Abstract
Uncontrolled inflammation of the periodontal area may arise when complex microbial communities transition from a commensal to a pathogenic entity. Communication among constituent species leads to polymicrobial synergy among metabolically compatible organisms that acquire functional specialization within the developing community. Keystone pathogens, even at low abundance, elevate community virulence and the resulting dysbiotic community targets specific aspects of host immunity to further disable immune surveillance while promoting an overall inflammatory response. Inflammophilic organisms benefit from proteinaceous substrates derived from inflammatory tissue breakdown. Inflammation and dysbiosis reinforce each other and the escalating environmental changes further select for a pathobiotic community. We have synthesized the polymicrobial synergy and dysbiotic components of the process into a new model for inflammatory diseases.
Periodontitis: An exemplar of polymicrobial synergy and dysbiosis
Recent years have witnessed a sea change in our perception of diseases of microbial origin. It has become apparent that the etiology of many of diseases that initiate on the skin and mucosal membranes does not involve monocultures of bacteria, but rather heterotypic communities of organisms. Organisms within these communities often display polymicrobial synergy (see Glossary) and the communities become dysbiotic, resulting in disruption of tissue homeostasis and normal immune responses. Periodontal diseases are an exemplar of an inflammatory disease that involves the concerted action of polymicrobial communities, and the pathogenicity of periodontal diseases can be explained through a Polymicrobial Synergy and Dysbiosis (PSD) model [1] (Figure 1). In this model, colonizing bacteria first assemble into physiologically compatible communities, and the organisms within these communities communicate through sophisticated signaling mechanisms. Overgrowth and overt pathogenicity are controlled by the host inflammatory response, and indeed a controlled immuno-inflammatory state is normal in a healthy gingiva. It is interesting to note here that the oral cavity is not unique in this regard and a similar homeostatic inflammatory state has been described in the gut [2]. In the mouse model of periodontitis, it has been established that pathogenicity is initiated by colonization with keystone pathogens, such as Porphyromonas gingivalis, which even in low numbers can elevate the virulence of the entire community [3]. Communication between P. gingivalis and organisms that are otherwise commensal, the accessory pathogens, facilitates synergy and the transition to pathogenicity [4]. The dysbiotic community continues to develop and stimulate inflammatory responses; however in susceptible hosts these are poorly controlled and are ineffective at constraining the community. Worse, frustrated and misdirected responses contribute to tissue destruction and shape a modified ‘inflammophilic’ community which sustains itself through inflammatory tissue breakdown-derived nutrients [5]. Pathobionts in the community become active and further exacerbate the disease process [6].
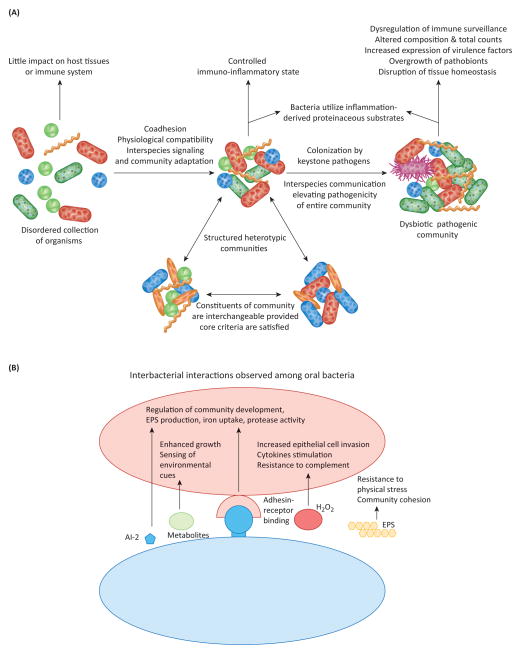
Figure 1. The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology.
A. Model overview. Community formation driven by co-adhesion and physiological compatibility initially leads to a balanced interaction between host immunity and the metabolism of the inflammophilic microbiota. The identities of individual species are less important than the presence of the appropriate complement of genes. As established in mouse models, colonization by keystone pathogens such as P. gingivalis enhances community virulence through interactive communication with accessory pathogens such as S. gordonii and disruption of immune surveillance. The dysbiotic community increases in number, pathobionts (green) overgrow and become more active, and tissue destruction ensues. B. Summary of synergistic interbacterial interactions that have been documented among oral bacteria. Both direct contact, primarily through adhesin-receptor binding, and sensing of compounds in solution can effect signaling and modulate the phenotypic properties of partner organisms (e.g. induction of adhesin/invasin, protease or complement-resistance genes). Interspecies signaling can also increase levels of extracellular polymeric substances (EPS) which contribute to community cohesion and resistance to physical stresses.
Interbacterial interactions
The central tenets of the PSD model are that communities of periodontal bacteria exhibit properties that are more than the sum of their constituent organism parts, and that pathogenicity is dictated by a subset of these bacteria. Initial colonizers of the periodontal area adhere to each other through a multiplicity of complementary adhesins, forming spatially distinct polymicrobial consortia [7]. Constituent organisms are generally metabolically compatible, and communities are thus physically and physiologically integrated, and through the progressive action of collective metabolic enzymes are capable of utilizing a wider range of nutritional substrates than possible for individual species [8]. Further development of heterotypic communities involves interspecies communication and adaptive responses which can occur through direct contact, soluble mediators and nutrient transfer (summarized in Figure 1). Within communities, bacteria are thus able to collectively regulate activities and functional specialization can arise. The composition of the communities varies over time, from person to person and even from site to site; however these communities are in a homeostatic equilibrium with the host [1]. Immune responses, characteristic of a healthy gingiva, limit bacterial overgrowth and neutralize toxic products such as proteases [9,10]. The delicately balanced host-microbe interaction changes upon colonization with keystone pathogens such as P. gingivalis.
Direct effects of keystone pathogens on other members of the community in combination with targeted subversion of specific components of the host responses, elevates the virulence of the community as a whole, and the now dysbiotic community increases in number causing further disruption of tissue homeostasis and destruction of periodontal tissues, mediated predominantly by pathobionts [6,11–13] (Figure 1). In human periodontitis, pathobionts are likely represented by previously underappreciated species (e.g. Filifactor alocis, Peptostreptococcus stomatis and species from the genera Prevotella, Megasphaera, Selenomonas, and Desulfobulbus) that have now been shown by metagenomic analyses to exhibit as good (or better) a correlation with disease as traditional pathogenic species, such as P. gingivalis, Tannerella forsythia, and Treponema denticola [14–17]. Consistent with this notion, a recent metatranscriptomic study revealed that the majority of virulence factors upregulated in the microbiome of periodontitis patients is primarily derived from previously underappreciated species that were not traditionally implicated in periodontitis [18]. It is possible that pathobionts might outcompete keystone or keystone-like pathogens such as P. gingivalis and T. forsythia in late stages of periodontal disease pathogenesis. Indeed, the relative abundance of P. gingivalis and T. forsythia shows a trend for negative correlation with the total bacterial load in human periodontitis, in contrast to certain newly recognized periodontitis-associated organisms probably acting as pathobionts [15]. This notion is reminiscent of the bacterial driver–passenger model for colorectal cancer, in which driver bacteria (i.e. bacteria which initiate the pathogenic process) are outcompeted in late stages of the disease by passenger bacteria (e.g. pathobionts which exacerbate the disease) [19].
Consistent with the PSD model, synergistic pathogenicity of combinations of periodontal organisms in animal models has been documented for some time [20]. Moreover, while considerable evidence implicates P. gingivalis as a major pathogen in chronic and severe manifestations of periodontal diseases, it is also clear that pathogenicity is only expressed in the context of a community, and P. gingivalis is unable to cause disease by itself in germ-free mice [3]. The full range of interactions between P. gingivalis and periodontal microbial community members is yet to be revealed; however, the molecular underpinnings of community pathogenicity are now beginning to emerge. Introduction of P. gingivalis into an otherwise health-associated community causes major transcriptomic and proteomic reverberations [21,22]. Post-translational cooperation is also evident as the cysteine protease interpain A (InpA) of Prevotella intermedia works in conjunction with the HmuY hameophore of P. gingivalis to acquire haem from haemoglobin [23]. P. gingivalis and T. denticola, which co-localize in subgingival biofilms in vivo, also exhibit symbiotic biofilm growth in vitro as well as synergistic virulence upon co-infection in animal models of alveolar bone loss. These organisms co-operate metabolically through the generation of a variety of metabolites by each species which can be used as a nutritional substrates by the other [24]. In addition, pathways that lead to the production of mutually beneficial nutrients are upregulated in co-culture of the organisms. Co-culture of the two species also results in increased production of virulence factors including dentilisin by T. denticola, and the RgpA, Kgp gingipains along with HagA in P. gingivalis. In combination, RgpA, Kgp and HagA increase nutrient acquisition and, through the Hag domains on all of these proteins, enhance binding of P. gingivalis to substrates including epithelial cells and Streptococcus gordonii [25].
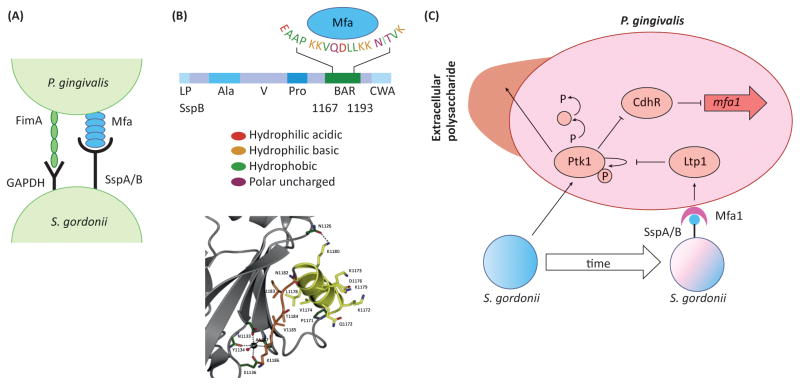
As a strict anaerobe, P. gingivalis colonization benefits from the activity of the antecedent host-controlled community to reduce the oxygen tension and also to provide an attachment substratum. The influence of the accessory pathogen S. gordonii on P. gingivalis extends beyond passive attachment, and when in combination with each other P. gingivalis and S. gordonii differentially expresses a large proportion of their proteomes, indicating that interspecies communication causes profound phenotypic changes [21]. P. gingivalis and S. gordonii avidly bind to each other through two sets of adhesin-receptor pairs, namely the FimA and Mfa1 component fimbriae of P. gingivalis that interact with streptococcal GAPDH and SspA/B surface proteins respectively (Figure 2). Community development between P. gingivalis and S. gordonii requires the soluble mediator AI-2 and is controlled by a tightly regulated signal transduction cascade within P. gingivalis based on protein tyrosine (de)phosphorylation [26–29] (Figure 2). In association with S. gordonii, the P. gingivalis tyrosine kinase Ptk1 acts to limit production of the transcription factor CdhR, which is a negative regulator of mfa1 and hence coadhesion of the organisms is favored. Over time, however, there is increased expression of a tyrosine phosphatase, Ltp1, which dephosphorylates Ptk1 thus lifting inhibition of CdrR, reducing mfa1 expression and constraining further community development. Moreover, tyrosine phosphorylation/dephosphorylation also regulates protease expression and capsule production by P. gingivalis, thus influencing pathogenic potential. This is reflected in murine models of alveolar bone loss in which dual infection with P. gingivalis and S. gordonii results in more bone loss compared to infection with either organism alone [30].
Figure 2. The molecular basis of synergy between P. gingivalis and S. gordonii.
A. The physical association between P. gingivalis and S. gordonii involves FimA–GAPDH and Mfa1–SspA/B adhesin–receptor pairs on the surfaces of the organisms. B. The upper panel shows the domain structure of the SspB protein and the amino acid residues involved in recognition of Mfa1. BAR spans aa residues 1167–1193, and the EAAP, KKVQDLLKK and NITVK sequences are involved in Mfa1 recognition. The lower panel shows the structure of the SspB C-terminal region with the protruding BAR domain stabilized by a calcium ion, and coordinated by three main chain and two side-chain oxygen atoms and a water molecule. Reproduced from [4] with permission. C. Signaling interactions between S. gordonii and P. gingivalis. S. gordonii induces autophosphorylation of the Ptk1 tyrosine kinase. Ptk1 activates a signaling cascade which converges on inactivation of the transcriptional repressor CdhR. As a result, expression of the minor fimbrial adhesin subunit Mfa1 is elevated and P. gingivalis is ‘primed’ for attachment to S. gordonii. Ptk1 is also a key component of the machinery for secretion of extracellular polysaccharide, and active Ptk1 increases the amount of polysaccharide material on the surface of P. gingivalis. Over time, however, direct contact mediated by Mfa1-SspA/B binding increases expression of the tyrosine phosphatase Ltp1, which dephosphorylates Ptk1, ultimately relieving repression of ChdR. Expression of the Mfa1 fimbrial adhesin is reduced and community development is constrained.
Abbreviations: Ala, alanine-rich repeats; BAR, SspB Adherence Region, the Mfa1-interacting domain; CWA, cell wall anchor; LP, leader peptide; Pro, proline-rich repeats; V, variable region.
S. gordonii can also increase the virulence of Aggregatibacter actinomycetemcomitans, a pathogen more closely associated with Localized Aggressive Periodontitis, an aggressive form of localized periodontitis. A. actinomycetemcomitans displays resource partitioning to favor carbon sources such as lactate generated by streptococcal metabolism [31]. Moreover, A. actinomycetemcomitans possesses two mechanisms, known as fight or flight responses, to avoid toxicity from H2O2, a metabolic by-product of S. gordonii. Detection of H2O2 induces upregulation of catalase (KatA) which degrades H2O2, and upregulation of Dispersin B (DspB) an enzyme that releases A. actinomycetemcomitans cells from biofilms [32]. In the murine abscess infection model, both of these responses are required for A. actinomycetemcomitans to successfully cross-feed with S. gordonii. Study of the spatial organization in a co-infection experiment shows that A. actinomycetemcomitans is positioned at an optimal distance (>4 μm) from S. gordonii, which allows cross-feeding but reduces exposure to inhibitory levels of H2O2 [32]. Growth of S. gordonii in abscesses is also favored in the presence of A. actinomycetemcomitans, which is known to express leukocyte killing toxins [33]. Such properties establish keystone pathogen credentials of A. actinomycetemcomitans in that it can impact the fitness of the community as a whole [32] and disable essential elements of immune surveillance.
Not all interactions among periodontal bacteria are synergistic. S. cristatus for example, induces downregulation of fimbrial gene expression in P. gingivalis through the signaling action of arginine deiminase [34]. Consequently, the distribution of P. gingivalis and S. cristatus in human subgingival plaque is negatively correlated, and S. cristatus attenuates P. gingivalis-induced bone loss in mice [35]. Such antagonistic interaction may lead to a ‘Red Queen’ effect, as P. gingivalis can induce cell death of another oral streptococcus, S. mitis [18]. Overall, however, the periodontal microbiota conforms more with the ‘Black Queen Hypothesis’ [36], whereby functions that are energetically costly and are discarded by ‘cheaters’, are nevertheless retained by a subset of community members as they benefit the entire community. Furthermore, The Black Queen Hypothesis predicts the emergence of keystone organisms: those that have maintained a function that provides an indispensable public good.
Interactions between bacterial communities and epithelial cells
When considering the multi-species aspects of periodontal microbiology, it is important to remember that microbial communities exist in and on gingival epithelial cells (GECs). In addition to hosting microbial communities, GECs constitute an interactive interface that senses organisms and signals their presence to the underlying cells of the immune system. The encounter between colonizing bacteria and GECs thus sculpts the early stages of both innate and acquired immunity. Many periodontal bacterial are capable of actively internalizing within GECs, a location that provides a nutrient-rich, generally reducing environment that is partially protected from immune effector molecules [1]. Intracellular bacteria disrupt host cell signaling pathways and gene expression patterns which can have profound effects on immune activity and cellular physiology [37]. Co-operative invasion among periodontal bacteria has been documented, for example consortia of P. gingivalis and F. nucleatum invade GECs in higher numbers than either organism alone [38]. Filifactor alocis and P. gingivalis also exhibit synergistic infection of epithelial cells and dual species invasion elicits a distinct pattern of host cell response [39]. Furthermore, transcriptional profiling of epithelial cell responses to several periodontal community members (P. gingivalis, A. actinomycetemcomitans, F. nucleatum and S. gordonii) revealed that the common core transcriptional response to all organisms was very limited, and organism-specific responses predominate. Overall, F. nucleatum and S. gordonii perturbed the transcriptome much less significantly than A. actinomycetemcomitans or P. gingivalis [40]. This supports the concept that microbial communities associated with health tread more lightly on host cells, as compared with the keystone pathogens that disrupt not only an ecologically balanced microbial community, but also epithelial cell physiology [40].
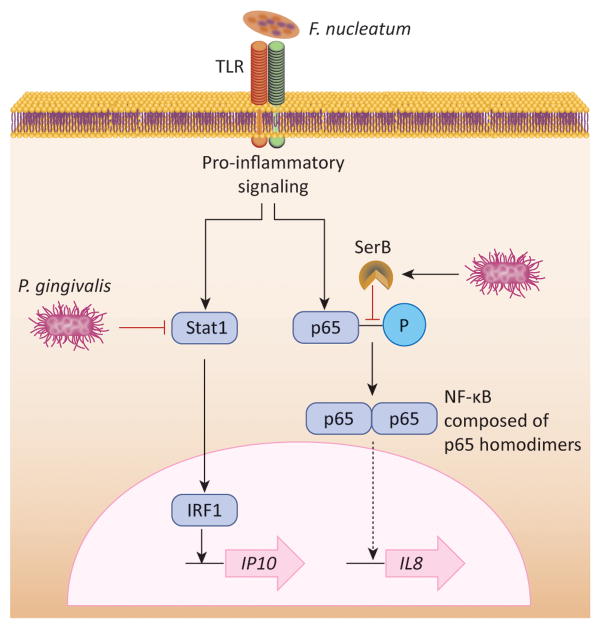
A major reaction of epithelial cells to bacterial challenge is the production and secretion of cytokines. Regulated recruitment of neutrophils, and to a lesser extent T and B cells, is an important feature of a healthy periodontium. Disruption of normal cytokine signaling is, therefore, a major contributor to microbially-induced dysbiotic host responses. Proteolytic degradation of primary and secondary cytokines by P. gingivalis, T. denticola and other pathogens is well established. However, P. gingivalis can selectively impinge upon the production of specific cytokines and chemokines, creating what is known as a localized chemokine paralysis which will favor the growth of the entire microbial community [12,41]. Indeed, studies in mice have shown that the colonization of P. gingivalis is associated with increased total counts and altered composition of the periodontal microbiota [3]. This dysbiotic effect of P. gingivalis may in part be attributed to suppression of the neutrophil chemokine CXCL8 (IL-8) production by GECs, even in the presence of otherwise stimulatory organisms such as F. nucleatum. Inhibition of CXCL8 production depends upon the capacity of P. gingivalis to invade GECs and secrete the bacterial serine phosphatase SerB, which specifically dephosphorylates the S536 residue on the p65 subunit of NF-κB. Dephosphorylation of p65 prevents nuclear translocation and ultimately the transcription of the IL8 gene which is predominantly controlled by NF-κB p65 homodimers [42] (Figure 3). Endothelial cells are additionally targeted by P. gingivalis whereby upregulation of E-selectin by other periodontal bacteria is diminished, thus impacting the leukocyte adhesion and transmigration aspects of neutrophil recruitment [43]. While this immune-evasive action may be transient, it appears to have long lasting consequences for the composition and virulence of the microbial community, and for host tissue damage, as a mutant of P. gingivalis incapable of producing SerB induces less bone loss in experimental animals compared to the parental strain [44].
Figure 3. Mechanisms of localized chemokine paralysis.
Many oral bacteria such as F. nucleatum engage TLRs on epithelial cell surfaces and activate pro-inflammatory signaling pathways. However, production of the neutrophil chemokine IL-8 (CXCL8) and the T-cell chemokine IP-10 (CXCL10) from epithelial cells is suppressed by P. gingivalis. Invasive P. gingivalis inactivates Stat1 which in turn reduces expression of IP10 promoted by the IRF1 transcription factor. Intracellular P. gingivalis also secretes the serine phosphatase SerB which specifically dephosphorylates the serine 536 residue of the p65 NF-κB subunit, and prevents the formation of p65 homdimers. Translocation of NF-κB into the nucleus is impeded, and transcription of the IL8 gene is reduced.
The dysbiotic influence of P. gingivalis on chemokines is not limited to CXCL8, and the organism also inhibits production of the T-helper 1 (Th1) chemokines CXCL10 (IP-10), CXCL9 (Mig) and CXCL11 (ITAC) from GECs, through downregulation of IRF-1 and Stat1 [45]. As with CXCL8, this inhibition supersedes the ability of pro-inflammatory organisms such as F. nucleatum to incite production of these chemokines. P. gingivalis may therefore reduce Th1 development, which arguably dictates cell-mediated immunity against periodontal bacteria [46], and allow Th17-mediated inflammation to flourish, as suggested by its ability to induce Th17-promoting cytokines (IL-6 and IL-23, but not the Th1-related IL-12) in antigen-presenting cells [47]. Th17 cells are abundant in periodontitis lesions where they can function as an osteoclastogenic subset that links T-cell activation to inflammatory bone loss.
Similar to P. gingivalis, T. denticola can incapacitate F. nucleatum-driven GEC IL-8 responses. T. denticola suppresses the fusion of internalized F. nucleatum with lysosomes, which may inhibit TLR9-dependent IL-8 secretion. In addition, T. denticola can reduce the induction of intracellular reactive oxygen species (ROS) in response to F. nucleatum which, besides inhibiting a killing mechanism, will also impede TLR responses that control IL-8 production [48].
Subversion of host immunity by dysbiotic microbial communities
In periodontitis, the periodontal pockets become a ‘breeding soil’ for periodontal bacteria that successfully escape host defense mechanisms. However, evasion of immune-mediated killing is not sufficient to ensure the persistence of periodontitis-associated microbial communities. Growth of these communities depends on nutrients derived from inflammatory tissue breakdown, for example degraded collagen peptides and haem-containing compounds [5]. The inflammophilic community, therefore, needs to subvert the host response in ways that interfere with bacterial killing while promoting inflammation. Several illustrative examples are given below.
Subversion of complement
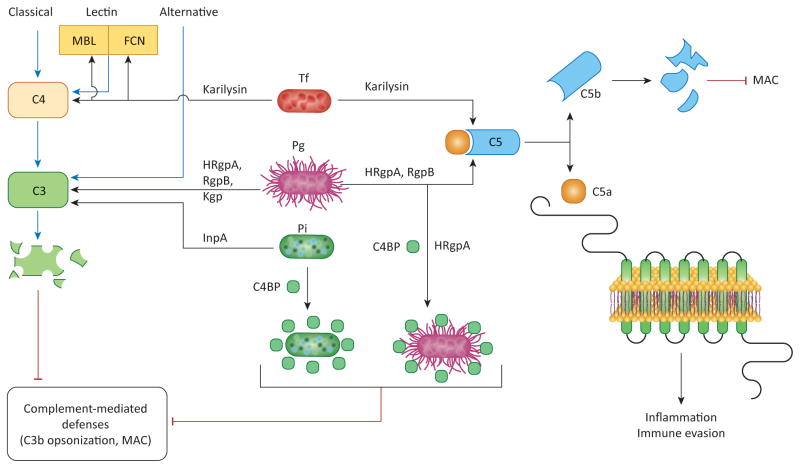
The activation of complement proceeds via distinct initiation mechanisms (classical, lectin, or alternative) which converge at the third complement component (C3), leading to the generation of effector molecules that mediate recruitment and activation of inflammatory cells (anaphylatoxins C3a and C5a), microbial opsonization and phagocytosis (opsonins such as C3b), and direct lysis of targeted microbes (C5b-9 membrane attack complex) [49]. Components of the complement system can be found in the gingival crevicular fluid (GCF) at up to 80% of their concentration in serum [50]. Periodontitis-associated bacteria, including P. gingivalis, T. forsythia, and P. intermedia, express proteases that effectuate avoidance of complement-mediated phagocytosis and killing. Whereas P. gingivalis gingipains and P. intermedia InpA degrade the central component C3 [51,52], T. forsythia karilysin degrades upstream components such as C4 or pattern-recognition molecules (mannose-binding lectin or ficolins) [53] (Figure 4). As a consequence of complement inhibition, the deposition of opsonins or the membrane attack complex on the bacterial surface is suppressed. Importantly, the gingipains act synergistically with karilysin or interpain A in inhibiting complement [51–53], suggesting that these synergistic proteases can interfere with complement activation even after their release and diffusion within the biofilm, thereby protecting the entire the microbial community.
Figure 4. Synergistic inhibition of complement-dependent antimicrobial activities.
P. gingivalis, P. intermedia, and T. forsythia can inhibit the classical, lectin, and alternative pathways of complement activation by degrading C3, C4, mannose-binding lectin (MBL), or ficolins (FCN) through the action of proteases, as indicated. These activities are synergistic and prevent the deposition of C3b opsonin or the membrane attack complex (MAC) on the surface of these pathogens as well as bystander bacterial species. Moreover, P. gingivalis and P. intermedia protect themselves against complement also by using surface molecules (HRgpA gingipain for P. gingivalis, undefined molecule for P. intermedia) to capture the circulating C4b-binding protein (C4BP), a physiological negative regulator of the classical and lectin pathways. Furthermore, P. gingivalis (via its Arg-specific gingipains HRgpA and RgpB) and T. forsythia (via its karilysin) can release biologically active C5a from C5, thereby stimulating inflammation through the activation of the C5a receptor (C5aR).
Abbreviations: InpA, interpain A; Kgp, Lys-specific gingipain.
Intriguingly, at low concentrations the gingipains of P. gingivalis and the InpA of P. intermedia activate, rather than inhibit, complement [51,52]. As alluded to above, complement-mediated inflammation is beneficial for the bacteria as it increases the flow of GCF and hence the amount of GCF-derived nutrients. Moreover, these organisms have alternative mechanisms for protection against activated complement. Indeed, both P. gingivalis and P. intermedia can capture and co-opt physiological soluble inhibitors of the complement cascade, such as the C4b-binding protein [53,54] (Figure 4). In a similar context, T. denticola expresses a protein which binds and exploits factor H, a major soluble inhibitor of complement [55].
An interesting hypothesis synthesizing these findings is the following: P. gingivalis and P. intermedia activate complement when present at low numbers (such as in the initial stages of colonization), resulting in a nutritionally favorable inflammatory response that does not eliminate them (or other bacteria that can similarly hijack host-derived complement inhibitors). However, complement activation can suppress complement-sensitive bacteria, which could otherwise compete for space and nutrients (these might be species associated with periodontal health). At advanced stages of biofilm development, the concentration of proteases is sufficient to inhibit complement activation by all three initiation mechanisms, thereby promoting the survival of the entire biofilm [56,57]. Notably, even under conditions that suppress the canonical activation of the complement cascade, P. gingivalis Arg-specific gingipains and T. forsythia karilysin were shown to act directly on C5 to release biologically active C5a (the most proinflammatory component of the entire cascade), thereby stimulating inflammatory serum exudate for acquisition of hemin and other nutrients [52,53,58]. Interestingly, the same proteases readily destroy the C5b component of C5, thus preventing the generation of the membrane attack complex [52,53] (Figure 4).
Manipulation of neutrophils
Neutrophils comprise the overwhelming majority (≥95%) of phagocytes recruited to the gingival crevice or periodontal pocket [59,60]. Biofilms used to be thought of as intrinsically resistant to phagocytosis because phagocytes were assumed to be unable to penetrate biofilms. However, biofilms are actually more like a porous hydrogel than a solid and rigid structure, and thus do not constitute an impermeable physical barrier to phagocytes [61,62]. Therefore, phagocytes do penetrate biofilms and if they fail to engulf the bacteria within, this may be attributed to mechanisms such as the operation of immune evasive strategies within the biofilm [61,62]. In periodontitis, neutrophils largely fail to control bacterial growth and prevent dysbiosis, despite maintaining viability and the capacity to elicit immune responses [6,63]. A number of studies have shown that P. gingivalis, T. denticola, T. forsythia and certain other periodontal bacteria can resist neutrophil killing through various mechanisms including degradation or inactivation of antimicrobial molecules or granular proteases (for review see [60]). However, it should be noted that in vitro resistance of periodontal bacteria to antimicrobial molecules might not necessarily apply to in vivo conditions, at least not with equal potency. This is because the activity of antimicrobial molecules and their susceptibility to bacterial proteases can be modified by other host molecules expected to be found at sites of infection or inflammation [64,65].
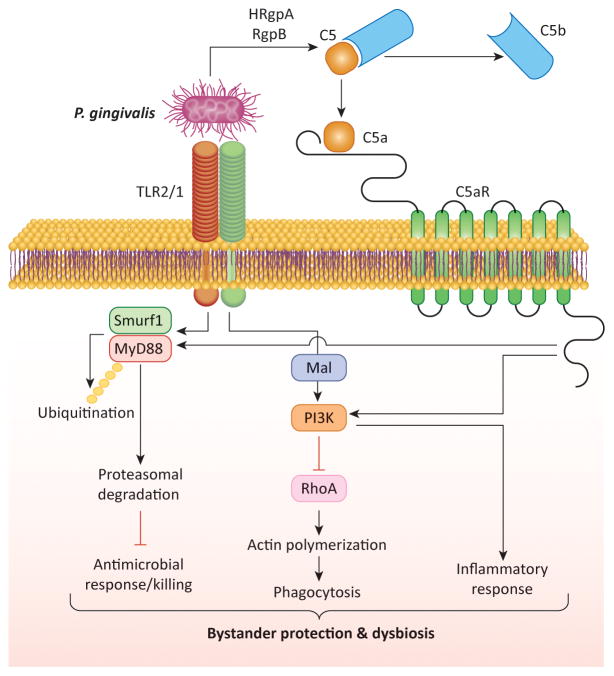
A novel mechanism for escaping human or mouse neutrophil killing was recently described for P. gingivalis. The pathogen was shown to block an antimicrobial Toll-like receptor (TLR)2-MyD88 pathway via proteasomal degradation of MyD88, whereas it activates a proinflammatory TLR2-Mal-phosphoinositide 3-kinase (PI3K) pathway that additionally blocks phagocytosis by interfering with RhoA GTPase-dependent actin polymerization [66] (Figure 5). Both subversive pathways require an intimate crosstalk between TLR2 and C5aR [66], which is directly controlled by P. gingivalis through the C5 convertase-like activity of its Arg-specific gingipains (HRgpA, RgpB) [52,58]. Moreover, the survival of the otherwise neutrophil-susceptible F. nucleatum is dramatically enhanced in the presence of P. gingivalis, unless C5aR, TLR2, or PI3K signaling in neutrophils is genetically or pharmacologically ablated [66]. Consistent with the in vitro findings, local treatments of P. gingivalis-colonized mice with inhibitors of C5aR, TLR2 (but not TLR4), or PI3K essentially eliminate P. gingivalis, reverse dysbiosis, and inhibit development of periodontitis [3,66,67]. Therefore, by dissociating inflammation from immune clearance that is disarmed, P. gingivalis is able to protect itself and also bystander community bacteria.
Figure 5. Subversion of neutrophil function and dysbiosis.
P. gingivalis co-activates TLR2 and C5aR in neutrophils, and the resulting crosstalk leads to E3 ubiquitin ligase Smurf1-dependent ubiquitination and proteasomal degradation of MyD88, thereby inhibiting a host-protective antimicrobial response. Moreover, the C5aR-TLR2 crosstalk activates PI3K, which prevents phagocytosis through inhibition of RhoA activation and actin polymerization, while stimulating an inflammatory response. In contrast to MyD88, another TLR2 adaptor, Mal, is involved in the subversive pathway and acts upstream of PI3K. The integrated mechanism provides ‘bystander’ protection to otherwise susceptible bacterial species and promotes polymicrobial dysbiotic inflammation in vivo. Reproduced from [61] with permission.
Whereas opsonization of bacteria with complement fragments (e.g. with C3b) or specific antibodies can enhance phagocytic killing by neutrophils, P. gingivalis was shown to effectively degrade both C3 and IgG [52,68–71]. Conversely, hyperimmune antisera to P. gingivalis proteases reverses their capacity to degrade C3 and IgG, thereby suppressing the organism’s resistance to neutrophil killing [68]. In principle, therefore, specific antibodies induced in the course of periodontitis against P. gingivalis could be protective, although in fact periodontitis can proceed despite the presence of antibodies to P. gingivalis and other periodontal bacteria [46]. In this regard, naturally induced antibodies to periodontal bacteria are thought to be of low affinity and poor functionality [10,46].
Inhibition of macrophage responses
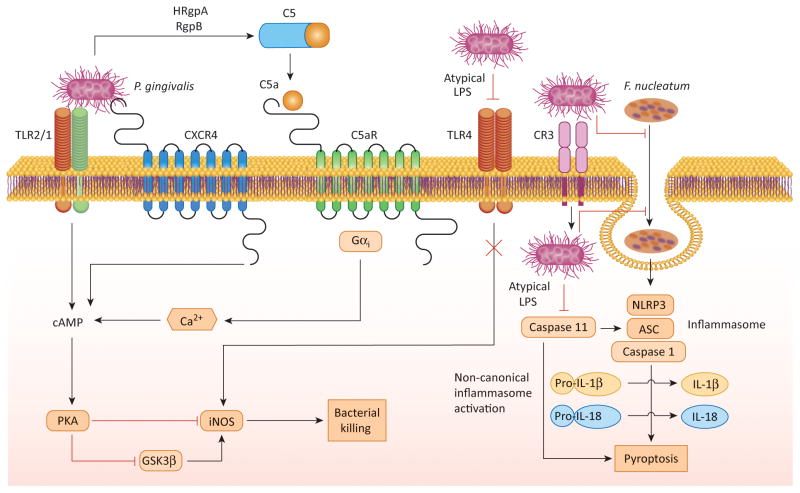
Although macrophages are minimally present in the gingival crevice (<3%), they can readily encounter periodontal bacteria upon their invasion into the gingival connective tissue [72]. In this context, nitric oxide is a major effector of the intracellular killing capacity of macrophages [73]. P. gingivalis can counteract this antimicrobial function by inhibiting the expression of the inducible nitric oxide synthase (iNOS) via a cAMP-dependent mechanism [74]. Maximal induction of the cAMP response requires the concurrent activation of TLR2, C5aR, and CXCR4 by P. gingivalis (Figure 6), whereas antagonistic blockade of either C5aR or CXCR4 diminishes the intracellular cAMP levels and promotes the killing of the microbe [74].
Figure 6. Mechanisms of inhibition of macrophage intracellular killing.
P. gingivalis interacts with the TLR2/1 receptor complex, whereas it evades or antagonizes TLR4 by expressing atypical lipopolysaccharide structures (with non-phosphorylated tetra-acylated lipid A or mono-phosphorylated tetra-acylated lipid A, respectively). The TLR2 response is proactively modified through crosstalk with other receptors that are under P. gingivalis control, C5aR and CXCR4. P. gingivalis induces C5aR activation by virtue of its Arg-specific gingipains (HRgpA and RgpB) which attack C5 and release biologically active C5a. C5a stimulates Gαi-dependent intracellular Ca2+ signaling which synergistically enhances the otherwise weak cAMP responses induced by TLR2 activation alone. Maximal cAMP induction is achieved through the participation of CXCR4, which is activated directly by the pathogen’s FimA fimbriae and coassociates with both TLR2 and C5aR in lipid rafts. The ensuing activation of the cAMP-dependent protein kinase A (PKA) pathway inactivates glycogen synthase kinase-3β (GSK3β) and impairs the inducible nitrogen synthase (iNOS)-dependent killing of the pathogen in macrophages. The expression of evasive or antagonistic lipid A moreover allows internalized P. gingivalis (possibly entering via complement receptor 3 [CR3] which promotes its intracellular survival) to prevent caspase 11–dependent non-canonical activation of the inflammasome. This mechanism is normally triggered by LPS of intracellular gram-negative bacteria and leads to pyroptosis, a proinflammatory mode of lytic cell death that protects the host against bacterial infection. P. gingivalis also inhibits F. nucleatum-induced NLRP3 inflammasome activation and cytokine secretion by inhibiting its endocytosis, although it is uncertain whether this is mediated by extracellular or intracellular P. gingivalis.
The expression of iNOS can also be induced by TLR4 signaling, although P. gingivalis can prevent the activation of this receptor (Figure 6). In this regard, the organism can enzymatically modify the lipid A moiety of its lipopolysaccharide (LPS) to either evade or antagonize TLR4 activation [9,12]. The shifting of the lipid A activity from TLR4 agonist (mono-phosphorylated penta-acylated lipid A) to TLR4 evasive (non-phosphorylated tetra-acylated lipid A) or antagonist (mono-phosphorylated tetra-acylated lipid A) is dependent upon lipid A phosphatase activity [75]. Genetic ablation of 4′-phosphatase activity leads to synthesis of lipid A with TLR4 agonist activity, whereas ablation of 1-phosphatase activity leads to lipid A that functions as a TLR4 antagonist [75,76]. In experiments using wild-type P. gingivalis and isogenic phosphatase mutants with a ‘locked’ lipid A profile, the expression of antagonistic or evasive (inert) lipid A was linked to increased bacterial survival in macrophages, as compared to the expression of TLR4 agonist lipid A [77]. Although it is likely that less nitric oxide is produced in macrophages in response to P. gingivalis strains expressing antagonistic or evasive lipid A, a documented mechanism associated with protection of these strains involves the suppression of non-canonical inflammasome activation [77] (Figure 6). The non-canonical activation of the inflammasome, which requires the participation of caspase 11 and can be triggered by the LPS of gram-negative bacteria independently of TLR4, is thought to discriminate cytosolic from vacuolar bacteria that can be detected by canonical inflammasome consisting of the NLRP3 protein (NOD-like receptor family pyrin domain-containing-3), the adaptor molecule ASC (Apoptosis-associated speck-like protein containing CARD), and caspase 1 [78]. In the non-canonical mechanism, cytosolic LPS directly binds and activates caspase 11 [79]. Although TLR4 antagonist lipid A structures can similarly bind caspase 11, they fail to induce caspase 11 oligomerization that is required for its activation [79], thereby explaining why P. gingivalis expressing antagonistic or evasive lipid A can prevent the activation of both TLR4 and the caspase 11-dependent non-canonical inflammasome. Importantly, the ability of P. gingivalis to modulate the lipid A structure of its LPS was recently shown to contribute to its oral colonization and augmentation of the commensal bacterial load in a rabbit model of experimental periodontitis [76].
In addition to blocking the non-canonical pathway, P. gingivalis can inhibit canonical inflammasome activation by other bacteria. Indeed, P. gingivalis was shown to suppress endocytic events required for F. nucleatum-induced NLRP3 inflammasome activation in macrophages [80]. This mechanism is consistent with the capacity of P. gingivalis to modulate actin cytoskeletal rearrangements [66,81] and can potentially promote the virulence of the entire microbial community.
Concluding remarks and future perspectives
Emerging trends in periodontal disease have established that no single microbe is adequately pathogenic by itself; what drives disease is the properties of the community as a whole. According to the PSD model, the periodontal host response is initially subverted by keystone pathogens, the colonization and metabolic activities of which are assisted by accessory pathogens, and is subsequently over-activated by pathobionts. This concept is increasingly acknowledged to be pertinent to other chronic diseases, such as inflammatory bowel disease or colon cancer, where no single or selected few species have been consistently associated with disease but, rather, microbial communities work in concert to cause pathology [19,82–84]. Moreover, keystone or keystone-like pathogens are considered as potentially important players in the gut ecosystem [82,85,86]. Due to inherent limitations and ethical considerations, human studies cannot normally address cause-and-effect relationships and, therefore, addressing the validity of the keystone-pathogen concept may be quite challenging. However, useful insights can be obtained by interventional studies that specifically target a keystone species such as P. gingivalis. In this regard, in non-human primates where P. gingivalis is a natural inhabitant of the subgingival biofilm, a gingipain-based vaccine causes a reduction both in P. gingivalis numbers and in the total subgingival bacterial load (as well as inhibiting bone loss) [87]. These findings suggest that the presence of P. gingivalis benefits the entire biofilm, as predicted by the keystone-pathogen hypothesis [88]. Indeed, periodontitis is an attractive study model of dysbiotic diseases, as it is readily accessible for obtaining both microbial and host tissue samples for longitudinal studies. For instance, study of the host-microbe interaction is facilitated by the availability of organisms recovered directly from specific microenvironments in the mouth, rather than those that have flowed through the ecosystem. The identification of organisms present in discrete clinical conditions at defined times has allowed the molecular dissection of interbacterial interactions, pathogenic mechanisms and host responses. What has emerged is a picture of an intricate synergistic network among organisms that maximizes persistence and metabolic activity through induction of a dysfunctional immune response. Moreover, ineffective immune responses actually stimulate the growth of the periodontal microbial community through provision of proteinaceous and haem-containing substrates derived from inflammatory tissue breakdown. Clearly the periodontal microbiota is well-adapted to the human host, and disease can been regarded as a side-effect of survival and long-term nutritional strategies. Of course, not all individuals are equally susceptible to disease, and although not addressed in this review, genetic and epigenetic variations along with risk factors such as smoking, make a major contribution to the initiation and progression of disease.
Although key questions still remain (Box 1), the PSD model has a number of sequelae that are consistent with clinical and microbiological observations of periodontal disease. For example, immunological rebound and the moderating influence of some community members on P. gingivalis will constrain the pathogenic activity of the microbial community and ameliorate tissue destruction, in concurrence with clinical reports of bursts of disease activity. Additionally, decades of microbiological study have failed to produce an inviolable association between any organism or group of organisms and periodontitis. According to the PSD model, however, the identities of individual organism are irrelevant so long as the appropriate genes and associated functions are operational. This notion is supported by a recent metatranscriptomic study which showed that disease-associated microbial communities in periodontitis patients exhibit highly conserved metabolic gene expression profiles, despite high inter-patient variability in terms of microbial composition [89]. Finally, the PSD model has implications for the development of novel therapeutics. Hitherto, most strategies are based on the elimination of P. gingivalis, which is difficult to achieve as the organism is embedded in biofilms and within epithelial cells. Instead, community manipulation may be a more viable option. Theoretically at least, favoring the growth of organisms that are antagonistic to P. gingivalis and reducing the levels of accessory pathogens would impede the transition to a pathogenic community. Another potentially effective measure could involve targeted modulation of immunity to limit destructive inflammation and reverse the microbial immune evasive strategies that fuel dysbiosis (e.g. chemokine paralysis, subversion of complement-TLR crosstalk, and TLR4 antagonism). For example, antagonizing complement pathways in the gingival tissues could reset host immunity to a state that is nonresponsive to the subversive activities of P. gingivalis, and help restore periodontal tissue homeostasis.
BOX 1. Outstanding questions.
In complex multispecies communities, how do organisms decipher and sort multiple, possibly conflicting, signals from different community members?
Can epithelial cell signaling be reprogrammed and ‘locked’ by colonization with health-associated organisms such that they are resistant to dysbiotic communities?
Can key targets (receptors, signaling molecules) of microbial immune subversion be confirmed by genetic association studies?
Can host modulation to counteract microbial immune subversion, or bacterial modulation to establish a dominant health-associated community, contribute to the treatment of human periodontitis?
Highlights.
Polymicrobial synergy and dysbiosis drives periodontitis in a susceptible host
Dysbiosis involves specialized accessory and keystone pathogens and pathobionts
Microbial immune subversion is central to the persistence of dysbiotic communities
The dysbiotic microbiota sustains itself by feasting on the ‘inflammatory spoils’
Acknowledgments
The authors’ research is supported by NIH grants; DE011111, DE012505, DE016690, DE017921, DE022867, DE023193 (RJL) and DE015254, DE017138, DE021685, AI068730 (GH). The authors regret that several important studies could only indirectly be acknowledged through comprehensive reviews owing to space and reference number limitations.
Glossary
- Accessory pathogen
an organism that, while commensal in a particular microenvironment, nonetheless supports or enhances the virulence of another organism
- Co-adhesion
binding between different bacterial species mediated by specific adhesin-receptor interactions
- Dysbiosis
a state of imbalance in the relative abundance or influence of species within a microbial community associated with inflammatory disease
- Gingival crevicular fluid (GCF)
serum exudate originating in the gingival capillaries and flowing into the gingival crevice containing also locally produced immune and inflammatory mediators
- Homeostasis
a condition of equilibrium or stability in a system which is maintained by adjusting physiological processes to counteract external changes; a balanced relation between a host tissue and the resident microbiota that prevents destructive inflammation or disease
- Keystone pathogen
a pathogen with a disproportionately large effect on its environment relative to its abundance (e.g. low-abundance P. gingivalis remodels a commensal microbial community into a dysbiotic and disease-provoking microbiota)
- Pathobiont
an organism that promotes pathology under conditions of disrupted host homeostasis
- Periodontal diseases
group of microbially induced inflammatory disorders of the tissues that surround and support the tooth including the gingiva (gum), periodontal ligament and alveolar bone. Uncontrolled periodontal disease leads to tissue destruction and eventually tooth loss
- Periodontal pocket
the pathologically deepened gingival crevice (narrow space between the tooth surface and the free gingiva)
- Polymicrobial synergy
the process by which one organism enhances the colonization and/or virulence of another
- Red Queen effect
an hypothesis in evolutionary biology which holds that organisms must continually adapt in order to survive in an environment with continuously changing competing organisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright CJ, et al. Disruption of heterotypic community development by Porphyromonas gingivalis with small molecule inhibitors. Mol Oral Microbiol. 2014;29:185–193. doi: 10.1111/omi.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 10.Schenkein HA. Host responses in maintaining periodontal health and determining periodontal disease. Periodontol 2000. 2006;40:77–93. doi: 10.1111/j.1600-0757.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Lamont RJ. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darveau RP, et al. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y, et al. Induction of bone loss by pathobiont-mediated nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffen AL, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar PS, et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duran-Pinedo AE, et al. The periodontal pathogen Porphyromonas gingivalis Induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infect Immun. 2014;82:3374–3382. doi: 10.1128/IAI.01976-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjalsma H, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 20.Kesavalu L, et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuboniwa M, et al. Insights into the virulence of oral biofilms: discoveries from proteomics. Expert Rev Proteomics. 2012;9:311–323. doi: 10.1586/epr.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frias-Lopez J, Duran-Pinedo A. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol. 2012;194:2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne DP, et al. Evidence of mutualism between two periodontal pathogens: co-operative haem acquisition by the HmuY haemophore of Porphyromonas gingivalis and the cysteine protease interpain A (InpA) of Prevotella intermedia. Mol Oral Microbiol. 2013;28:219–229. doi: 10.1111/omi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan KH, et al. Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses. PLoS Pathog. 2014;10:e1003955. doi: 10.1371/journal.ppat.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meuric V, et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28:40–53. doi: 10.1111/omi.12004. [DOI] [PubMed] [Google Scholar]
- 26.Wright CJ, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright CJ, et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen. 2014;3:383–394. doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda K, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chawla A, et al. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. 2010;78:1510–1522. doi: 10.1111/j.1365-2958.2010.07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daep CA, et al. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey MM, et al. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kachlany SC. Aggregatibacter actinomycetemcomitans leukotoxin: from threat to therapy. J Dent Res. 2010;89:561–570. doi: 10.1177/0022034510363682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang BY, et al. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47:3902–3906. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie H, et al. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. 2012;47:578–583. doi: 10.1111/j.1600-0765.2012.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris JJ, et al. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012:3. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handfield M, et al. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 38.Saito A, et al. Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts. Microb Pathog. 2012;53:234–242. doi: 10.1016/j.micpath.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aruni W, et al. Filifactor alocis: The newly discovered kid on the block with special talents. J Dent Res. 2014;93:725–732. doi: 10.1177/0022034514538283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handfield M, et al. Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87:203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darveau RP, et al. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi H, et al. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog. 2013;9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darveau RP, et al. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun. 1995;63:1311–1317. doi: 10.1128/iai.63.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bainbridge B, et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect Immun. 2010;78:4560–4569. doi: 10.1128/IAI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jauregui CE, et al. Suppression of T-cell chemokines by Porphyromonas gingivalis. Infect Immun. 2013;81:2288–2295. doi: 10.1128/IAI.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gemmell E, et al. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol 2000. 2007;43:14–40. doi: 10.1111/j.1600-0757.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 47.Moutsopoulos NM, et al. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J Autoimmun. 2012;39:294–303. doi: 10.1016/j.jaut.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin JE, et al. A periodontal pathogen Treponema denticola hijacks the Fusobacterium nucleatum-driven host response. Immunol Cell Biol. 2013;91:503–510. doi: 10.1038/icb.2013.35. [DOI] [PubMed] [Google Scholar]
- 49.Ricklin D, et al. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hajishengallis G. Complement and periodontitis. Biochem Pharmacol. 2010;80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potempa M, et al. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316. doi: 10.1371/journal.ppat.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popadiak K, et al. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 53.Jusko M, et al. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J Immunol. 2012;188:2338–2349. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potempa M, et al. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–5544. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDowell JV, et al. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417–1425. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krauss JL, et al. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol 2000. 2010;52:141–162. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo Y, et al. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang S, et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186:869–877. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sima C, Glogauer M. Neutrophil dysfunction and host susceptibility to periodontal inflammation: Current state of knowledge. Curr Oral Health Rep. 2014;1:95–103. [Google Scholar]
- 60.Nussbaum G, Shapira L. How has neutrophil research improved our understanding of periodontal pathogenesis? J Clin Periodontol. 2011;38:49–59. doi: 10.1111/j.1600-051X.2010.01678.x. [DOI] [PubMed] [Google Scholar]
- 61.Archer NK, et al. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leid JG, et al. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryder MI. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol 2000. 2010;53:124–137. doi: 10.1111/j.1600-0757.2009.00327.x. [DOI] [PubMed] [Google Scholar]
- 64.Sol A, et al. Actin enables the antimicrobial action of LL-37 in the presence of microbial proteases. J Biol Chem. 2014;289:22926–22941. doi: 10.1074/jbc.M114.579672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gutner M, et al. Saliva enables the antimicrobial activity of LL-37 in the presence of proteases of Porphyromonas gingivalis. Infect Immun. 2009;77:5558–5563. doi: 10.1128/IAI.00648-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maekawa T, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15:768–778. doi: 10.1016/j.chom.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abe T, et al. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol. 2012;189:5442–5448. doi: 10.4049/jimmunol.1202339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cutler CW, et al. Inhibition of C3 and IgG proteolysis enhances phagocytosis of Porphyromonas gingivalis. J Immunol. 1993;151:7016–7029. [PubMed] [Google Scholar]
- 69.Sundqvist G, et al. Degradation of human immunoglobulins G and M and complement factors C3 and C5 by black-pigmented Bacteroides. J Med Microbiol. 1985;19:85–94. doi: 10.1099/00222615-19-1-85. [DOI] [PubMed] [Google Scholar]
- 70.Grenier D, et al. Further studies on the degradation of immunoglobulins by black-pigmented Bacteroides. Oral Microbiol Immunol. 1989;4:12–18. doi: 10.1111/j.1399-302x.1989.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 71.Vincents B, et al. Cleavage of IgG1 and IgG3 by gingipain K from Porphyromonas gingivalis may compromise host defense in progressive periodontitis. FASEB J. 2011;25:3741–3750. doi: 10.1096/fj.11-187799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- 73.Nathan C. Role of iNOS in human host defense. Science. 2006;312:1874–1875. doi: 10.1126/science.312.5782.1874b. author reply 1874–1875. [DOI] [PubMed] [Google Scholar]
- 74.Wang M, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coats SR, et al. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 2009;11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zenobia C, et al. Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infect Immun. 2014;82:650–659. doi: 10.1128/IAI.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slocum C, et al. Distinct lipid A moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathog. 2014;10:e1004215. doi: 10.1371/journal.ppat.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 80.Taxman DJ, et al. Porphyromonas gingivalis mediates inflammasome repression in polymicrobial cultures through a novel mechanism involving reduced endocytosis. J Biol Chem. 2012;287:32791–32799. doi: 10.1074/jbc.M112.401737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hasegawa Y, et al. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stecher B, et al. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 83.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bornigen D, et al. Functional profiling of the gut microbiome in disease-associated inflammation. Genome Med. 2013;5:65. doi: 10.1186/gm469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Page RC, et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol Immunol. 2007;22:162–168. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 88.Hajishengallis G, et al. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jorth P, et al. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5:e01012–01014. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]