
| Human | Skin Tissue | 122.5 | unreported | Newborn - 76 | 8 - 11 (C) (X) | 8 - 12 (C) | Gardner et al 2007 | ||
| Human (L) | Leukocytes | 122.5 | Both | 20–48.2 | 7.3 (C) | 40.7 | Aviv et al 2009 | ||
| Human (L) | PBMCs | 122.5 | unreported | 27–31 (Z) | 8.5 (Z) | 50–60 (Z) | Feng et al 1999 | ||
| Human (L) | Leukocytes | 122.5 | Both | 51–76 | 7.1- 8.9 (C) | 45.5 | Ehrlenbach et al 2009 | ||
| Human (L) | Leukocytes | 122.5 | Both (male biased) | 55–83 (C) | 4.5–7.5 (C) | 42 | Farzaneh-Far et al 2010 | ||
| Orangutan (Pongo pygmaeus) | Cultured skin fibroblasts | 59 | unreported | unreported | 5.9–10.9 | 151 (bp/PD) | Steinert et al 2002 | ||
| Pig tailed monkey (Macaca nemestrina) | PBMCs | 37.6 | unreported | 1.7–9.0 | 14.9 | 440 | Shibata et al 1999 | ||
| Rhesus monkey (Macaca mulatta) | PBMCs | 40 | unreported | unreported | 16.3 | Bhatnagar et al 1995 | |||
| Rhesus monkey (Macaca mulatta) | Cultured skin fibroblasts | 40 | unreported | unreported | 9.4–16 | 120 (bp/PD) | Steinert et al 2002 | ||
| Ring-tailed lemur (Lemur catta) | Cultured skin fibroblasts | 37.3 | unreported | unreported | 17–24.4 | 96 (bp/PD) | Steinert et al 2002 | ||
| Squirrel monkey (Saimiri sciureus) | Cultured skin fibroblasts | 30.2 | unreported | unreported | 3.8-8.7 | 49 (bp/PD) | Steinert et al 2002 |
Notes:
1)Table adapted from Table 2 of Haussmann et al 2003; Maximum life span from AnAge database http://genomics.senescence.info/species/
2)Unless otherwise noted single numbers are averages when ranges weren't provided
3)Recent human publications prioritized, though older human data from groups who have also investigated nonhuman primates are included
4)When possible for studies that included very young individuals, TL and TROC are recalculated excluding those samples
Values reported only for healthy individuals (particularly non-HIV/SIV infected individuals)
Telomere lengths for the qPCR and flow-FISH methods were calculated from regression curves of TRF with T/S and kMESF respectively, assuming linearity of the regressions
PBMCs – Peripheral Blood Mononuclear Cells L – The study used longitudinal data C – Calculated/Approximated from published data or graphs
X – qPCR T/S values were converted from regression equations in Figure 5 for 13 monkeys with unknown age and tissue and for 32 human kidney tissues with donor age ranging from 0.1–71.4 years
Z – Two individuals were followed 8 and 10 years and had TROC of 50 and 60 nt/year respectively; TL for one individual was calculated from Fig 2 at ~8.5 and the other was unreported
We hypothesized that as somatic aging appears to be faster in chimpanzees than in humans, telomere biology might differ as well. Explicitly, chimpanzee telomeres might be shorter or their attrition rate faster than similar measures in humans. We investigate TL and TROC in a cross-sectional data set of similarly aged cohorts of captive female chimpanzees and human females from the Utah Centre pour les Etudes du Polymorphisme Humaine (CEPH) family pedigrees (Dausset et al., 1990). Table 2 summarizes our sample of 65 female chimpanzees and 43 female humans. Histograms of participant numbers per 5-year age cohort are shown in Supporting Information Figure 1. The average age of the chimpanzees in this study was 28.19 years, similar to the human average age of 31.38 years. We use a monochrome multiplex quantitative polymerase chain reaction (qPCR) method (Cawthon, 2009) to directly assay canonical telomere repeats in whole-blood DNA samples using the same qPCR conditions and same loci for the telomere (T) and single-copy gene (S) signals in both species. All telomere measurements are quantified across multiple qPCRs and, for chimpanzees, multiple extractions (METHODS section). This project avoids the cross-study variance associated with TRF smear analysis differences and the problems introduced by subtelomeric sequence length and restriction site polymorphisms in cross-species comparisons (Cawthon, 2002; Horn et al., 2010; Nakagawa et al., 2004). It represents the largest available telomere data set for chimpanzees.
TABLE 2.
Descriptive Statistics of the Primary Chimpanzee and Human Data Used in this study
| Chimpanzees (n = 65) |
Humans (n = 43) |
|||||
|---|---|---|---|---|---|---|
| Parameter | Mean (95% CI) | Min | Max | Mean (95% CI) | Min | Max |
| T/S Ratio | 2.70 (2.66, 2.74) | 1.11 | 4.33 | 1.26 (1.22, 1.30) | 0.33 | 2.93 |
| Basepairs | 9,005 (8,873, 9,137) | 3,697 | 14,419 | 4,201 (4,069, 4,333) | 1,099 | 9,757 |
| Age (years) | 28.19 (25.07, 31.32) | 6.22 | 56.68 | 31.38 (27.18, 35.59) | 7.40 | 57.30 |
METHODS
Chimpanzee samples and DNA extraction
Blood was drawn from female chimpanzees (Pan troglodytes) during routine health checks of captive populations at the Southwest National Primate Research Center hosted by Texas Biomed (formerly Southwest Foundation for Biomedical Research) in San Antonio, Texas and at the Yerkes National Primate Research Center at Emory University Atlanta, Georgia. Frozen EDTA-treated whole blood was then shipped to the University of Utah for genomic DNA extraction. Samples were chosen to maximize the age range of the chimpanzee population (6.2– 56.7 years; Histogram shown in Supporting Information Fig. 1). A Qiagen QIAamp DNA Blood Mini Kit was used with the Spin Protocol for Blood; RNase A was added during lysis with Qiagen Protease and the amount of ethanol added for binding was increased to 230 ml. A total of 65 individuals were processed and for 57 samples (88%) a second DNA extraction was performed, on a different day, from the same blood draw. DNA concentration was quantified with a NanoDrop 1000 Spectrophotometer.
Of the 41 chimpanzees analyzed from Southwest, 24 pairs had a kinship coefficient of ≥12.5%. In total, 29 individuals were exposed to some combination of HBV, HCV, and/or HIV, and 26 of those individuals tested serology positive for the virus(es) based on the documented medical records for each individual. Pedigrees for the chimpanzees from Yerkes were limited to the second generation and, of the 24 females analyzed, nine pairs had a kinship coefficient of ≥12.5%. All individuals tested serology negative for the above viruses, again based on the medical records. We assayed the chimpanzees from the two centers independently and determined that our TL/TROC conclusions are not altered although the relatedness and infectious disease load differs between the two sample sets (Supporting Information Section 1). We did not explicitly control for relatedness after this check, which may result in underestimated confidence intervals for the chimpanzee sample.
Human DNA samples
Genomic DNA was available from a long-term storage panel of samples from the Utah CEPH collection. These individuals were sampled in the 1980s, were of northern and western European descent, and were selected so that each family was represented by three living generations. The grandparents were typically sampled in their 60s and there were typically 10 siblings in generation 3. DNA was phenol–chloroform extracted from whole blood and stored at ~100 ng/μl in TE−24. Some individuals were later resampled 9.8–20.8 years later. These second samples were extracted with the GentraSystems PureGene kit and were stored at 200 ng/ml in TE24. We requantified a subset of the DNA samples via Nanodrop prior to analysis. No serological tests had been performed on the blood samples prior to DNA extraction. We selected females from the CEPH panel to (1) match the age range of the chimpanzee samples and (2) guarantee that only unrelated females were sampled based on the available three-generation pedigrees for each CEPH family. For individuals with two blood draws, we selected only one DNA sample based on the criteria just listed. For the human samples used in most analyses, a total of 43 females from 7.4 to 57.3 years of age were chosen in this manner (histogram, Supporting Information Fig. 1). A total of 24 DNA samples were selected from original blood draws and 19 DNA samples were selected from the secondary blood draws. Where noted, qPCR and TRF measurements were also available from 47 CEPH female subjects from six families assayed earlier (Cawthon, 2009). Six individuals overlapped between the two sets (Supporting Information Section 2). In all, 35 females from the Cawthon runs fell within the desired age range (5.2–48.2 years), whereas 12 females were older (61.2–84.4 years) (Supporting Information Sections 2 and 3).
Monochrome multiplex qPCR for T/S measurements
PCR reactions were set up as described in Cawthon (2009) but with some modifications. In brief, 96-well plates were run with 25 μl reactions in Bio-Rad iQ5 real-time PCR detection systems. The same conditions were used for both the human and the chimpanzee DNA samples. The working concentrations of reagents in the PCR were as described in Cawthon (2009), excluding the primers. Albumin was picked as the single-copy gene, but newly developed primers albugcr2 and albdgcr2 were used to amplify the target. The albumin primers were changed owing to a four-base overlap with perfect complementarity between the 3’ ends of one albumin primer and one telomere primer. The telomere primer sequences remained the same. Primer sequences and final concentrations in the PCR are listed in Supporting Information Table 1. Mismatches to true telomere or albumin sequence are printed in lowercase. Primer-Blast results confirm that the same sized albumin amplicon should be produced in both species, with no nonspecific amplification, and the panTro-2.1.4 assembly obtained from Ensembl release 73 does not indicate any known copy number variants of this gene in the chimpanzee genome. These databases are not from populations of chimpanzees and hence SNP or CNV variants at this locus might be present at some unknown frequency in this species.
The thermal cycling profile was as follows: Stage 1: 15 min at 95°C; Stage 2: 2 cycles of 2 s at 98°C, 30 s at 49°C; and Stage 3: 34 cycles of 2 s at 98°C, 30 s at 59°C, 15 s at 74°C with signal acquisition, 30 s at 84°C, 15 s at 85°C with signal acquisition. All runs had an attached melting curve analysis to aid in troubleshooting. For each qPCR plate, independently for both the telomere and the single-copy signal amplification curves, the same baseline threshold was applied to all wells of the plate and a crossing threshold was chosen to get an amplification efficiency value for the standard curve between 85 and 100% and a R2-value of >0.990.
The same reference DNA pool, “Standard DNA,” from Cawthon (2009) was used spanning the same 81-fold serial dilution series. This reference is a pooling of clinical DNA samples and not a purified commercial sample. Specimen samples were input from Nanodrop determined concentrations to fall within the range of the standard curve (50 to ~0.6 ng). Any sample whose telomere or single-copy signal fell outside the standard curves was repeated. Three “reference” samples were run in triplicate on all qPCR plates. These samples had T/S measurements of 0.5, 1.0, and 1.3. They were used to determine proper functioning of the assay. All unknown samples were run in triplicate and on two different days. T/S ratios used in analysis were the average values of these measurements for each individual.
Reproducibility criteria for T/S measurements
Reproducibility was assayed on different PCR plates, on different days, and with multiple DNA extractions for the chimpanzee T/S measurements. This corresponds to a target observation number of 12 measurements. For eight chimpanzees, only one extraction was performed, and hence they are represented by six measurements. For one chimpanzee, a PCR triplicate failed, and hence she is represented by nine measurements. For a final chimpanzee one T/S measurement failed, and hence she is represented by 11 T/S ratios. Reproducibility in humans was assayed similarly as described above, but on six measurements. For CEPH samples, only DNA was available and hence a duplicate extraction was impossible. For two human females, one T/S measurement failed, and hence they are represented by five measurements.
For both chimpanzee and human T/S ratios, strict reproducibility criteria were followed. PCR plates of 24 samples were generally set up with the same samples on two different days. Individual PCR plate results were accepted only if the mean coefficient of variation (CV) was 10% or lower across the PCR triplicates on each plate. Once both days were collected, the mean CV of the replicates (typically 12 observations for chimpanzees and 6 observations for humans) across each individual analyzed those 2 days also had to fall at or below 10% (for human samples this is then a check on average interassay CV). For chimpanzees with two extractions, the mean CV of the T/S measurements across each extraction (typically six measurements) had to fall at or below 10% (again an average interassay CV). These criteria allowed certain individual CVs to be above 10% if by chance the rest of the samples on the shared plate(s) gave very precise T/S measurements. In other words, CV criteria were judged based on the averages for the plates and not on an individual basis. Individuals (should they be obvious failures) were combined into new plates and/or whole plates were rerun if these criteria were not met.
The average CV for the chimpanzee samples was 9.76% and the average CV for the human samples was 9.66% (note the lower # of human T/S observations). CV values for all 108 samples are shown in Supporting Information Figure 2a, b; these CV values were based on all observations that met the reproducibility criteria. Two individuals had outlier CVs of ~22 and ~25% for the human and chimpanzee females, respectively. The human sample was from a 30-year-old female from six T/S measurements and the chimpanzee sample was from a 20-year-old female also from six T/S measurements (no duplicate extraction was run). As the outliers fell within plates that met the criteria, the results of both samples were left in the analysis.
Converting T/S ratios to TRF base pair lengths
We used the regression formula from Cawthon (2009) to translate our T/S ratios to base pair (bp) lengths and compare our TLs and attrition rates to those published using only TRF measurements. Unless noted, we did not incorporate the y-intercept term and therefore we report only true TL and not additional sub-TL. This means that a sample with a relative T/S ratio of 1.0 would have a cellular average TL of ~3,330 bp. This also means that the assay is not sensitive to sub-TL variation between the two species, which justifies our application of the same equation to the chimpanzee results although we did not run a TRF assay on those samples (Caveats section and Supporting Information Material). In any plot of mean TRF length versus T/S, we assume that the average length of the subtelomeric segment does not change across the plotted range of mean TRF length, and that the regressions remain linear beyond the measured range. We used the same Standard DNA sample as Cawthon (2009), but as our assay is a modified version of that used earlier we evaluated the use of this conversion factor (a concern raised by Horn et al., 2010) by comparing the T/S ratios of the human samples from both studies. They gave similar results (TL and TROC) regardless of the assay that was used (Supporting Information Section 2).
Statistical methods
Using R version 2.15.0 (R Development Core Team 2012), we constructed linear regressions for the relationship between age and T/S ratio, assuming normally distributed error. We justify this approach in Supporting Information Section 4.
RESULTS
Human and chimpanzee age-related change in Telomere Length
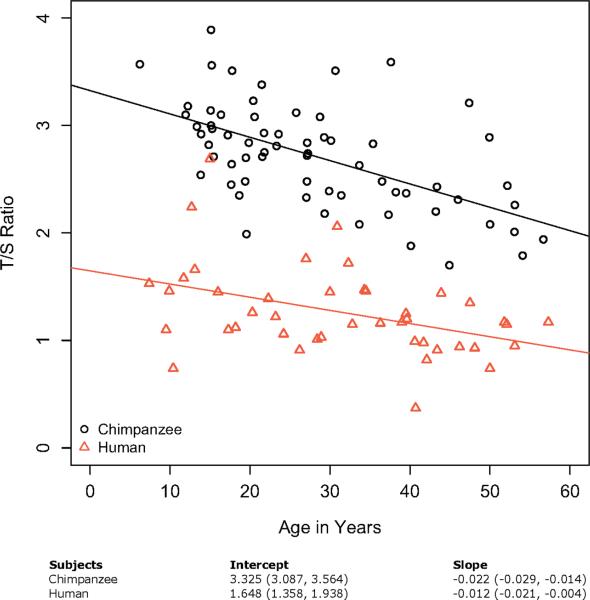
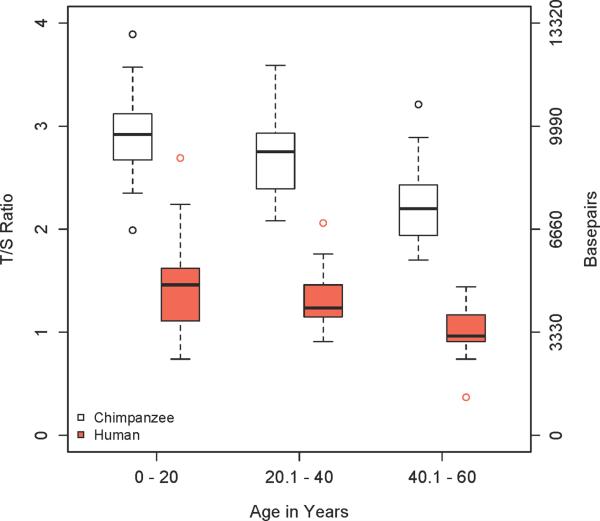
We estimated the influence of age on TL for our sample of female chimpanzees and female humans (Table 2) with a linear regression of relative TL (measured as a T/S ratio) on age (measured in years; Fig. 1). This estimates telomere attrition as approximately 0.022 T/S per year for the female chimpanzees and 0.012 T/S per year for the female humans. These rates convert to a chimpanzee TROC of ~73 nt/year and a human TROC of ~40 nt/year though both values have 95% confidence intervals that overlap. An alternative view of telomere attrition in these two species is shown in box plots of TL for our samples divided arbitrarily into three cohorts of 20 years each (Fig. 2 ).
Fig. 1.
TL change as a function of age for cross-sectional data sets of chimpanzees and humans measured with a modified (METHODS section) monochrome multiplex quantitative PCR method (Cawthon, 2009). All T/S measurements made relative to the same standard DNA curve and the same primer sets. Estimates of the slope and y-intercept of the two regression lines reported as: mean value (95% confidence interval). An ANCOVA regression shows that while the intercepts are meaningfully different (P << 0.001), the slopes are not (P = 0.102).
Fig. 2.
Modified box plots of TL variation when each species is separated into three age cohorts of 20 years. Whiskers represent 1.5× IQD and any data points outside of these maximal values are plotted; the top of the box signifies the 3rd quartile/75th percentile and the bottom of the box signifies the 1st quartile/25th percentile; the dark line is the median. Bp values on the right vertical axis were calculated using the conversion factor of 3,330 from Cawthon (2009) (METHODS section). Measurements were calculated across individuals.
Human and chimpanzee TL
Averaging across all replicates for all individuals shows that chimpanzees had a mean T/S ratio of 2.70 (95% confidence interval: 2.66, 2.74)—more than double the human mean T/S ratio of 1.26 (1.22, 1.30). Using the conversion factor of 3,330 of mean TRF (bps) to relative T/S ratio from Cawthon (2009) gives an average TL (not including subtelomeric regions; see METHODS and DISCUSSION sections) of 9.0 kb for chimpanzees and 4.2 kb for humans (Table 2; summary statistics in this table are based on all replicates for all samples). Both species showed a range of TLs between similarly aged individuals and across all the ages of the study, with some overlap between species, particularly around a boundary T/S of 2.0 or ~6.6 kb (short for chimpanzees and long for humans; Fig. 1).
DISCUSSION
We initially hypothesized that blood cell telomeres would shorten faster in chimpanzees than in humans in a chronological age-matched study. Chimpanzee somatic aging is faster and we expected their telomere biology to reflect that. Instead, for our samples covering the age range of 6–57 years, telomere attrition rates are very similar, but TL is approximately twice as long in chimpanzees as in humans. Table 1 lists the recent human TL/TROC measurements and all such measurements made for non- human primates up to this point. Upon converting our qPCR results to equivalent TRF results, we confirmed that both chimpanzees and humans share short (< 20 kb) telomeres, but humans carry lengths clearly on the short side for the known primate order as a whole. The TL measurements for our Utah CEPH samples fall well within that reported in the literature for expected human leukocyte lengths. Details of length measurements and comparisons to the published literature are in Supporting Information Section 5. Although estimated telomere attrition rates agree with the previously published human data sets, the lack of age-matched samples complicates attrition data for nonhuman primates. Our chimpanzee cross-sectional TROC represents the best estimate so far for a lifelong chimpanzee telomere attrition rate. A discussion of these measurements is in Supporting Information Section 6.
This chimpanzee–human comparison is motivated by our interest in the evolution of human longevity. As all the living hominids (humans and nonhuman great apes) share similar latest ages of female fertility, and all but humans have similar longevity (Robson et al., 2006), we use the best studied of the great apes, chimpanzees, to represent the condition ancestral to our own genus. The importance of such a phylogenetic perspective on telomere biology is demonstrated in Gomes and colleagues’ (2011) examination of TL and telomerase expression in cultured fibroblast cells from more than 60 mammalian species (with nonhuman primate data in Gomes et al. taken from the earlier Steinert et al., 2002). They found significant inverse correlations between TL and lifespan and between telomerase expression and body mass, which itself is strongly correlated with lifespan. Intriguingly, the slope of TL across lifespan was relatively flat and uninformative for mammals with short telomeres (Gomes et al., 2011), an issue we will return to below. On these phylogenetic grounds, we do not draw inferences about humans from published literature on mice and birds (Supporting Information Section 7).
We focus on females because the similarity in ovarian aging between humans and chimpanzees makes the difference in somatic aging especially striking; because female fertility constrains population growth rates—a key evolutionary consideration; and because the evolution of human postmenopausal longevity has been of particular interest to life-history theoreticians (Hamilton, 1966; Hawkes, 2003; Hawkes and Coxworth, 2013; Hawkes et al., 1998). Eisenberg (2011) recently developed a different set of arguments to propose the special importance of males for understanding human telomere biology (for more on paternal age effects, see Aviv and Susser, 2013). Eisenberg suggested that fathers’ age reflects the adult mortality experience of a lineage and that older fathers indicate lower adult mortality. According to his Thrifty Telomere Hypothesis, longer telomeres are optimal when adult mortality is lower. Though our subjects include no males, Eisenberg's hypothesis would predict longer telomeres in humans than chimpanzees as adult mortality is lower and fathers are older in humans compared to chimpanzees (Langergraber et al., 2012). Our interspecific findings show the opposite pattern.
Telomeres and life-history traits
As noted above, telomere integrity plays a role in organismal aging and longevity, and hence telomere biology should be important to those interested in life-history tradeoffs. Likelihood of the future survival of an individual might depend, in part, on their TL and TROC. Evolutionary expectations about selection on these features assume that if background mortality goes down and/or the contributions individuals can make to their fitness at older ages go up, then variants that increase somatic maintenance would be favored. If relatively shorter telomeres or faster attrition than the population mean were the cause of quicker cell-cycle arresT/Senescence, organ failure, aging, and/or death in that individual, then selection would shift population means to longer telomeres or slower attrition. Alternatively, though not mutually exclusively, shorter than average telomeres could be an indicator of compromised health as telomere loss might be accelerated by a variety of stresses over the lifespan (Monaghan, 2010).
At the cellular level, long-lived primates must minimize DNA damage while maintaining tissue homeostasis and avoiding cancer. As all known primates use replicative aging, telomere dysfunction must be a critical cell-cycle checkpoint, with species differences arising in telomere damage and DNA repair or in the rate of cellular senescence and apoptosis. Varied patterns of stem cell division between species are also likely, which might explain the uninformative relationship between TL and longevity in those mammals utilizing replicative aging (Gomes et al., 2011).
Quantitative data are lacking on when and to what degree senescent cells appear with age in humans (Campisi, 2013) and in other great apes, though senescent cells with damaged telomeres do accumulate in baboon dermal fibroblasts (Jeyapalan et al., 2007). Controlling for body mass and phylogeny, and different from other vertebrate homeotherms, increased longevity in primates does not correlate with decreased fibroblast cellular reactive oxygen species (ROS) production at baseline or under stress (Csiszar et al., 2012). The long-lived great apes do tend to show reduced mitochondrial ROS production at baseline and under stress compared to other primates. Humans, the longest lived of all, cluster with the great apes and the great apes as a whole do not differ enough from other primates to support the ROS production–longevity correlation (Csiszar et al., 2012).
Human cells with similar ROS production, similar TROC, but shorter telomeres should be hitting cell-cycle arrest more often than in comparable chimpanzee tissue. Yet, human and chimpanzee tissues express apoptotic pathway genes differently, and human fibroblasts show reduced apoptotic function (greater cell viability) relative to both chimpanzees and a macaque outgroup, which cannot be attributed simply to differential expression of DNA damage repair genes (Arora et al., 2009, 2012; but see Weis et al., 2008). Fibroblasts from mammals with shorter TL and increased maximum lifespan show decreased sensitivity to oxidative stress independent of body mass (Gomes et al., 2011); controlling for mass and phylogenetic confounders, Csiszar and colleagues (2012) found that the degree of H2O2-induced apoptosis and maximum lifespan are inversely correlated in primates. It is thus unclear whether reduced apoptotic function in human cells is due to increased damage repair and/or decreased sensitivity to shortened telomeres.
Selection for longer lifespan in humans required increased investment in somatic maintenance. Perhaps, this also involved reallocation among maintenance mechanisms. Humans can maintain tissue homeostasis with decreased TLs because we evolved less sensitivity to apoptosis, increased DNA repair, and/or decreased cell division rates. Lacking these adaptations, and all else being equal, chimpanzee cells cannot afford shorter TL. The similarity in telomere attrition rates that we observed between chimpanzees and humans suggests that both species use replicative senescence in equal measure though this remains to be determined. Our very short telomeres might represent an asymptote of TL for replica-tive senescent mammals, leading humans with even shorter telomeres to suffer increased mortality and morbidity.
Caveats
Our results speak only to telomere biology in females of either species from blood DNA samples, and therefore from the in vivo leukocyte population. We assume that this is an appropriate tissue type to investigate telomere evolution between the two species; it need not be (Eisenberg, 2011; Hornsby, 2006). When we have compared our results to nonblood DNA samples from the literature, we have noted and/or tried to correct for differences in tissue type (Supporting Information Sections 5 and 6). Leukocytes are also a heterogeneous, and fluctuating, population of cells and TL changes appear to be cell subtype specific (Supporting Information Section 6); our experiments do not specify cell subpopulation.
As our samples are cross-sectional, we are not accurately representing the attrition rate of individuals (Supporting Information Section 6) and cohort effects apply. It is rare for chimpanzees to live to the ages of our oldest subjects, and hence those subjects may be individuals whose telomeres were always longer or shortening more slowly. The same mortality selection may bias the human comparative sample although we are sampling from a very low mortality European population (CEPH Utah families). When we include an older cohort of humans instead of restricting our comparison to age-matching the chimpanzees, the telomere attrition rate is less steep but clearly remains within our confidence intervals (Supporting Information Section 3).
Our chimpanzees were captive and most had undergone medical testing in the past. Hence their diets differ and they likely suffer from different life stresses and diseases than wild chimpanzees. Our conclusions do not change if we restrict our analysis to those chimpanzees from one center with no detectable HBV/HCV/HIV infection (METHODS section; Supporting Information Section 1). Most of the other primate studies we have consulted (Table 1), averaged across ages to give species-level statistics, whereas we provide cross-sectional data at several ages for chimpanzees. The 50-year age range that we sample from each species gives our study the appropriate power to assess species telomere attrition rates. Our combined 108 samples might, however, fall short in assessing attrition rate differences of less than threefold (Aviv et al., 2006).
Concerns over the use of the qPCR assay for telomere research have been raised in the literature (Aviv et al., 2006; Horn et al., 2010), and we have addressed many of these critiques in our METHODS section and Supporting Information. Our multiplex quantitative PCR assay measures the mean TL in the DNA sample and hence would not detect differences between these species in the shortening of a critical subset of telomeres. Additionally, interstitial telomeric repeats (ITRs) are amplified by the assay and hence would contribute to a falsely high telomere signal (though ITRs also affect mean TRF length measurements, causing an underestimate of average TL, owing to the average length of ITR-containing restriction fragments being shorter than the average length of the true TRFs). However, ITRs are not expected to change in length during aging, and therefore none of the telomere shortening that we observed should result from ITR differences between the species (discussions in Supporting Information Sections 5 and 6).
One advantage of the qPCR method over the Southern blot method is that TRF lengths are affected by any species-specific single-nucleotide substitutions altering restriction enzyme(s) cut sites in the long subtelomere regions of each chromosome, as well as by large species-specific insertions or deletions between the cut site and the beginning of the true telomere repeats. Should those regions not be well characterized/sequenced, this source of error would be problematic. Alternatively, the lengths determined by qPCR can be negatively influenced by SNPs or CNVs at the chosen single-copy locus. Our albumin locus is well characterized in both species and we have not identified any such issues based on the available sequence data (METHODS section). An unknown frequency of PCR-inhibiting variants in our chimpanzee albumin locus would have the effect of masking any signal of telomere attrition over age as random individuals would have greatly increased T/S ratios. The clear age attrition signal in our data suggests that unknown albumin variants are not at issue here unless they are preferentially present in all of the younger chimpanzees in our study. We have investigated the comparability of our assay to the human- and chimpanzee-extracted DNA samples and our single-copy gene primers behave similarly in both species. Previously reported concerns with the linearity of the qPCR method or extraction technique biases do not undercut the chimpanzee results (Supporting Information Section 8).
CONCLUSIONS
The average leukocyte TL estimated by our methods in humans is half that of chimpanzees, though both species share the ancestral mammalian state of short (<20 kb) telomeres. Both species show age-related decline in leukocyte TL, and our cross-sectional data suggest that this rate is similar in both (<100 nt/year). Telomere dynamics vary in other homeotherms, and differences from rodents are especially notable (Gomes et al., 2011), making laboratory mice inappropriate models to represent these processes in humans (Bolker, 2012). Previous literature suggests that all known primates use replicative aging, and because chimpanzees are both longer-lived than monkeys and phylogenetically closer to humans, chimpanzee comparisons are of special relevance for understanding the evolution of human telomere aging. Shorter telomeres in humans, the longest lived of the great apes, suggest that this is a derived state in our own lineage. Potential adaptations to DNA repair and cellular apoptosis sensitivity have allowed human cells to maintain function with shorter telomeres. By investing less in TL upkeep (Eisenberg, 2011), humans could invest more in other mechanisms of somatic maintenance. Individuals with yet further shortened telomeres would still be expected to show increased mortality/morbidity as humans might have reached a lower limit of TL. Much work will be required to identify the differences in telomere dysfunction response and effects across the primate order though our results present a clearer picture of what is distinctive in human telomere dynamics.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Silvia Smith for assistance and logistics early in this project and the keepers, veterinarians, and biological material procurement staff at both Yerkes and Southwest National Primate Research Centers. Access to the Utah CEPH DNA samples was provided by Dr. Mark Leppert. Finally, the authors are in debt to the laboratory of Dr. Vicente Planelles, and in particular Dr. Alberto Bosque, who generously provided time and space in their BSL-2 research facilities for the chimpanzee DNA extractions and the storage of primate blood samples.
Contract grant sponsor: National Science Foundation (grant number BCS 0717886 to Hawkes); Contract grant sponsor: National Institutes of Health (grant number P51 RR000165 to Yerkes National Primate Research Center; grant numbers P5 1RR013986 and OD P51 OD011133 to Southwest National Primate Research Center).
Footnotes
AUTHOR CONTRIBUTIONS
JT, RMC, and KH conceived and designed the experiments. JT and RMC performed the experiments. JEC analyzed the data. RMC, JEC, and KH contributed reagents/ materials/analysis tools. JT, JEC, and KH wrote the paper.
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Alberts SC, Altmann J, Brockman DK, Cords M, Fedigan LM, Pusey A, Stoinski TS, Strier KB, Morris WF, Bronikowski AM. Reproductive aging patterns in primates reveal that humans are distinct. Proc Natl Acad Sci USA. 2013;110:13440–13445. doi: 10.1073/pnas.1311857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Alder JK, Parry EM, Karim B, Strong MA, Greider CW. Short telomeres are sufficient to cause the degenerative defects associated with aging. Am J Hum Genet. 2009;85:823–832. doi: 10.1016/j.ajhg.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G, Mezencev R, McDonald JF. Human cells display reduced apoptotic function relative to chimpanzee cells. PLoS One. 2012;7:e46182. doi: 10.1371/journal.pone.0046182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G, Polavarapu N, McDonald JF. Did natural selection for increased cognitive ability in humans lead to an elevated risk of cancer? Med Hypotheses. 2009;73:453–456. doi: 10.1016/j.mehy.2009.03.035. [DOI] [PubMed] [Google Scholar]
- Aviv A, Susser E. Leukocyte telomere length and the father's age enigma: Implications for population health and for life course. Int J Epidemiol. 2013;42:457–462. doi: 10.1093/ije/dys236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv A, Valdes AM, Spector TD. Human telomere biology: pitfalls of moving from the laboratory to epidemiology. Int J Epidemiol. 2006;35:1424–1429. doi: 10.1093/ije/dyl169. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Rice K, Vulto I, Lansdorp PM. Longitudinal data on telomere length in leukocytes from newborn baboons support a marked drop in stem cell turnover around 1 year of age. Aging Cell. 2007;6:121–123. doi: 10.1111/j.1474-9726.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Bolker J. Model organisms: there's more to life than rats and flies. Nature. 2012;491:31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant MA, Johnstone RA. Reproductive conflict and the separation of reproductive generations in humans. Proc Natl Acad Sci. 2008;105:5332–5336. doi: 10.1073/pnas.0711911105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Maley CC. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol. 2011;26:175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- Csiszar A, Podlutsky A, Podlutskaya N, Sonntag WE, Merlin SZ, Philipp EE, Doyle K, Davila A, Recchia FA, Ballabh P. Testing the oxidative stress hypothesis of aging in primate fibroblasts: is there a correlation between species longevity and cellular ROS production? J Gerontol A Biol Sci Med Sci. 2012;67:841–852. doi: 10.1093/gerona/glr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniali L, Benetos A, Susser E, Kark JD, Labat C, Kimura M, Desai K, Granick M, Aviv A. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013;4:1597. doi: 10.1038/ncomms2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel J-M, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- Eisenberg DT. An evolutionary review of human telomere biology: the thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am J Hum Biol. 2011;23:149–167. doi: 10.1002/ajhb.21127. [DOI] [PubMed] [Google Scholar]
- Ellison PT. Life historical perspectives on human reproductive aging. Ann N Y Acad Sci. 2010;1204:11–20. doi: 10.1111/j.1749-6632.2010.05611.x. [DOI] [PubMed] [Google Scholar]
- Gomes N, Ryder OA, Houck ML, Charter SJ, Walker W, Forsyth NR, Austad SN, Venditti C, Pagel M, Shay JW. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurven M, Kaplan H. Longevity among hunter-gatherers: a cross-cultural examination. Popul Dev Rev. 2007;33:321–365. [Google Scholar]
- Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- Haussmann MF, Marchetto NM. Telomeres: linking stress and survival, ecology and evolution. Curr Zool. 2010;56:714–727. [Google Scholar]
- Hawkes K. Grandmothers and the evolution of human longevity. Am J Hum Biol. 2003;15:380–400. doi: 10.1002/ajhb.10156. [DOI] [PubMed] [Google Scholar]
- Hawkes K, Coxworth JE. Grandmothers and the evolution of human longevity: a review of findings and future directions. Evol Anthropol. 2013;22:294–302. doi: 10.1002/evan.21382. [DOI] [PubMed] [Google Scholar]
- Hawkes K, O'Connell JF, Jones NB, Alvarez H, Charnov EL. Grandmothering, menopause, and the evolution of human life histories. Proc Natl Acad Sci. 1998;95:1336–1339. doi: 10.1073/pnas.95.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K, Smith KR, Robson SL. Mortality and fertility rates in humans and chimpanzees: how within-species variation complicates cross-species comparisons. Am J Hum Biol. 2009;21:578–586. doi: 10.1002/ajhb.20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Robertson BC, Gemmell NJ. The use of telomere length in ecology and evolutionary biology. Heredity. 2010;105:497–506. doi: 10.1038/hdy.2010.113. [DOI] [PubMed] [Google Scholar]
- Hornsby PJ. Short telomeres: cause or consequence of aging? Aging Cell. 2006;5:577–578. doi: 10.1111/j.1474-9726.2006.00249.x. [DOI] [PubMed] [Google Scholar]
- Hou L, Zhang X, Gawron AJ, Liu J. Surrogate tissue telomere length and cancer risk: shorter or longer? Cancer Lett. 2012;319:130–135. doi: 10.1016/j.canlet.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Walker L, Anderson D, Lacreuse A, Robson SL, Hawkes K. Depletion of ovarian follicles with age in chimpanzees: similarities to humans. Biol Reprod. 2007;77:247–251. doi: 10.1095/biolreprod.106.059634. [DOI] [PubMed] [Google Scholar]
- Kaplan H, Gurven M, Winking J, Hooper PL, Stieglitz J. Learning, menopause, and the human adaptive complex. Ann N Y Acad Sci. 2010;1204:30–42. doi: 10.1111/j.1749-6632.2010.05528.x. [DOI] [PubMed] [Google Scholar]
- Kim PS, Coxworth JE, Hawkes K. Increased longevity evolves from grandmothering. Proc R Soc Lond B Biol Sci. 2012;279:4880–4884. doi: 10.1098/rspb.2012.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Hjelmborg JvB, Gardner JP, Bathum L, Brimacombe M, Lu X, Christiansen L, Vaupel JW, Aviv A, Christensen K. Telomere length and mortality: a study of leukocytes in elderly Danish twins. Am J Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond B Biol Sci. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- Langergraber KE, Prüfer K, Rowney C, Boesch C, Crockford C, Fawcett K, Inoue E, Inoue-Muruyama M, Mitani JC, Muller MN. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Sociality, selection, and survival: simulated evolution of mortality with intergenerational transfers and food sharing. Proc Natl Acad Sci. 2008;105:7124–7128. doi: 10.1073/pnas.0710234105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RD. Rethinking the evolutionary theory of aging: transfers, not births, shape senescence in social species. Proc Natl Acad Sci. 2003;100:9637–9642. doi: 10.1073/pnas.1530303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitis DA, Burger O, Lackey LB. The human post-fertile lifespan in comparative evolutionary context. Evol Anthropol. 2013;22:66–79. doi: 10.1002/evan.21332. [DOI] [PubMed] [Google Scholar]
- Monaghan P. Telomeres and life histories: the long and the short of it. Ann N Y Acad Sci. 2010;1206:130–142. doi: 10.1111/j.1749-6632.2010.05705.x. [DOI] [PubMed] [Google Scholar]
- Morton RA, Stone JR, Singh RS. Mate choice and the origin of meno-pause. PLoS Comput Biol. 2013;9:e1003092. doi: 10.1371/journal.pcbi.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Gemmell NJ, Burke T. Measuring vertebrate telomeres: applications and limitations. Mol Ecol. 2004;13:2523–2533. doi: 10.1111/j.1365-294X.2004.02291.x. [DOI] [PubMed] [Google Scholar]
- Oeppen J, Vaupel JW. Broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavcan JM. Body size, size variation, and sexual size dimorphism in early Homo. Curr Anthropol. 2012;53:S409–S423. [Google Scholar]
- Robson SL, Van Schaik CP, Hawkes K. The derived features of human life history. In: Paine RL, Hawkes K, editors. The evolution of human life history. School of American Research Press; Santa Fe: 2006. pp. 17–44. [Google Scholar]
- Sahin E, DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol. 2012;13:397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Blount JD, Nussey DH, Speakman JR. Oxidative damage, ageing, and life-history evolution: where now? Trends Ecol Evol. 2012;27:570–577. doi: 10.1016/j.tree.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Hallmarks of telomeres in ageing research. J Pathol. 2007;211:114–123. doi: 10.1002/path.2090. [DOI] [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp Cell Res. 2002;272:146–152. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Tuljapurkar SD, Puleston CO, Gurven MD. Why men matter: mating patterns drive evolution of human lifespan. PLoS One. 2007;2:e785. doi: 10.1371/journal.pone.0000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis E, Galetzka D, Herlyn H, Schneider E, Haaf T. Humans and chimpanzees differ in their cellular response to DNA damage and non-coding sequence elements of DNA repair-associated genes. Cytogenet Genome Res. 2008;122:92–102. doi: 10.1159/000163086. [DOI] [PubMed] [Google Scholar]
- Wiemann SU, Satyanarayana A, Buer J, Kamino K, Manns MP, Rudolph KL. Contrasting effects of telomere shortening on organ homeostasis, tumor suppression, and survival during chronic liver damage. Onco-gene. 2005;24:1501–1509. doi: 10.1038/sj.onc.1208308. [DOI] [PubMed] [Google Scholar]
- Wood B. Reconstructing human evolution: achievements, challenges, and opportunities. Proc Natl Acad Sci. 2010;107:8902–8909. doi: 10.1073/pnas.1001649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.