Abstract
Activation of the Pax2 gene marks the intermediate mesoderm shortly after gastrulation, as the mesoderm becomes compartmentalized into paraxial, intermediate, and lateral plate. Using an EGFP knock-in allele of Pax2 to identify and sort cells of the intermediate mesodermal lineage, we compared gene expression patterns in EGFP positive cells that were heterozygous or homozygous null for Pax2. Thus, we identified critical regulators of intermediate mesoderm and kidney development whose expression depended on Pax2 function. In cell culture models, Pax2 is thought to recruit epigenetic modifying complex to imprint activating histone methylation marks through interactions with the adaptor protein PTIP. In kidney organ culture, conditional PTIP deletion showed that many Pax2 target genes, which were activated early in renal progenitor cells, remained on once activated, whereas Pax2 target genes expressed later in kidney development were unable to be fully activated without PTIP. In Pax2 mutants, we also identified a set of genes whose expression was up-regulated in EGFP positive cells and whose expression was consistent with a cell fate transformation to paraxial mesoderm and its derivatives. These data provide evidence that Pax2 specifies the intermediate mesoderm and renal epithelial cells through epigenetic mechanisms and in part by repressing paraxial mesodermal fate.
Keywords: Pax2, PTIP, kidney development, intermediate mesoderm, epigenetic
INTRODUCTION
The vertebrate Pax gene family was identified based on sequence homology to a subset of Drosophila segmentation genes that encode a conserved DNA binding domain called the paired-box. In mammals, Pax genes control the specification of particular cells and tissues, such as the eye, the vertebral column, the thymus, and parts of the nervous system, and have also been linked to cancer and human congenital malformations (Chi and Epstein, 2002; Robson et al., 2006). The Pax2 gene is crucial for the development of the kidney and the reproductive tract, both of which are derived from the intermediate mesoderm (Dressler, 2006; Dressler, 2009). Pax2 is among the earliest markers for the intermediate mesoderm, along with the related gene Pax8 (Bouchard et al., 2002) and the homeodomain protein Lhx1 (Tsang et al., 2000). This Pax2 positive intermediate mesoderm generates the nephric, or Wolffian, duct, an outgrowth of the duct called the ureteric bud (UB), and the surrounding metanephric mesenchyme (MM). Kidney development starts when the UB invades the MM and transmits inductive signals, such as Wnt9b (Carroll et al., 2005), to promote condensation of the MM around the UB tips. These UB tip associated Cap mesenchyme cells (CM) continue to express Pax2 and are the stem cells of the nephron that generate all of the epithelial derivatives, including distal, proximal and glomerular epithelium (Kobayashi et al., 2008; Mugford et al., 2008). The CM undergoes a mesenchymal- to-epithelial transition to generate all the epithelial cells of the developing nephron. However, Pax2 expression is down-regulated in the podocyte precursor cells and in the mature epithelial cells of the nephron as development comes to an end (Ryan et al., 1995).
Loss of Pax2 function results in complete renal and reproductive tract agenesis in mice (Torres et al., 1995; Soofi et al., 2012). In humans, multiple Pax2 mutations have been identified in patients with renal coloboma syndrome, characterized by dysplasia, hypoplasia, and vesicoureteral reflux (Sanyanusin et al., 1995; Bower et al., 2012). The reactivation of Pax2 expression is also observed in adult kidneys after acute injury, suggesting a critical role for Pax2 in regenerating the epithelia (Imgrund et al., 1999; Humphreys et al., 2008; Kusaba et al., 2014). Ectopic or deregulated expression of Pax2 is also seen in Wilms' tumor (Dressler, G.R. and Douglass, E.C., 1992), renal cell carcinoma (Gnarra and Dressler, 1995), and polycystic kidney disease (Ostrom et al., 2000), where it is thought to promote proliferation and/or survival. Despite its central role in kidney development and renal disease, the biochemistry of Pax2 and its affects on gene regulation are not well characterized in a developing tissue. Several target genes have been identified, including Gata3 (Grote et al., 2006) and Gdnf (Brophy et al., 2001), but these are likely to represent only a fraction of the total genes that may be regulated by Pax2. A more recent report showed that in the pro/mesonephros a core transcriptional network formed by Pax2, Gata3 and Lhx1 regulates the expression of downstream effectors of kidney development (Boualia et al., 2013).
In cell culture systems, Pax2 is known to recruit epigenetic complexes to DNA that can positively or negatively regulate gene expression. The adaptor protein PTIP, links Pax2 to an Mll3/4 histone H3K4 methyltransferase complex that imprints activating epigenetic marks on chromatin (Patel et al., 2007). Similarly, PTIP was shown to be required for H3K4me of immunoglobulin switch regions that depend on long-range chromatin interactions between Pax5, the 3’ immunoglobulin heavy chain enhancer, and the new transcription start sites (Daniel et al., 2010; Schwab et al., 2011). Yet, Pax2 can also recruit Polycomb repressor complexes to chromatin, depending upon the availability of the co-repressor Groucho/Grg4/Tle4 (Patel et al., 2013). In the presence of Grg4, Pax2 is dephosphorylated (Cai et al., 2003), PTIP and the MLL complex are displaced, and the arginine methyltransferase PRMT5 and the H3K27 methyltransferase Ezh2 are recruited to silence gene expression (Patel et al., 2012). Still, it is not clear whether Pax2 is primarily an activator or a repressor within the developing intermediate mesoderm and kidney.
In this report, we identify the genes regulated by Pax2 in renal progenitor cells using novel Pax2EGFP alleles that allow for the sorting of cells from embryonic intermediate mesoderm and its early derivatives. Microarray analyses determined the complete expression profiles of Pax2 null and Pax2 positive renal progenitor cells. The data confirm that many critical kidney developmental genes require Pax2 for expression. Furthermore, the data suggest that in the absence of Pax2, some genes normally expressed in the paraxial mesoderm and in interstitial stromal cells are now expressed within the intermediate mesodermal cells. Moreover, to address the role of PTIP during kidney development, we developed an organ culture system in which the gene encoding PTIP (Paxip1) could be deleted from all cells of the developing kidney at a specific stage of development. The data indicate that Pax2 regulated genes can be PTIP dependent or PTIP independent and confirm a central role for Pax2 in regulating critical developmental genes and pathways in the kidney.
MATERIALS AND METHODS
Animals
Mice were kept according to NIH guidelines. All procedures were approved by the University Committee on Use and Care of Animals at the University of Michigan. The Egfp/+ mice were generated as previously described (Soofi et al., 2012). R26CP mice were obtained by crossing the Ptipf/f mouse strain (Kim et al., 2007) with B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J mice (Jackson Labs, Stock No. 8463). The progeny was then crossed with Ptipf/f mice to generate R26CP mice carrying two copies of the Ptipf/f locus. R26CP mice were then bred to the reporter R26TdTomato strain (Jackson Labs, Stock No. 7576) to generate R26CPTG mice. For genotyping, genomic DNA was prepared by a standard method and amplified by PCR using the following primer pair sets: Egfp – CCCACCGTCCCTTCCTTTTCTCCTCA, GAAAGGCCAGTGTGGCCTCTAGGGTG; Ptip – GGTTCTCTTGCAGCATCTCC, GGGAACTGATCTTCGATGAGG; Cre – CGAGTGATGAGGTTCGCAAG, TGATGTAACGAACCTGGTCG; TdTomato – CTCTGCTGCCTCCTGGCTTCT, CGAGGCGGATCACAAGCAATA, TCAATGGGC GGGGGTCGTT.
Primers were sequenced at the University of Michigan DNA Sequencing Core Facility.
Fluorescent Activated Cell Sorting
The entire intermediate mesoderm regions, comprising the mesonephric tubules, nephric duct and metanephric mesenchyme, dissected from embryonic day (E) 11.5 Egfp/+ and Egfp/Egfp embryos, was dissociated using 0.25% trypsin-EDTA for 5 minutes. Cells were resuspended in 1% FBS in PBS and filtered through a 70μm cell strainer (BD Falcon) to remove cell clumps. Cells expressing EGFP were isolated using a FACSAria III (BD) cell sorter.
Microarray expression analysis
Total RNA was extracted from EGFP positive FACS-sorted cells using TRIzol reagent (Life Technologies) and RNeasy Mini Kit (Qiagen). Microarray expression analysis was performed using three independent cohorts of embryos and carried out by the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility using a Mouse 430 2.0 Affymetrix GeneChip 3 expression arrays (Affymetrix), as described (Zhang et al., 2012). Expression values for each gene was calculated using the robust multi-array average (RMA) method and fitted to weighted linear models in R using the Affymetrix package of bioconductor (Irizarry et al., 2003; Zhang et al., 2012). The complete dataset and original Cel files can be accessed at GEO repository (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64129).
Bioinformatics analysis
Gene ontology and functional expression analyses of the genes whose expression was altered in Pax2 null embryos, compared to controls was carried out using Genomatix Genome Analyzer (Genomatix Software GmbH) and ToppGene Suite (Chen et al., 2009). Analysis parameters were kept as default.
RNA reverse transcription and real-time PCR
Real time PCR analysis of EGFP positive FACS-sorted cells was carried out using the same RNA used for microarray expression analysis. In details, 100ng of total RNA was reverse transcribed into cDNA using SuperScript II Reverse Transcriptase (Life Technologies). cDNA templates were amplified with iTaq Universal SYBR Green Supermix (Bio-Rad) in a StepOne Plus real-Time PCR System (Life Technologies). Real time PCR analysis of organ cultures was performed using 100ng of total RNA extracted from 5 days ex vivo cultured R26PTG and R26CPTG kidney rudiments as described above. The following primer pairs were used: Bmp7 –CAGCCAGAATCGCTCCAAGA, GCAATGATCCAGTCCTGCCA; Cited1 – CTCTGGGAAGGAGGATGCC, CCAGAGGAGCTAGTGGGAAC; Eya1 – ATCTAACCAGCCCGCATAGC, CTGCTTCCGAGAGCTGAACC; Gdnf – CCAGTGACTCCAATATGCCTGA, TGCCGATTCCTCTCTCTTCG; Hnf4a – TACTCCTGCAGGTTTAGCCG, CAGCCCGGAAGCACTTCTTA; Hprt – GTTGGGCTTACCTCACTGCT, TCATCGCTAATCACGACGCT; Msx1 – GCCCCGAGAAACTAGATCGG, GGACTCAGCCGTCTGGC; Osr1 – TTCGTTTGCAAGTTCTGTGG, TGTAGCGTCTTGTGGACAGC; Six2 – GCAAGTCAGCAACTGGTTCA, AACTGCCTAGCACCGACTTG; Ret – AGAGCAGAGACTACTTGGACCT, AGTAAATGCATGTGAAATTCTACCA; Twist2 – GTCTCAGCTACGCCTTCTCC, CAGGTGGGTCCTGGCTTG; Wnt4 – GCAGGAAGGCCATCTTGACA, CACGTCTTTACCTCGCAGGA.
In situ hybridization
Whole mount RNA in situ hybridization was performed on E11.5 Egfp/+ and Egfp/Egfp embryos, as described (Soofi et al., 2012). In situ hybridization was also performed on 5 days ex vivo cultured R26CP and control embryonic kidney rudiments. In details, the samples were fixed in 4% PFA in PBS for 1 hour at 4C, then washed three times in PBS + 0.1% Tween-20 (PBT) for 10 minutes at room temperature and de-hydrated through a PBT/methanol series. Templates for digoxigenin (DIG)-labeled riboprobes were generated by PCR amplification of E11.5-E15.5 mouse embryo cDNAs and sequenced. Primer pairs used to prepare riboprobe templates are listed in Table S6. Morphometric analysis of whole mount RNA in situ hybridization performed on kidney rudiments was carried out using ImageJ software.
Organ culture
E11.5 R26CPTG and control kidneys were cultured on 0.4μm Transwell filter inserts (Costar) in Dulbecco's Modified Eagle Medium supplemented with 10% FCS and 1X penicillin/streptomycin for 5 days. To induce Cre-mediated recombination, kidneys were cultured in the presence of 500nM 4-hydroxytamoxifen (Sigma) for the initial 48 hours of culture.
Western blot analysis
Single R26CPTG kidneys cultured for 5 days were carefully detached from the membranes and lysed in 2X SDS buffer (20% glycerol, 4% sodium dodecyl sulfate, 0.2M dithiothreitol, 125mM Tris, pH 6.8), as described (Zhang et al., 2012). Total proteins were separated on 10% SDS-PAGE gel, transferred to PVDF membranes and immunoblotted with a rabbit anti-Pax2 antibody (Dressler, G. R. and Douglass, E. C., 1992) and a rabbit anti-PTIP antibody (Patel et al., 2007).
RESULTS
Pax2 controls the expression of critical regulators in kidney development
Recently, we described two new Pax2 alleles that mark the Pax2 positive cells with EGFP in developing embryos (Soofi et al., 2012). Loss of Pax2 function resulted in abnormal nephric duct development, characterized by epithelial cells with increased motility and reduced tight and adherent junctions. In the Pax2 null mutants, the EGFP positive metanephric mesenchyme was also unable to respond to inductive signals. To identify potential targets of the transcription factor Pax2 in renal progenitor cells, we compared the whole expression profile of Pax2 positive and Pax2 null cells after fluorescent activated cell sorting (FACS) from mouse embryos carrying the Pax2Egfp alleles. Mice carrying one Egfp allele (Pax2Egfp/+) are viable and fertile and express EGFP in all Pax2 expressing cells. The presence of two Egfp alleles (Pax2Egfp/Egfp) completely abrogates Pax2 expression, however EGFP is still present in all cells of the embryos that would otherwise express Pax2 (Fig. 1A). In E11.5 Pax2 null embryos (Fig. 1A), the nephric duct is present albeit the epithelial integrity is compromised, as reported previously (Soofi et al., 2012). There are also many Pax2 positive cells adjacent to the mutant nephric duct that would correspond to Pax2 positive mesenchyme (Fig. 1A). Thus, we could insure that EGFP positive cells from similar time points represented cells of similar origin, the only difference being the presence or absence of Pax2 protein.
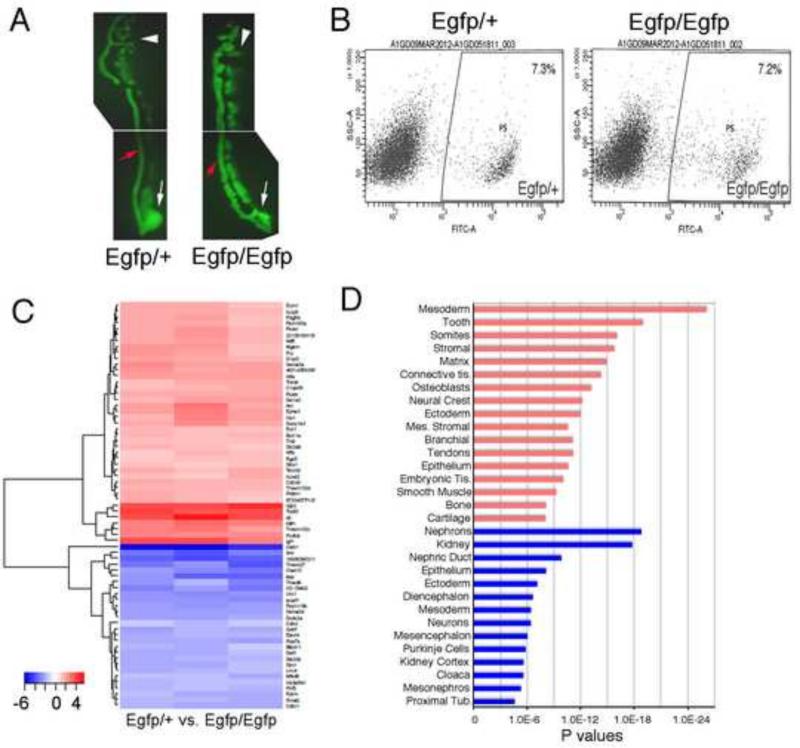
Figure 1. Expression Analyses of Pax2 Mutant Intermediate Mesoderm.
A) Microdissected E11.5 IMs are shown from heterozygous Pax2Egfp/+ and homozygous Pax2Egfp/Egfp embryos, prior to single cell isolation. The nephric duct (red arrow), the mesonephros (white arrowhead) and the presumptive metanephric regions (white arrow) are marked. Note EGFP positive mesenchymal cells all along the nephric duct in Pax2Egfp/Egfp embryos. B) Fluorescent activated cell sorting of populations from either Pax2Egfp/+ or Pax2Egfp/Egfp embryos. Note the nubmer of EGFP positive cells is not significaintly different. C) Heat map of Affymetrix microarrray expression analyses comparing EGFP positive cells from Pax2Egfp/+ to Pax2Egfp/Egfp isolated from E11.5 IMs. Red indicates increased expression in the mutants, whereas blue idicateds decreaed expression in mutants compared to heterozygotes. D) Genomatix tissue expression analyses from candidate genes that activated in Pax2Egfp/Egfp embryos are in red and genes decreased in Pax2Egfp/Egfp are in blue. P values reflect the probability of finding the number of genes in each tissue at random.
At E11.5, EGFP positive cells from both genotypes were clearly distinguishable from the EGFP negative fractions, and could be easily separated using FACS analysis (Fig. 1A, B). There was no significant difference in the numbers of EGFP positive cells between tissues from Pax2Egfp/+ and Pax2Egfp/Egfp embryos. After total RNA isolation, gene expression profile analysis by Affymetrix microarrays (Fig. 1C) identified 496 unique genes whose expression was altered by more than 2-fold in Pax2 null renal progenitors, compared to controls. The entire dataset is available from the Gene Expression Omnibus (GEO accession number GSE64129). Among them, 286 genes were upregulated and 210 downregulated in Pax2 null renal progenitor cells (See Table S1 for all significant Pax2 regulated genes). Strikingly, among the downregulated genes we identified many known regulators of kidney development, such as Six2, Gdnf, Eya1 and Lhx1 (See Table 1 for a select list of genes).
Table 1.
Renal Developmental Control Genes Suppressed in Pax2 Mutants
| Gene | Expression | Mutant Phenotype | Reference |
|---|---|---|---|
| BMP7 | MM, ND, UB | Dysplasia, arrested development | (Dudley et al., 1995) |
| Eya1 | MM | Agenesis, absence of UB outgrowth | (Xu et al., 1999) |
| Gata3 | ND, UB | Ectopic UB, kidney dysplasia | (Grote et al., 2006) |
| Gdnf | MM | Agenesis due to lack of UB growth | (Moore et al., 1996; Sanchez et al., 1996) |
| Hnf4a | SSB, PT | (Heliot et al., 2013) | |
| Irx1 | SSB | Defects in Xenopus pronephros | (Alarcon et al., 2008) |
| Irx3 | CS | Defects in Xenopus pronephros | (Reggiani et al., 2007) |
| Lhx1 | ND, UB, CS | Agenesis due to ND defects | (Tsang et al., 2000) |
| Npnt | UB | Agenesis or hypoplasia | (Linton et al., 2007) |
| Ret | ND, UB | Agenesis due to lack of UB outgrowth | (Schuchardt et al., 1996) |
| Sal1 | MM, RV | Agenesis due to defect in UB | (Nishinakamura et al., 2001) |
| Six2 | CM | Depletion of progenitor cells, dysplasia | (Self et al., 2006) |
| Vsnl1 | UB tips | (Ola et al., 2011) |
MM, metanephric mesenchyme; ND, nephric duct; UB, ureteric bud; CM, cap mesenchyme; RV, renal vesicle; Cs, comma shaped body; SSB, s-shaped body.
To further annotate the genes and networks whose expression levels significantly decreased in renal progenitor cells in the absence of Pax2, we utilized Genomatix and Toppgene informatics software to correlate gene expression, tissue distribution, and gene ontologies. Using Genomatix, the most significant tissues and terms for genes whose expression was decreased in Pax2 mutants included: nephrons, kidney, nephric duct, and epithelium (Fig. 1D). Additional tissues and terms for this gene set included components of the developing nervous system, many parts of which also express Pax2. These data suggest some common targets for Pax2 in the kidney and nervous system, which could include genes such as Gdnf, Gfra1, Hdc, Lhx1, Ret, Sim1, Trpv4, and Vsnl1 that are expressed in both tissues. The entire Genomatix tissue expression analysis dataset and the associated genes that are down regulated in Pax2 mutants are listed in Table S2.
Using Toppgene gene ontology and tissue expression analyses, genes down regulated in Pax2 mutants also were associated with mesonephric and metanephric development and were expressed in intermediate mesoderm derivatives like the nephric duct, ureteric bud, and metanephric mesenchyme (Table 2, and Table S3)
Table 2.
TOPPGENE Analyses of Pax2 Target Genes
| Upregulated Genes in Pax2 Mutants | |||
|---|---|---|---|
| Co-Expression Atlas | Source | p Value | No. (input/annotated) |
| E13.5 testis, interstitial | Gudmap | 8.2E-78 | 105/778 |
| E13.5 ovary, mesenchyme | Gudmap | 1.6E-56 | 88/798 |
| E11.5 female gonad, mesenchyme | Gudmap | 3.0E-55 | 69/424 |
| E11.5 male gonad, mesenchyme | Gudmap | 3.9E-55 | 88/828 |
| Adult kidney, renal capsule | Gudmap | 2.5E-38 | 70/780 |
| E15.5 kidney, pelvic mesenchyme | Gudmap | 3.3E-36 | 47/305 |
| E15.5 kidney, peripheral blastema | Gudmap | 4.0E-34 | 67/819 |
| E8.5 paraxial mesoderm | Facebase | 2.9E-32 | 70/970 |
| Gene Ontology, Biological Processes | |||
| Mesenchymal development | GO:0060484 | 2.4E-14 | 22/179 |
| Mesenchyme cell differentiation | GO:0048762 | 1.1E-8 | 18/152 |
| Epithelial to mesenchyme transition | GO:0001837 | 4.4E-7 | 10/84 |
| Downregulated Genes in Pax2 Mutants | |||
|---|---|---|---|
| Co-Expression Atlas | Source | p Value | No. (input/annotated) |
| E11.5 kidney, ureteric bud | Gudmap | 4.6E-75 | 92/813 |
| E15.5 kidney, cortical col. duct | Gudmap | 1.0E-59 | 79/775 |
| P4 kidney, cap mesench., renal vesicle | Gudmap | 1.9E-41 | 64/785 |
| E15.5 kidney, S-shaped body | Gudmap | 2.4E-33 | 44/406 |
| E15.5 kidney, prox. tubules | Gudmap | 1.2E-31 | 54/759 |
| Gene Ontology, Biological Processes | |||
| Renal development | GO:0001656 | 3.9E-10 | 12/94 |
| Ureteric bud development | GO:0001657 | 2.5E-9 | 12/110 |
| Nephron development | GO:0072006 | 1.1E-7 | 11/100 |
Although the expression of most of the kidney markers was significantly reduced in Pax2 null renal progenitor cells, the levels of Cited1 transcripts, which are expressed in a sub-compartment of the cap mesenchyme before induction (Brown et al., 2011), increased in the absence of Pax2 (Table S1). This seems to indicate that Pax2 is required to transition the early renal progenitor cells into a slightly more differentiated cell population, which maintain the expression of Six2 but loses the expression of Cited1 (Brown et al., 2011).
Increased Expression of Interstitial Stroma and Paraxial Mesodermal genes in Pax2 Mutants
In the Pax2 mutant IMs, we also detected more than 280 genes whose expression levels increased in the EGFP positive cells in the absence of Pax2. As above, two independent bioinformatics software packages were used to analyze the ontologies and functions of genes whose expression is increased in Pax2 null EGFP positive cells from E11.5 IM. Genomatix analyses of this gene data set found many genes associated with the paraxial mesoderm and its derivatives, the ectoderm and neural tissues, or stromal cells (Fig. 1D). Among the most common tissues associated with the upregulated gene data set were: mesoderm, tooth, somites, bone, connective tissue, and muscle. The complete gene ontology and expression sets and their associated tissue are listed in Table S4.
Using Toppgene for candidate prioritization, we also found that genes upregulated in Pax2 mutants were primarily associated with stromal and interstitial cells of the gonads and kidney (Table 2). Gene ontology terms included: regulation of development, organ morphogenesis, cell adhesion, and skeletal system development. Cellular component analyses pointed to the extracellular matrix as a significant component upregulated in Pax2 mutants. By comparing to known gene expression atlases, such as Gudmap and Facebase, genes up-regulated in Pax2 mutants are associated with the interstitial cells in the gonads, the kidney capsule and the cells of the kidney interstitium, and the paraxial mesoderm (Table S5). These associations are based on microarray expression data sets from laser capture dissection of specific regions in embryonic kidneys, reproductive tracts, or whole embryos.
Given that paraxial mesoderm and the somites eventually generate the axial skeleton and muscles, the data suggest that Pax2 can suppress genes normally expressed in axial mesoderm and its derivatives.
Confirmation of Expression Patterns
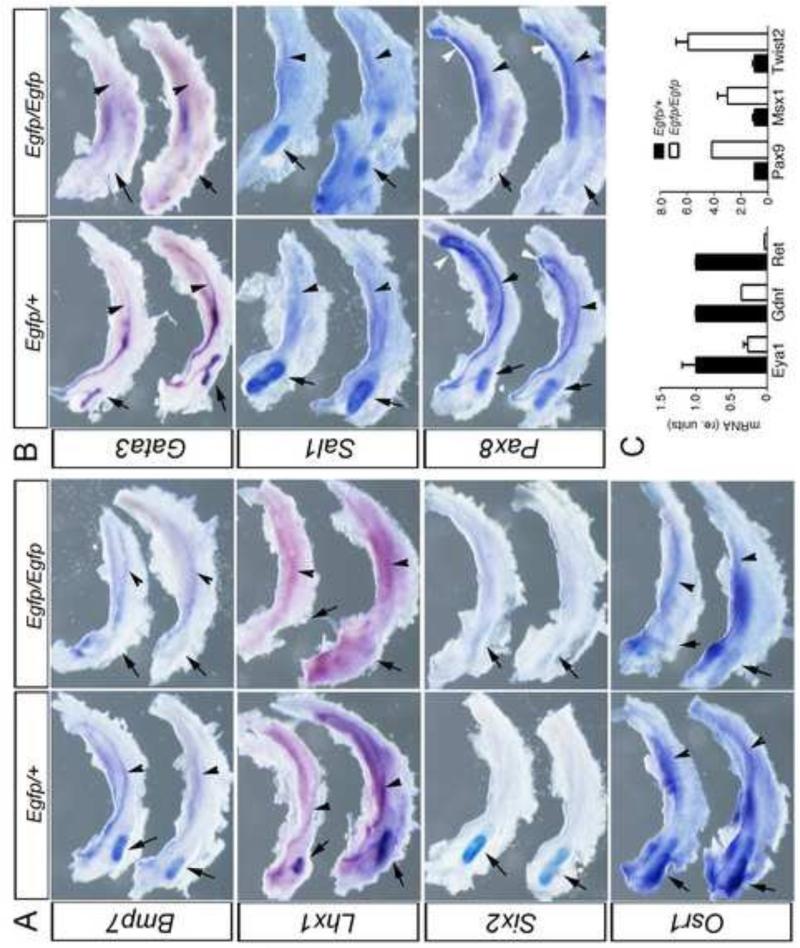
To validate our microarray results, we performed whole mount in situ hybridization (WISH) and/or quantitative RT-PCR on E11.5 Pax2 null and control embryos (Fig. 2). We analyzed the expression pattern of multiple genes important for kidney development whose expression significantly decreased in renal progenitor cells in the absence of Pax2. For kidney regulators, such as Bmp7, Lhx1, Six2, Gata3, Osr1, Pax8 and Sall1, we dissected the entire intermediate mesoderm region, including the mesonephros and developing metanephros, from the embryos after whole mount staining of intact embryos. Control Pax2Egfp/+ embryos displayed the correct expression pattern for all probes tested. For example, Six2 mRNA was detected in the cap mesenchyme surrounding the tips of the UB, the expression of Bmp7 was noticeable along the nephric duct, in the UB and MM, and Lhx1 was expressed along the nephric duct and the UB (Fig. 2A). Ureteric bud, nephric ducts and some mesonephric tubules also expressed the transcription factors Gata3 and Pax8, whereas Sall1 mRNA was present only in the MM (Fig. 2B). The expression levels of all genes identified as decreased in Pax2Egfp/Egfp mutants by microarray analyses also showed dramatic reductions in expression as determined by WISH. These results validate the microarray analysis and further confirm that Pax2 positively regulates the expression of genes important for kidney development.
Figure 2. Confirmation of Gene Expresssion Differences of Kidney Developmental Regulators.
Intermediate mesoderm was dissected free from surrounding tissues and includes the posterior metanephros (arrow) the nephric duct (arrowhead). A) Whole mount in situ hybridization of genes suppressed in Pax2Egfp/Egfp mutant IMs show decreased Bmp7, Lhx1, and Six2 in the developing metanephros (arrow) at E11.5. B) Whole mount in situ hybridization of Gata3 and Sall1 show decrease expression in Pax2Egfp/Egfp in the metanephric mesenchyme (arrow) and in the nephric duct epithelia (arrowhead). White arrowheads indicate mesonephric tubules. C) Quantitative RT-PCR for mRNAs of Eya1, Gdnf, and c-Ret all show significaintly decreased levels of transcripts in Pax2Egfp/Egfp IMs at E11.5. All qRT-PCRs were done in triplicate and normalized to Hprt expression levels with error bars representing one standard deviation from the mean.
Our microarray analysis also revealed that the kidney markers Pax8 and Osr1 were both downregulated in Pax2 null renal progenitor cells, however the reduction of the expression level was less than two-fold compared to the controls. Thus, we decided to assess the mRNA expression pattern of these genes by WISH. As expected, in control embryos the expression of Pax8 mRNA was present in the nephric duct, UB and mesonephric tubules, but reduced in the Pax2 mutants (Fig. 2B). Osr1 expression was detectable in the MM but was also significantly reduced in the Pax2 mutants (Fig. 2A). Furthermore, real time RT-PCR confirmed that the expression levels of the kidney markers Eya1, Gdnf and Ret were reduced by 73%, 64% and 97%, respectively, in Pax2 null renal progenitor cells, compared to controls (Fig. 2C). We also tested a select group of genes that were upregulated in the Pax2 mutants (Fig. 2C). The paraxial genes Pax9 and Msx1 were increased 4 and 3 fold respectively, whereas the mesenchymal expressed gene Twist2 was upregulated 6 fold in Pax2 mutants. In our experience, the Affymetrix microarrays are robust and reproducible, but routinely underestimate the degree of differences in expression levels, as both WISH and qRT-PCR routinely demonstrate greater differences in mRNA expression levels compared to microarrays.
PTIP dependent and PTIP independent expression of kidney developmental genes
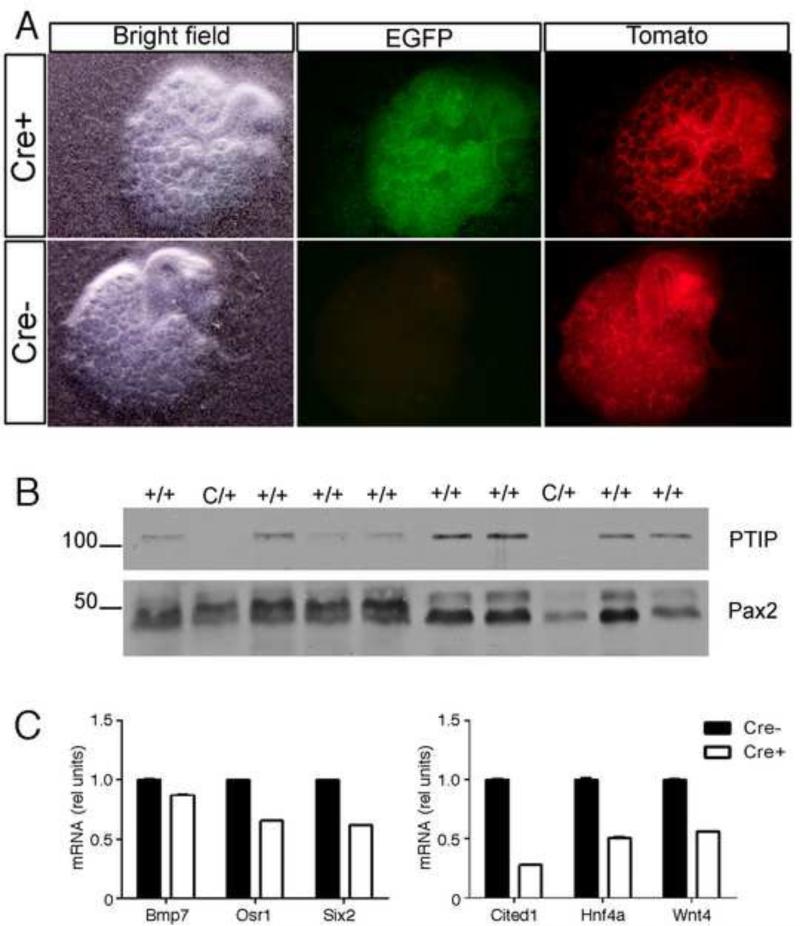
Pax2 interacts with the nuclear protein PTIP to recruit a histone methyltransferase complex to a DNA binding site, resulting in increased levels of H3K4me3 (Patel et al., 2007). The PTIP protein is encoded by the Paxip1 gene and is required for early embryonic development, as Paxip1−/− embryos are developmentally arrested at E9.5 (Cho et al., 2003). Loss of PTIP results in spontaneous differentiation of embryonic stem (ES) cells (Kim et al., 2009), altered arrhythmias in adult murine cardiomyocytes (Stein et al., 2011) and chronic glomerular disease in adult mice (Lefevre et al., 2010). To investigate the role of PTIP during kidney development, we made use of an ex vivo organ culture system to delete Paxip1 in E11.5 R26CPTG (Cre+) kidneys by adding 4-hydroxytamoxifen (4-OHT) to the culture medium at day 0 of culture. R26PTG (Cre-) kidneys were cultured in the presence of 4-OHT and used as control. Both Cre+ and Cre- kidneys also carried one R26TdTomato allele, which enabled us to monitor Cre-mediated recombination, as the ubiquitously expressed TdTomato protein is replaced by EGFP upon recombination (Muzumdar et al., 2007). EGFP expression was detectable in Cre+ kidneys after 24 hours of culture in the presence of 4-OHT (data not shown). After 5 days of culture EGFP was present in most of the cells of Cre+ kidneys, whereas Cre-kidneys didn't show EGFP expression (Fig. 3A). TdTomato was still present in Cre+ kidneys, albeit in much fewer cells compared to Cre- kidneys. Western blot analysis confirmed the absence of PTIP in Cre+ kidneys (Fig. 3B). After 5 days of culture there were no substantial differences in branching morphogenesis and size between Cre+ and Cre- kidneys (Fig. 3 and data not shown). Quantitative RT-PCR demonstrated that in PTIP null kidneys Bmp7 was slightly downregulated compared to control kidneys, whereas the expression level of both Osr1 and Six2 was reduced by 35% and 39%, respectively (Fig. 3C).
Figure 3. Deletion of PTIP in Embryonic Kidney Organ Cultures.
A) Tamoxifen induction of the Rosa26-Cre-ER transgene activates EGFP expression and suppresses dT-Red in the kidney mesenchyme. B) Western blotting of protein lysates from individual kidney rudiments that carry the Rosa26-Cre-ER transgene (C/+) and wild-type controls cultured with tamoxifen. Note that the 120 kD PTIP protein is absent in kidney culures that are positive for Cre-ER. C) qRT-PCR of selected Pax2 target genes show significant down regulation of Cited1, HNF4a, and Wnt4 upon deletion of PTIP. All qRT-PCRs were done in triplicate and normalized to Hprt expression levels with error bars representing one standard deviation from the mean.
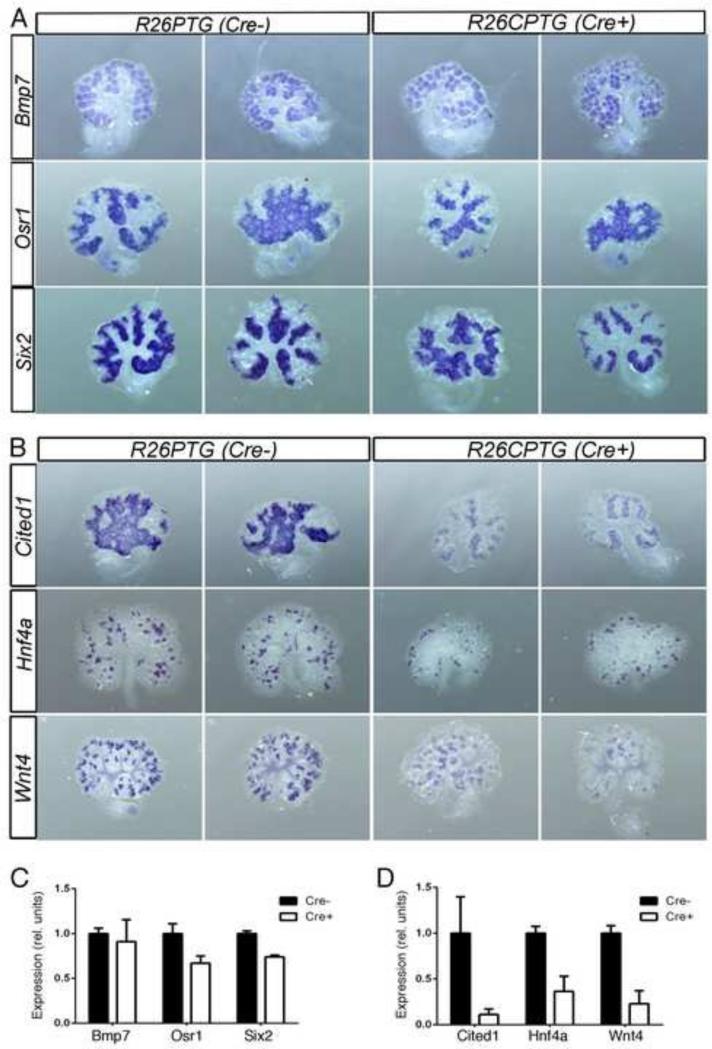
To identify the Pax2 target genes that may be affected in the absence of PTIP during kidney development, we performed WISH on 5 days cultured Cre+ and Cre- kidneys. We started to analyze the expression pattern of the early markers of kidney development Bmp7, Osr1 and Six2. In Cre negative kidneys WISH staining recapitulated the correct expression pattern of these genes. In the absence of PTIP both the expression level and pattern of these genes did not seem to be altered qualitatively (Fig. 4A). The surface area of staining and the intensity of staining were measured by quantitative morphometry to generate a relative measure of expression as determined by WISH (Fig. 4C). As observed, the expression of Bmp7 did not change significantly upon PTIP deletion, whereas the expression levels of Osr1 and Six2 mRNA were down slightly to 75% of wild-type (Fig. 4C). We then assessed the expression of two late markers of kidney development, Hnf4a and Wnt4. By WISH staining, the expression levels of both genes was reduced in PTIP null kidneys compared to control explants, however the spatial pattern of gene expression was not perturbed (Fig. 4B, D). Quantitative RT-PCR showed that in the absence of PTIP, Hnf4a and Wnt4 expression was reduced by 50% and 44%, respectively (Fig. 3C). Quantitation of the WISH staining was consistent with the qRT-PCR data and showed a reduction of approximately 60% and 75% respectively. Surprisingly, both RT-PCR and WISH showed that Cited1, whose expression is found in early renal progenitors of the cap mesenchyme (Brown et al., 2011), was significantly downregulated in PTIP null kidneys compared to the controls (Fig. 3C, 4B, D). Since the Cited1 cells are thought to represent a sub-population of true renal progenitor cells, the data suggest that PTIP is essential for maintaining at least part of the transcriptome of renal stem cells.
Figure 4. Whole Mount in situ hybridization of select Pax2 target genes in kidney organ cultures with or without PTIP.
Represenative kidneys are shown hybridized with the indicated probes; wild-type (Cre-) rudiments are on the left and PTIP deleted rudiments (Cre+) are on the right. Genes that are minimally affected by the loss of PTIP are show in panel A, whereas B shows three genes whose expression is lower upon PTIP deletion. C & D) Morphometric analyses of whole mount staining, taking both surface area and intensity into account. Genes unaffected by the loss of PTIP are in C, whereas genes suppressed by the loss of PTIP are in D.
Discussion
Gene regulatory networks are thought to establish and maintain the differentiated state. The Pax genes expression is activated early in development, when tissues are first specified with respect to their eventual fates, and are critical for cell lineage decisions. Prior studies implicate Pax2 in a core regulatory network, which includes Gata3 and Lhx1, that defines the intermediate mesoderm and the renal epithelial cells derived from this mesoderm (Boualia et al., 2013). In this report, we define a complete data set of genes affected by the loss of Pax2 in the intermediate mesoderm and its early derivatives, the nephric duct, the mesonephros and the metanephric mesenchyme. The genes include many known kidney developmental regulators, such as Gdnf, c-ret, Six2, Sal1, and Lhx1, but also affected are genes and proteins associated with glycosylation, cell membranes, cell-cell signaling, and cell adhesion.
We utilized gene ontology and tissue expression software to analyze and cluster the genes affected by the loss of Pax2 in the intermediate mesoderm. Genomatix software utilizes available literature to associate gene expression with particular tissues and processes. Toppgene analyses are similar but also include comparisons to gene expression atlases and tissue expression databases. As expected, Pax2 mutants showed decreased expression of early kidney developmental genes, many of which had been previously identified. More surprising was our finding that in Pax2 mutants the intermediate mesodermal cells, as marked by EGFP expression, had increased expression of interstitial stromal genes and genes associated with paraxial mesoderm. For example, Pax9 expression is observed in paraxial mesoderm and the sclerotomes, which will contribute to the vertebral column (Neubuser et al., 1995; Peters et al., 1999; Rodrigo et al., 2003). Yet EGFP positive cells from the IM of Pax2 mutants show a four fold upregulation of Pax9. This suggests a cellular fate transformation from renal epithelial progenitor cell to a stromal or mesenchymal phenotype and a fate more consistent with paraxial mesoderm and its derivatives. Thus Pax2 may be specifying a boundary between paraxial and intermediate mesoderm along the medio-lateral body axis. Similarly, Pax6 and Pax2 function within the central nervous system to define the boundaries between the developing forebrain and midbrain (Matsunaga et al., 2000). Given that Pax genes were originally identified based on homology to Drosophila pair-rule segmentation genes, which define the para-segments in the fly embryo, the specification of boundaries between developing tissue compartments may be an evolutionarily conserved feature of Pax genes.
Despite the genetic evidence for cell lineage specification in development, the biochemistry of Pax protein function remains poorly characterized. The conserved 128 amino acid paired box binds a bipartite DNA sequence that spans nearly two turns of the DNA helix. (Czerny et al., 1993; Epstein, J. A. et al., 1994; Czerny and Busslinger, 1995; Xu et al., 1995; Phelps and Dressler, 1996) The crystal structure shows three amino-terminal α-helices that resemble a homeo-domain, followed by a carboxyl terminal region with three smaller α-helices. The amino terminal α-helices contact the 3’ part of the bipartite target sequence, whereas the carboxyl terminal tail recognizes the 5’ end of the DNA sequence. Furthermore, interactions between the paired-domain and the DNA recognition sequences can change the conformation of the Pax protein and the target DNAs (Chalepakis et al., 1994; Epstein, J. et al., 1994). The carboxy-terminus of Pax2/5/8 can bind to the adaptor protein PTIP/Paxip1 to recruit the Mll3 or Mll4 histone methyltransferases to imprint the activating H3K4me1/2/3 epigenetic marks (Patel et al., 2007; Daniel et al., 2010; Schwab et al., 2011). Yet, associations with the co-repressor Grg4/Tle4 displace PTIP and the Mll complex and subsequently recruit an arginine methyltransferase and the Polycomb repressor 2 complex (Patel et al., 2012). Thus, Pax2 targets could be subject to activation or repression depending on the availability of co-factors. Such cofactor interactions may also depend on the local DNA sequence context. Our screens in Pax2Egfp/Egfp mutants identified both positive and negatively regulated genes.
Because of the prior evidence linking PITP to Pax2 and gene activation, we investigated the expression of potential Pax2 targets in PTIP null embryonic kidneys. Using an organ culture system to delete a conditional PTIP allele, we show that a subset of genes whose expression is reduced in Pax2 mutants are also affected in PTIP mutants kidneys. However, many genes that are presumptive early targets of Pax2, those already expressed at the time of kidney explant in our organ cultures, do not appear to require PTIP for maintaining expression. These include Six2, Osr1 and Bmp7. This underscores the epigenetic effects of Pax2 within the intermediate mesoderm, as genes already expressed remain on, whereas certain genes that are activated after explant and after PTIP deletion are affected. However, Cited1, a marker of early cap mesenchyme is affected in PTIP deletions, despite being expressed early in kidney cultures, before deletion of PTIP. Whether similar results will be observed with temporal Pax2 deletions awaits the analyses of a conditional allele.
In summary, we have defined a genetic network dependent on Pax2 and, at least in part, dependent on PTIP. Such a network helps define the intermediate mesoderm and the renal epithelial lineage. In the absence of Pax2, the IM cells assume a pattern of gene expression more consistent with paraxial mesoderm and its derivatives. The data suggest that epigenetic mechanisms underlie early lineage decisions and compartmentalization of the mesoderm after gastrulation.
Supplementary Material
Highlights.
The effects of Pax2 loss on gene expression were examined in embryos.
Many kidney specific regulators were suppressed in Pax2 mutant cells.
Paraxial mesoderm and interstitial stromal genes were increased in Pax2 mutants.
Some kidney specific Pax2 target genes required PTIP for full expression.
The data suggest that Pax2 and epigenetic mechanisms imprint a renal epithelial fate.
Acknowledgments
We thank J. Washburn and C. Johnson of the University of Michigan Microarray core Facility, B. Aronow of Cincinnati Children's Hospital for help with TOPPGENE analyses, I. Levitan for help with WISH, and the University of Michigan Flow Cytometry Core for help with FACS analyses. This work was funded in part by NIH grants DK054740 and DK073722 to G.R.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon P, Rodriguez-Seguel E, Fernandez-Gonzalez A, Rubio R, Gomez-Skarmeta JL. A dual requirement for Iroquois genes during Xenopus kidney development. Development. 2008;135:3197–3207. doi: 10.1242/dev.023697. [DOI] [PubMed] [Google Scholar]
- Boualia SK, Gaitan Y, Tremblay M, Sharma R, Cardin J, Kania A, Bouchard M. A core transcriptional network composed of Pax2/8, Gata3 and Lim1 regulates key players of pro/mesonephros morphogenesis. Dev. Biol. 2013a;382:555–566. doi: 10.1016/j.ydbio.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower M, Salomon R, Allanson J, Antignac C, Benedicenti F, Benetti E, Binenbaum G, Jensen UB, Cochat P, DeCramer S, Dixon J, Drouin R, Falk MJ, Feret H, Gise R, Hunter A, Johnson K, Kumar R, Lavocat MP, Martin L, Moriniere V, Mowat D, Murer L, Nguyen HT, Peretz-Amit G, Pierce E, Place E, Rodig N, Salerno A, Sastry S, Sato T, Sayer JA, Schaafsma GC, Shoemaker L, Stockton DW, Tan WH, Tenconi R, Vanhille P, Vats A, Wang X, Warman B, Weleber RG, White SM, Wilson-Brackett C, Zand DJ, Eccles M, Schimmenti LA, Heidet L. Update of PAX2 mutations in renal coloboma syndrome and establishment of a locus-specific database. Hum Mutat. 2012;33:457–466. doi: 10.1002/humu.22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- Brown AC, Adams D, de Caestecker M, Yang X, Friesel R, Oxburgh L. FGF/EGF signaling regulates the renewal of early nephron progenitors during embryonic development. Development. 2011;138:5099–5112. doi: 10.1242/dev.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Brophy PD, Levitan I, Stifani S, Dressler GR. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. The EMBO journal. 2003;22:5522–5529. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Wijnholds J, Gruss P. Pax-3-DNA interaction: flexibility in the DNA binding and induction of DNA conformational changes by paired domains. Nucleic Acids Res. 1994;22:3131–3137. doi: 10.1093/nar/22.15.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell. Biol. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different seqnence recognition of Pax-6 and BSAP (Pax-5). Molecular and Cell Biology. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP Promotes Chromatin Changes Critical for Immunoglobulin Class Switch Recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR, Douglass EC. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1179–1183. doi: 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994a;269:8355–8361. [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994b;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Dressler GR. Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res. 1995;55:4092–4098. [PubMed] [Google Scholar]
- Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133:53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- Heliot C, Desgrange A, Buisson I, Prunskaite-Hyyrylainen R, Shan J, Vainio S, Umbhauer M, Cereghini S. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development. 2013;140:873–885. doi: 10.1242/dev.086538. [DOI] [PubMed] [Google Scholar]
- Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Imgrund M, Grone E, Grone HJ, Kretzler M, Holzman L, Schlondorff D, Rothenpieler UW. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice 1. Kidney Int. 1999;56:1423–1431. doi: 10.1046/j.1523-1755.1999.00663.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kim D, Patel SR, Xiao H, Dressler GR. The role of PTIP in maintaining embryonic stem cell pluripotency. Stem Cells. 2009;27:1516–1523. doi: 10.1002/stem.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Wang M, Cai Q, Brooks H, Dressler GR. Pax transactivation-domain interacting protein is required for urine concentration and osmotolerance in collecting duct epithelia. J. Am. Soc. Nephrol. 2007;18:1458–1465. doi: 10.1681/ASN.2006060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010;6:e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton JM, Martin GR, Reichardt LF. The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–2509. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H. Pax6 defines the di-mesencephalic boundary by repressing En1 and Pax2. Development. 2000;127:2357–2365. doi: 10.1242/dev.127.11.2357. [DOI] [PubMed] [Google Scholar]
- Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryans AM, Carver-Moore K, Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Koseki H, Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128:3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- Ola R, Jakobson M, Kvist J, Perala N, Kuure S, Braunewell KH, Bridgewater D, Rosenblum ND, Chilov D, Immonen T, Sainio K, Sariola H. The GDNF target Vsnl1 marks the ureteric tip. Journal of the American Society of Nephrology : JASN. 2011;22:274–284. doi: 10.1681/ASN.2010030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom L, Tang MJ, Gruss P, Dressler GR. Reduced Pax2 gene dosage increases apoptosis and slows the progression of renal cystic disease. Dev. Biol. 2000;219:250–258. doi: 10.1006/dbio.2000.9618. [DOI] [PubMed] [Google Scholar]
- Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol. Cell. 2012;45:185–195. doi: 10.1016/j.molcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Ranghini E, Dressler GR. Mechanisms of gene activation and repression by Pax proteins in the developing kidney. Pediatr. Nephrol. 2013 doi: 10.1007/s00467-013-2603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Wilm B, Sakai N, Imai K, Maas R, Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- Phelps DE, Dressler GR. Identification of novel Pax-2 binding sites by chromatin precipitation. J. Biol. Chem. 1996;271:7978–7985. doi: 10.1074/jbc.271.14.7978. [DOI] [PubMed] [Google Scholar]
- Reggiani L, Raciti D, Airik R, Kispert A, Brandli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Rodrigo I, Hill RE, Balling R, Munsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130:473–482. doi: 10.1242/dev.00240. [DOI] [PubMed] [Google Scholar]
- Ryan G, Steele-Perkins V, Morris J, Rauscher FJ, III, Dressler GR. Repression of Pax-2 by WT1 during normal kidney development. Development. 1995;121:867–875. doi: 10.1242/dev.121.3.867. [DOI] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont MEM, Sullivan MJ, Dobyns WB, Eccles MR. Mutation of the Pax2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat. Genet. 1995;9:358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- Schuchardt A, D'Agati V, Pachnis V, Costantini F. Renal agenesis and hypodysplasia in ret-k- mutant mice result from defects in ureteric bud development. Development. 1996;122:1919–1929. doi: 10.1242/dev.122.6.1919. [DOI] [PubMed] [Google Scholar]
- Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Mol. Cell. Biol. 2011;31:1503–1511. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soofi A, Levitan I, Dressler GR. Two novel EGFP insertion alleles reveal unique aspects of Pax2 function in embryonic and adult kidneys. Dev. Biol. 2012;365:241–250. doi: 10.1016/j.ydbio.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, Dressler GR. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J. Clin. Invest. 2011;121:2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Tsang TE, Shawlot W, Kinder SJ, Kobayashi A, Kwan KM, Schughart K, Kania A, Jessell TM, Behringer RR, Tam PP. Lim1 activity is required for intermediate mesoderm differentiation in the mouse embryo. Dev. Biol. 2000;223:77–90. doi: 10.1006/dbio.2000.9733. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- Zhang P, Cai Y, Soofi A, Dressler GR. Activation of Wnt11 by transforming growth factor-beta drives mesenchymal gene expression through non-canonical Wnt protein signaling in renal epithelial cells. J. Biol. Chem. 2012;287:21290–21302. doi: 10.1074/jbc.M112.357202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.