Abstract
The effect of proinflammatory cytokines on the expression and activity of soluble guanylyl cyclase (sGC) and cGMP–phosphodiesterases (PDEs) was determined in intestinal longitudinal smooth muscle. In control muscle cells, cGMP levels are regulated via activation of sGC and PDE5; the activity of the latter is regulated via feedback phosphorylation by cGMP-dependent protein kinase. In muscle cells isolated from muscle strips cultured with interleukin-1β (IL-1β) or tumor necrosis factor α (TNF-α) or obtained from the colon of TNBS (2,4,6-trinitrobenzene sulfonic acid)-treated mice, expression of inducible nitric oxide synthase (iNOS) was induced and sGC was S-nitrosylated, resulting in attenuation of nitric oxide (NO)–induced sGC activity and cGMP formation. The effect of cytokines on sGC S-nitrosylation and activity was blocked by the iNOS inhibitor 1400W [N-([3-(aminomethyl)phenyl]methyl)ethanimidamide dihydrochloride]. The effect of cytokines on cGMP levels measured in the absence of IBMX (3-isobutyl-1-methylxanthine), however, was partly reversed by 1400W or PDE1 inhibitor vinpocetine and completely reversed by a combination of 1400W and vinpocetine. Expression of PDE1A was induced and was accompanied by an increase in PDE1A activity in muscle cells isolated from muscle strips cultured with IL-1β or TNF-α or obtained from the colon of TNBS-treated mice; the effect of cytokines on PDE1 expression and activity was blocked by MG132 (benzyl N-[(2S)-4-methyl-1-[[(2S)-4-methyl-1-[[(2S)-4-methyl-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]carbamate), an inhibitor of nuclear factor κB activity. NO-induced muscle relaxation was inhibited in longitudinal muscle cells isolated from muscle strips cultured with IL-1β or TNF-α or obtained from the colon of TNBS-treated mice, and this inhibition was completely reversed by the combination of both 1400W and vinpocetine. Inhibition of smooth muscle relaxation during inflammation reflects the combined effects of decreased sGC activity via S-nitrosylation and increased cGMP hydrolysis via PDE1 expression.
Introduction
The relaxant transmitter nitric oxide (NO) plays an important role in gastrointestinal smooth muscle relaxation. The effects of NO are mediated by activation of soluble guanylyl cyclase (sGC), which catalyzes the conversion of GTP to cGMP (Friebe et al., 1997; Mayer et al., 1998; Krumenacker et al., 2004; Francis et al., 2010; Derbyshire and Marletta, 2012). The majority of cGMP effects are mediated by activation of cGMP-dependent protein kinase, which phosphorylates several proteins regulating Ca2+ mobilization and myosin light-chain phosphatase (MLCP) activity, key determinants of smooth muscle contraction (Murthy and Makhlouf, 1998; Kamm and Stull, 2001; Somlyo et al., 2004; Murthy, 2006; De Godoy and Rattan, 2011). The strength and duration of cGMP signaling are regulated by the balance between sGC and cGMP-specific phosphodiesterase (PDE) activity (Soderling and Beavo, 2000; Francis et al., 2001; Rybalkin et al., 2003; Corbin et al., 2009).
sGC, a heterodimer protein with α (α1 and α2) and β (β1 and β2) subunits, consists of amino terminal heme-binding, central dimerization, and carboxyterminal catalytic domains (Mayer et al., 1998; Krumenacker et al., 2004; Derbyshire and Marletta, 2012). Binding of NO to the heme greatly increases the catalytic activity of sGC. Expression of α1, α2, or β1 alone is not sufficient for sGC activity, and heterodimerization is a prerequisite for sGC activity. The subunits α1, α2, and β1 are functional, but the function of the β2 subunit is not clear, and thus only α1β1 and α2β1 isoforms are active (Pyriochou and Papapetropoulos, 2005; Derbyshire and Marletta, 2012). sGC α1β1 heterodimer expression is most abundant in mammalian tissues including smooth muscle, whereas expression of α2β1 heterodimer expression is highest in the brain. sGC heterodimer containing the β2 subunit has not been identified.
Expression of sGC subunits has been shown to be regulated by both transcriptional and post-transcriptional mechanisms in response to cytokines and growth factors (Krumenacker et al., 2001; Pyriochou and Papapetropoulos, 2005; Xia et al., 2007; Derbyshire and Marletta, 2012). Activity of sGC has been shown to be regulated by post-translational mechanisms, such as phosphorylation, S-nitrosylation, and protein–protein interactions (Murthy, 2001; Rybalkin et al., 2003; Sayed et al., 2007; Zhou et al., 2008; Mayer et al., 2009; Su, 2014).
PDEs, which hydrolyze cyclic nucleotides cAMP and cGMP to the inactive noncyclic 5′-AMP and 5′-GMP, respectively, are classified based on their regulatory and catalytic properties into 11 families. PDE5, -6, and -9 are specific for cGMP, whereas PDE1, -2, and -3 hydrolyze both cAMP and cGMP, albeit with different affinity (Sharma et al., 2006; Ahmad et al., 2012). PDE5 is a dimer with cGMP-binding sites in its regulatory N-terminal and catalytic C-terminal domains (Soderling and Beavo, 2000; Francis et al., 2001; Rybalkin et al., 2003; Corbin et al., 2009). PDE5 activity is stimulated via binding of cGMP and cGMP-dependent protein kinase (PKG)–mediated phosphorylation of PDE5 at a conserved serine residue in the N-terminal region (Francis et al., 2001; Rybalkin et al., 2003). PDE5 expression is abundant in smooth muscle and plays an important role in the regulation of cGMP levels and smooth muscle relaxation (Murthy, 2001; Mahavadi et al., 2013). PDE1 (PDE1A and PDE1B) hydrolyzes cGMP with higher affinity and is regulated by Ca2+/calmodulin (Sharma et al., 2006). Both PDE2 and PDE3 (PDE3A and PDE3B) hydrolyze cAMP and cGMP with similar affinity, but due to a higher catalytic rate of PDE3 for cAMP than cGMP, cGMP acts as a competitive inhibitor, and PDE3 is known as cGMP-inhibited cAMP phosphodiesterase (Ahmad et al., 2012).
Recent studies have determined the changes in the expression and/or activity of adenylyl cyclase 5/6 (AC5/6) and cAMP-specific PDE4D5 in response to inflammatory cytokines [interleukin-1β (IL-1β) and tumor necrosis factor α (TNF-α)] in vitro and TNBS (2,4,6-trinitrobenzene sulfonic acid)-induced colonic inflammation in vivo in longitudinal smooth muscle (Mahavadi et al., 2014). Inflammation induced an increase in PDE4D5 activity and a decrease in AC5/6 activity, leading to a decrease in cAMP formation and a decrease in smooth muscle relaxation. The aim of the present study was to characterize the expression and activity of sGC and cGMP-PDE isoforms by cytokines in vitro and inflammation in vivo that result in altered smooth muscle relaxation. Our results demonstrate that sGC and PDE5 were expressed in normal longitudinal smooth muscle. In response to cytokines in vitro and during inflammation, inducible nitric oxide synthase (iNOS) expression was induced and sGC activity was decreased as a result of S-nitrosylation of sGC. Concurrently, PDE1A expression was induced via the nuclear factor κB (NF-κB) pathway. The combined effects of iNOS-mediated S-nitrosylation of sGC and NF-κB–dependent PDE1 expression decreased cGMP formation and longitudinal smooth muscle relaxation during inflammation.
Materials and Methods
[125I]cGMP, [α-32P]GTP, and [3H]cGMP were obtained from PerkinElmer Life Sciences (Boston, MA); collagenase and soybean trypsin inhibitor were from Worthington Biochemical Inc. (Freehold, NJ); Western blot, chromatography material, and protein assay kit were from Bio-Rad Laboratories (Hercules, CA); and antibody to sGC β1 subunit from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to PDE1A, PDE2A, PDE3A, and PDE5A, and phospho-antibody to PDE5 (Ser92) were obtained from FabGennix Inc. (Frisco, TX); vinpocetine and MG132 (benzyl N-[(2S)-4-methyl-1-[[(2S)-4-methyl-1-[[(2S)-4-methyl-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]carbamate) were purchased from Enzo Life Sciences, Inc. (Farmingdale, NY); the S-nitrosylation detection kit was from Cayman Chemical (Ann Arbor, MI); and cGMP, Crotalus atrox snake venom, and all other chemicals were from Sigma-Aldrich (St. Louis, MO).
All animal treatments were performed according to a protocol approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University.
Induction of Colonic Inflammation and Preparation of Dispersed Colonic Smooth Muscle Cells.
Colonic inflammation in mice was induced with TNBS as described previously (Hazelgrove et al., 2009; Alkahtani et al., 2013; Al-Shboul et al., 2014; Mahavadi et al., 2014; Nalli et al., 2014). Adult male mice (C57BL/6J; 6–8 weeks old) were anesthetized, and 100 µl of TNBS [2.5% in 50% ethanol (v/v)] was instilled intrarectally; mice were euthanized 3 days after the induction of inflammation. Control mice were treated with vehicle. Body weight and stool consistency were evaluated daily. Colonic tissue from mice treated with TNBS exhibited typical histologic characteristics of colitis (Alkahtani et al., 2013; Al-Shboul et al., 2014; Mahavadi et al., 2014; Nalli et al., 2014). Distal colonic segments 2–3 cm long were obtained from control and TNBS-treated mice, and muscle cells from the longitudinal muscle layer were obtained as described previously (Murthy et al., 2002, 2003; Mahavadi et al., 2013, 2014). Briefly, colonic longitudinal muscle strips were dissected and incubated at 31°C for 30 minutes in HEPES medium containing 120 mM NaCl, 4 mM KCl, 2.6 mM KH2PO4, 0.6 mM MgCl2, 25 mM HEPES, 14 mM glucose, 2.1% Eagle’s essential amino acid mixture, 0.1% collagenase, and 0.1% soybean trypsin inhibitor. Partly digested strips were washed twice with 50 ml of enzyme-free medium, the muscle cells were allowed to disperse spontaneously for 30 minutes, and the cells were harvested by filtration through 500 μm Nitex (Tetko Inc., Briarcliff Manor, NY) and centrifuged twice at 350g for 10 minutes. For some experiments, muscle cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum until they attained confluence, and were then passaged once for use.
Expression of PDE1 by Reverse-Transcription Polymerase Chain Reaction.
Total RNA was isolated from freshly dispersed smooth muscle cells with TRIzol reagent (Invitrogen, Carlsbad, CA) and cultured longitudinal muscle cells using ULTRASPEC reagent (Biotecx Laboratories, Houston, TX), and then treated with TURBO DNase (Ambion, Carlsbad, CA). RNA was reverse-transcribed using the SuperScript II system (Life Technologies, Carlsbad, CA) containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphate, 2.5 μM random hexamers, and 200 units of reverse transcriptase in a 20-μl reaction volume. The reactions were carried out at room temperature for 10 minutes and at 42°C for 50 minutes, and terminated by heating at 70°C for 15 minutes. Two microliters of the reverse-transcribed cDNA was amplified in a final volume of 20 μl by polymerase chain reaction (PCR) in standard conditions with specific primers for PDE1A (forward primer 5′-CTAAAGATGAACTGGAGGGATCTTCGGAAC-3′ and reverse primer 5′-TGGAGAAAATGGAAGCCCTAATTCAGC-3′), PDE1B (forward primer 5′-CCTCCACCTTCACCCAGCAG-3′ and reverse primer 5′-CACTGTGGGAATCTTGAAGCGGCTGATG-3′), and PDE1C (forward primer 5′-ATGGTTGGGCTGAGCTATCCACC-3′ and reverse primer 5′-CCAGTTTGCCACTCCTGTCTTATAAAGGAGG-3′). Real-time PCR was performed on cDNA samples synthesized from total RNA isolated from cultured muscle cells with the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster city, CA) and the intercalating dye SYBR green. PCR conditions were optimized on the gradient thermal cycler on the StepOne Plus. Real-time PCR reactions were performed in triplicate. Each primer set generated only one PCR product, and the identity and integrity of these products were confirmed by electrophoresis on 1.5% agarose gel containing 0.1 μg/ml ethidium bromide and sequencing of the individual bands. The fluorescent threshold value was calculated using the StepOne Plus. Glyceraldehyde-3-phosphate dehydrogenase was selected as a reference gene. Cycle threshold (Ct) values were obtained, and the relative fold change in gene expression was calculated as 2−△△Ct (Al-Shboul et al., 2013).
Assay for sGC Activity.
sGC activity was measured by using [α-32P]GTP as substrate as described previously (Murthy, 2001). Homogenates of muscle cells were incubated for 15 minutes at 37°C in 50 mM Tris-HCl (pH 7.4), 2 mM cGMP, 0.1 mM GTP, 1 mM IBMX (3-isobutyl-1-methylxanthine), 5 mM MgCl2, 100 mM NaCl, 5 mM creatine phosphate, 50 U/ml creatine phosphokinase, and 0.5 mM [α-32P]GTP (0.2 µCi). The reaction was terminated by addition of 2% SDS, 45 mM GTP, and 1.5 mM cGMP. [32P]cGMP was separated from [32P]GTP by sequential chromatography on Dowex AG50W-4X (Bio-Rad Laboratories) and alumina columns. The results were expressed as picomoles of cGMP per milligram of protein per minute.
Assay for PDE Activity.
PDE5 and PDE1A activity was measured in immunoprecipitates of PDE5 and PDE1A as described previously (Wyatt et al., 1998; Murthy, 2001; Mahavadi et al., 2013). One-milliliter aliquots (3 × 106 cells/ml) of muscle cells were incubated with S-nitroso-N-acetylpenicillamine (SNAP) for 5 minutes. Immunoprecipitates were washed in a medium containing 50 mM Tris (pH 7.5), 200 mM NaCl, and 5 mM EDTA, and then incubated for 15 minutes at 30°C in a medium containing 100 mM Mes (2-[N-morpholino] ethanesulfonic acid; pH 7.5), 10 mM EDTA, 0.1 M Mg acetate, 0.9 mg/ml bovine serum albumin, 20 μM cGMP, and [3H]cGMP. The samples were boiled for 3 minutes, chilled for 3 minutes, and then incubated at 30°C for 10 minutes in 20 mM Tris (pH 7.5) medium containing 10 μl of C. atrox snake venom (10 μg/μl). The samples were added to DEAE-Sephacel A-25 columns (Bio-Rad Laboratories), and the radioactivity in the effluent was counted. The results were expressed as counts per minute per milligram of protein.
Phosphorylation of PDE5 by PKG.
Phosphorylation of PDE5 was measured by immunoblot analysis using phospho-specific antibody (Ser92) as described previously (Murthy, 2001). One-milliliter aliquots (3 × 106 cells/ml) of samples were incubated with different concentrations of SNAP for 5 minutes, and the reaction was terminated with an equal volume of lysis buffer and placed on ice for 30 minutes. The cell lysates were separated from the insoluble material by centrifugation at 13,000g for 15 minutes at 4°C, precleared with 40 μl of protein A-Sepharose, and incubated with antibody to PDE5A for 2 hours at 4°C, and with 40 μl of protein A-Sepharose for another hour. The immunoprecipitates were washed five times with 1 ml of wash buffer (0.5% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.4), extracted with Laemmli sample buffer, and boiled for 15 minutes, then separated on 10% SDS-PAGE followed by transfer to polyvinylidene difluoride membranes. The membranes were incubated for 12 hours with phospho-specific antibodies to PDE5A (Ser92) and then for 1 hour with a horseradish peroxidase–conjugated secondary antibody. The bands were identified by enhanced chemiluminescence.
Assay for S-Nitrosylation.
The S-nitrosylated sGC was detected using a detection assay kit (Cayman Chemical) as described previously (Mahavadi et al., 2014). In brief, sGC was immunoprecipitated with anti-sGC conjugated with protein A/G plus agarose beads. Immunoprecipitated sGC was released by boiling the beads for 5 minutes, the free thiols were blocked, then S-NO bonds were cleaved. The proteins were labeled with biotin by biotinylation of the newly formed SH groups. The labeled proteins were then analyzed by SDS-PAGE and transferred to polyvinylidene difluoride membranes for Western blot.
Radioimmunoassay for cGMP.
Cyclic GMP production was measured by radioimmunoassay as described previously (Murthy, 2001; Mahavadi et al., 2013). In brief, muscle cells (3 × 106 cells) were treated with SNAP for 5 minutes, and the reaction was terminated with 10% trichloroacetic acid. After extraction with water-saturated diethyl ether, the lyophilized aqueous phase was reconstituted in 500 μl of 50 mM Na acetate (pH 6.2). The samples were acetylated with triethylamine/acetic anhydride (2:1) for 30 minutes, and cGMP was measured in duplicate using 100-μl aliquots. The results were expressed as picomoles per milligram of protein.
Measurement of Contraction and Relaxation in Smooth Muscle Cells.
Contraction in freshly dispersed colonic longitudinal muscle cells was determined by scanning micrometry (Murthy and Makhlouf, 1998; Murthy, 2001; Mahavadi et al., 2013, 2014). An aliquot (0.4 ml) of cells containing approximately 104 cells/ml was treated with 100 µl of medium containing different concentrations of SNAP for 5 minutes followed by carbachol (1 µM) for 30 seconds, and then the reaction was terminated with 1% acrolein at a final concentration of 0.1%. The resting cell length was determined in control experiments in which muscle cells were incubated with 100 μl of 0.1% bovine serum albumin without SNAP or carbachol. The mean lengths of 50 muscle cells treated with various agonists were measured by scanning micrometry and were compared with the mean lengths of untreated cells. The contractile response to carbachol was expressed as the percent decrease in mean cell length from control cell length. Relaxation was measured as a decrease in response to carbachol in the presence of SNAP. Relaxation was expressed as percent decrease in contractile response to carbachol.
Statistical Analysis.
All values are expressed as means ± S.E.M.; n represents the number of animal studies. Regression analysis was performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). Statistical analysis was performed by unpaired t test, and P < 0.05 was considered statistically significant.
Results
Regulation of cGMP Levels in Mouse Longitudinal Smooth Muscle Cells.
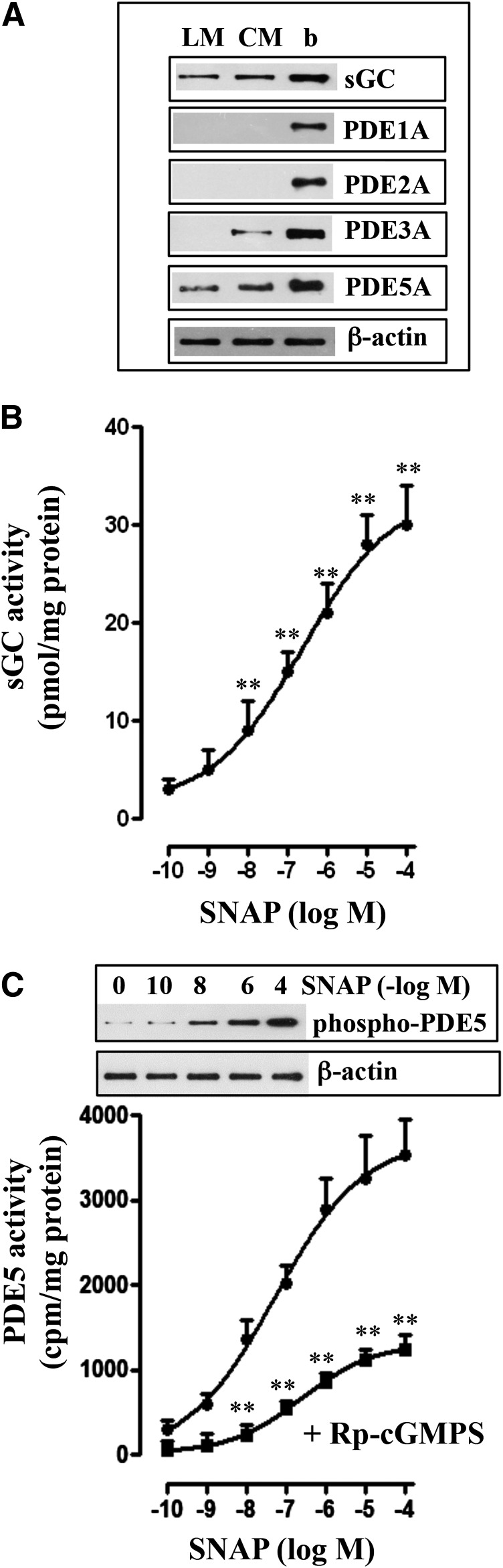
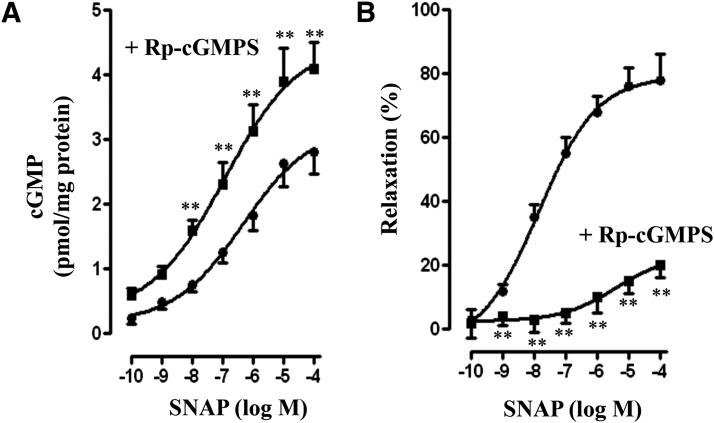
Studies in circular smooth muscle have shown the presence of sGC and PDE5, and cGMP levels in response to NO donors are regulated by stimulation of both sGC and PDE5 activities. Western blot analysis in the dispersed smooth muscle cells of colonic longitudinal muscle detected the presence of sGC and cGMP-hydrolyzing PDE5A, but not PDE1A, PDE2A, or PDE3A (Fig. 1A). In contrast, as shown previously, circular muscle cells expressed both PDE3A and PDE5A isoforms (Fig. 1A) (Murthy, 2001; Murthy et al., 2002; Mahavadi et al., 2014). Treatment of muscle cells with SNAP stimulated sGC and PDE5 activity in a concentration-dependent manner (Fig. 1, B and C). SNAP-stimulated PDE5 activity was accompanied by an increase in phosphorylation of PDE5A at Ser92 in a concentration-dependent manner (Fig. 1C). SNAP-stimulated PDE5 activity was attenuated in the presence of a selective PKG inhibitor, guanosine-3′,5′-cyclic monophosphorothioate (Rp-cGMPS) (Fig. 1C). Previous studies have shown that PDE5 activity is stimulated via binding of cGMP and PKG-mediated phosphorylation of PDE5 (Francis et al., 2001; Murthy, 2001; Rybalkin et al., 2003). Consistent with the activation of sGC, SNAP stimulated cGMP levels in a concentration-dependent manner, and the increase was augmented in the presence of Rp-cGMPS (Fig. 2A). The increase in cGMP levels could be due to attenuation of PDE5 activity in the presence of Rp-cGMPS. SNAP also caused a concentration-dependent inhibition of carbachol (1 µM)-induced contraction (i.e., relaxation) (Fig. 2B). SNAP-induced relaxation was inhibited by Rp-cGMPS, suggesting relaxation was mediated via PKG (Fig. 2B). Previous studies have shown that muscle relaxation in response to PKG activating agents results from inhibition of the initial increase in cytosolic Ca2+ leading to inhibition of myosin light-chain kinase and/or stimulation of MLCP activity (Murthy and Makhlouf, 1998; Murthy et al., 2003; Murthy, 2006).
Fig. 1.
Expression of sGC and cGMP-hydrolyzing PDEs in circular and longitudinal smooth muscle and SNAP-induced stimulation of sGC and PDE5 activity. (A) Expression of sGC β1 subunit and PDE isoforms were analyzed in freshly prepared dispersed circular (CM) and longitudinal (LM) smooth muscle of colon and brain (b). Lysates containing equal amounts of total proteins were separated with SDS-PAGE, and expression of sGC β1 subunit, PDE1A, PDE2A, PDE3A, and PDE5A was analyzed using selective antibody. Membranes were reblotted to measure β-actin. Protein bands were visualized with enhanced chemiluminescence. (B) Longitudinal muscle cells were treated with different concentrations of SNAP for 5 minutes. sGC activity was measured as described in Materials and Methods. Results are expressed as pmol cGMP/mg protein above basal levels (2.82 pmol/mg protein). Values are the means ± S.E.M. of five experiments. **P < 0.01, significant increase in sGC activity. (C) Longitudinal muscle cells were treated with different concentrations of SNAP in the presence or absence of PKG inhibitor, Rp-cGMPS (10 µM), for 5 minutes, and PDE5 activity was measured in PDE5A immunoprecipitates as described in Materials and Methods and expressed as cpm/mg protein above basal values (295 ± 42 cpm/mg protein). Values are the means ± S.E.M. of five experiments. **P < 0.01, significant inhibition of SNAP-stimulated PDE5 activity by Rp-cGMPS. Inset: Representative immunoblot of four different experiments. Longitudinal muscle cells were treated with different concentrations of SNAP, and PDE5A immunoprecipitates were separated on SDS-PAGE and analyzed with phospho-specific (Ser92) antibody in the Western blot.
Fig. 2.
SNAP-induced cGMP generation and muscle relaxation. (A) Longitudinal muscle cells were treated with different concentrations of SNAP in the presence or absence of Rp-cGMPS (10 µM) for 5 minutes. cGMP was measured in the presence of 100 µM IBMX by radioimmunoassay and expressed as pmol/mg protein above basal levels (0.22 ± 0.04 pmol/mg protein). Values are the means ± S.E.M. of four experiments. **P < 0.01, significant increase in SNAP-induced cGMP generation by Rp-cGMPS. (B) Longitudinal muscle cells isolated from colon were treated with different concentrations of SNAP in the presence or absence of Rp-cGMPS (10 µM) for 5 minutes followed by carbachol for 30 seconds to measure initial Ca2+-dependent contraction. Smooth muscle cell contraction was measured by scanning micrometry, and relaxation was expressed as the percent inhibition of carbachol-induced contraction (basal cell length: 95 ± 4 µm; carbachol-induced contraction: 31 ± 3% decrease in cell length). Values are the means ± S.E.M. of five to six experiments. **P < 0.01 significant inhibition in SNAP-induced relaxation by Rp-cGMPS.
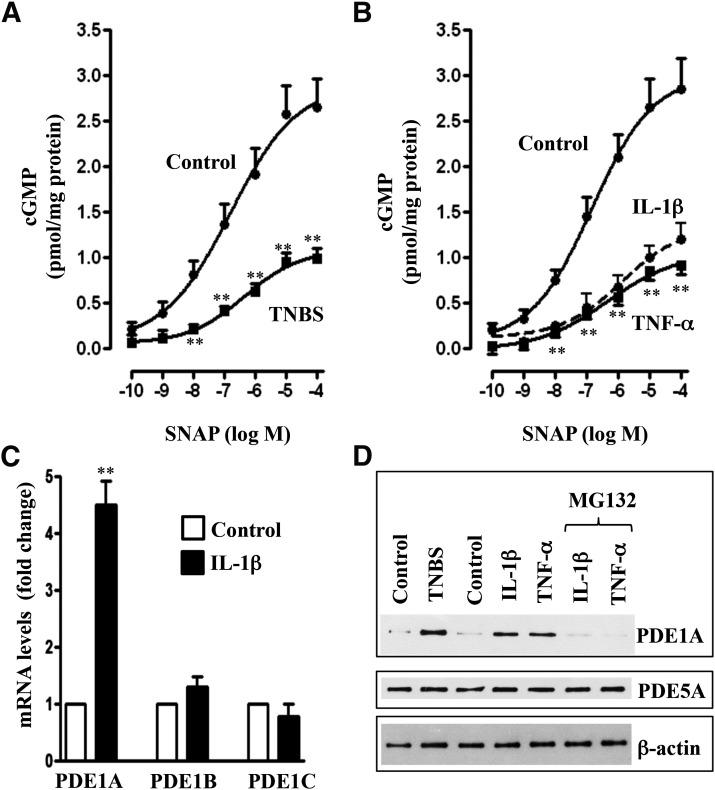
Inhibition of SNAP-Stimulated sGC Activity in Colonic Longitudinal Muscle Cells Isolated from TNBS-Treated Mice or from Control Muscle Strips Treated with IL-1β or TNF-α.
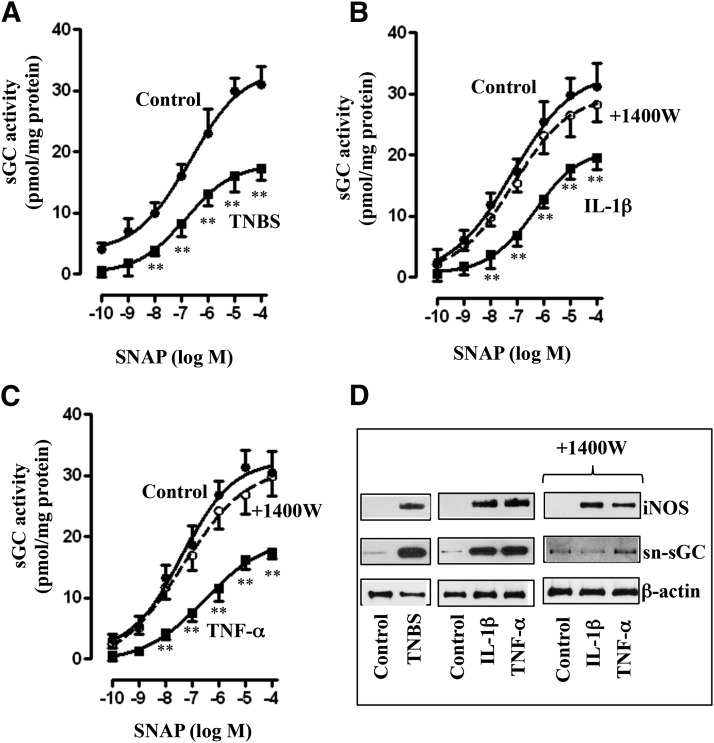
SNAP stimulated sGC activity in colonic longitudinal muscle cells isolated from control and TNBS-treated mice. Increase in sGC activity was attenuated, and the concentration-response curve was shifted to the right in muscle cells isolated from TNBS-treated mice (Fig. 3A). Similarly, SNAP-stimulated sGC activity was attenuated and the concentration-response curve was shifted to the right in muscle cells isolated from muscle strips treated for 48 hours with IL-1β (10 ng/ml) or TNF-α (1 nM) (Fig. 3, B and C).
Fig. 3.
Suppression of SNAP-induced sGC activity via iNOS-mediated nitrosylation of sGC. Longitudinal muscle cells isolated from colons of control and TNBS-treated mice (A) or from muscle strips cultured in the presence of IL-1β (10 ng/ml) (B) or TNF-α (1 nM) (C) for 48 hours were treated with different concentrations of SNAP, and sGC activity was measured as described in Materials and Methods. In some experiments, muscle strips were cultured in the presence of IL-1β (10 ng/ml) (B) or TNF-α (1 nM) (C) plus iNOS inhibitor 1400W (10 µM) for 48 hours. Basal sGC activity was not significantly different between control and TNBS-treated mice (2.61 ± 0.32 versus 2.54 ± 0.36 pmol/mg protein) or between control and cytokine-treated muscle strips (2.48 ± 0.36 versus 2.62 ± 0.36 pmol/mg protein). Values are the means ± S.E.M. of five experiments. **P < 0.01, significant inhibition of SNAP-induced sGC activity. (D) Representative immunoblot of four to five different experiments. Longitudinal muscle cells isolated from colons of control and TNBS-treated mice or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours. In some experiments, muscle strips were cultured with IL-1β (10 ng/ml) or TNF-α (1 nM) in the presence of iNOS inhibitor 1400W (10 µM) for 48 hours. Lysates were used to measure iNOS expression and S-nitrosylation of sGC (sn-sGC) as described in Materials and Methods.
As shown in previous studies (Kuemmerle, 1998; Mahavadi et al., 2014; Nalli et al., 2014), NOS-III (iNOS) was induced in colonic longitudinal smooth muscle isolated from the colon of TNBS-treated mice or control muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 3D). sGC was S-nitrosylated in colonic longitudinal muscle cells isolated from TNBS-treated mice or from muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 3D). S-Nitrosylation of sGC, but not iNOS expression, was blocked in cells isolated from muscle strips treated with IL-1β or TNF-α in the presence of iNOS inhibitor 1400W [N-([3-(aminomethyl)phenyl]methyl)ethanimidamide dihydrochloride], suggesting that S-nitrosylation was mediated by iNOS (Fig. 3D). It has been suggested that S-nitrosylation of cysteine residues in sGC causes inhibition of its activity (Mayer et al., 1998, 2009; Sayed et al., 2007, 2008). Consistent with this notion, sGC activity was restored to control levels in cells isolated from muscle strips treated with IL-1β or TNF-α in the presence of 1400W (Fig. 3, B and C).
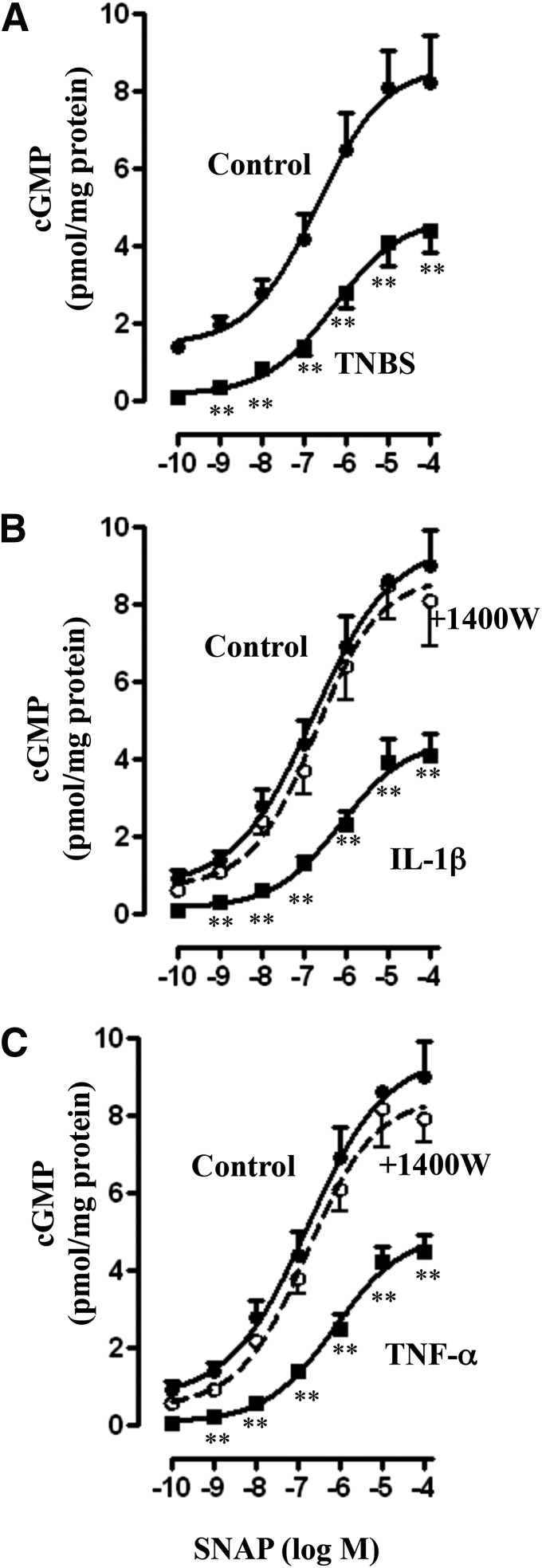
Consistent with the inhibition of sGC activity, SNAP-stimulated cGMP formation, measured in the presence of IBMX so as to eliminate the confounding effect of changes in PDE activity, was also inhibited in colonic longitudinal smooth muscle isolated from the colon of TNBS-treated mice or control muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 4, A–C). Treatment of muscle strips with IL-1β or TNF-α in the presence of 1400W blocked the inhibitory effect of IL-1β or TNF-α on SNAP-stimulated cGMP formation (Fig. 4, B and C), implying the inhibition of cGMP by proinflammatory cytokines is due to inhibition of sGC activity.
Fig. 4.
Suppression of SNAP-induced cGMP levels via iNOS. Longitudinal muscle cells isolated from colons of control and TNBS-treated mice (A) or from muscle strips cultured in the presence of IL-1β (10 ng/ml) (B) or TNF-α (1 nM) (C) for 48 hours were treated with different concentrations of SNAP. In some experiments, muscle strips were cultured with IL-1β (10 ng/ml) or TNF-α (1 nM) in the presence of iNOS inhibitor 1400W (10 µM) for 48 hours. cGMP levels were measured in the presence of 100 µM IBMX as described in Materials and Methods. Basal cGMP levels were not significantly different between control and TNBS-treated mice (0.24 ± 0.05 versus 0.21 ± 0.04 pmol/mg protein) or between control and cytokine-treated muscle strips (0.22 ± 0.04 versus 0.26 ± 0.05 pmol/mg protein). Values are the means ± S.E.M. of five experiments. **P < 0.01, significant inhibition in SNAP-induced cGMP formation.
Upregulation of PDE1A Expression and Activity in Colonic Longitudinal Muscle Cells Isolated from TNBS-Treated Mice or from Control Muscle Strips Treated with IL-1β or TNF-α.
SNAP-stimulated cGMP formation, measured in the absence of IBMX, was inhibited in colonic longitudinal smooth muscle cells isolated from the colon of TNBS-treated mice or control muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 5, A and B). Real-time reverse-transcription PCR analysis revealed selective upregulation of PDE1A, but not PDE1B, and PDE1C in colonic longitudinal smooth muscle cells treated with IL-1β for 48 hours (Fig. 5C). Western blot analysis also showed upregulation of PDE1A, but not PDE5A expression, in muscle cells isolated from the colon of TNBS-treated mice or control muscle strips treated with IL-1β or TNF-α for 48 hours (Fig. 5D). Expression of PDE1A was blocked in the presence of a NF-κB inhibitor, MG132 (10 µM) (Fig. 5D). Previous studies have shown that inflammation in vivo or treatment of muscle strips with IL-1β or TNF-α caused activation of NF-κB via the canonical pathway (Hu et al., 2008; Nalli et al., 2014).
Fig. 5.
Upregulation of PDE1A expression via NF-κB and suppression of SNAP-induced cGMP levels. Longitudinal muscle cells isolated from colons of control and TNBS-treated mice (A) or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) (B) for 48 hours were treated with different concentrations of SNAP. Cyclic GMP levels were measured in the absence of IBMX as described in Materials and Methods. Basal cGMP levels were not significantly different between control and TNBS-treated mice or between control and cytokine-treated muscle strips. Values are the means ± S.E.M. of five experiments. **P < 0.01, significant inhibition of SNAP-induced cGMP formation. (C) Longitudinal muscle cells in culture were treated with IL-1β for 48 hours, and expression of PDE1 isoforms was measured by real-time PCR as described in Materials and Methods. Values are the means ± S.E.M. of five experiments. **P < 0.01, significant increase in PDE1A expression. (D) Representative immunoblot of four to five different experiments. Longitudinal muscle cells were isolated from colons of control and TNBS-treated mice or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours. In some experiments, muscle strips were cultured with IL-1β (10 ng/ml) or TNF-α (1 nM) in the presence of NF-κB inhibitor MG132 (10 µM) for 48 hours. Expression of PDE1A and PDE5 was analyzed by Western blot.
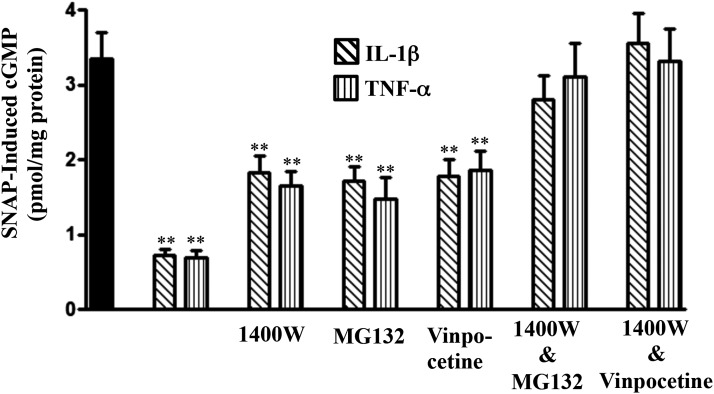
Although 1400W completely reversed the inhibition of sGC activity and inhibition of cGMP formation (measured in the presence of IBMX) (Fig. 4, B and C), it only partly reversed the inhibition of cGMP formation measured in the absence of IBMX, suggesting the residual inhibition of cGMP formation could be due to augmentation of cGMP-hydrolyzing PDE activity by proinflammatory cytokines (Fig. 6). Consistent with the blockade of PDE1A expression, vinpocetine, a selective PDE1 inhibitor, or MG132 partly reversed the inhibition of cGMP and completely reversed the inhibition in combination with 1400W (Fig. 6).
Fig. 6.
Suppression of SNAP-induced cGMP formation by proinflammatory cytokines via iNOS- and NF-κB–mediated PDE1A activity. Longitudinal muscle cells were isolated from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours. In some experiments, muscle strips were cultured with IL-1β (10 ng/ml) or TNF-α (1 nM) in the presence of NF-κB inhibitor MG132 (10 µM), iNOS inhibitor 1400W (10 µM), PDE1 inhibitor vinpocetine (50 µM), or 1400W in combination with MG132 or vinpocetine for 48 hours. cGMP levels in response to SNAP (10 µM) were measured in the absence of IBMX by radioimmunoassay. Values are the means ± S.E.M. of five experiments. **P < 0.01, significant inhibition compared with control SNAP-induced cGMP formation (solid bar).
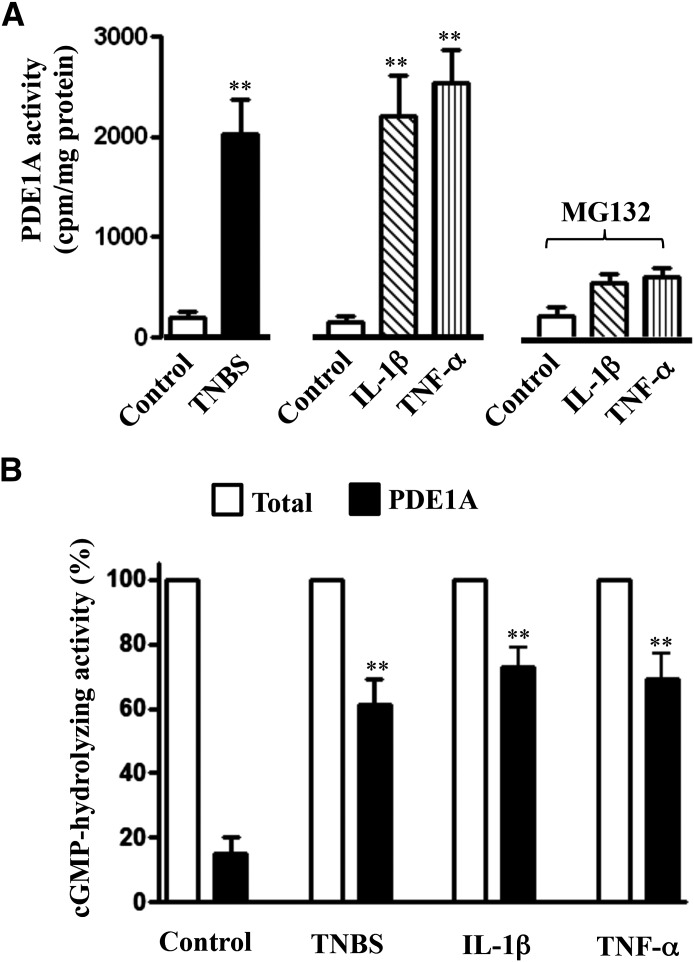
PDE1A activity was also augmented in colonic longitudinal smooth muscle isolated from the colons of TNBS-treated mice or control muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 7A). Increase in PDE1A activity in response to cytokines was blocked in the presence of a NF-κB inhibitor, MG132 (10 µM), and the effect of MG132 on PDE1A activity was consistent with its effect on PDE1A expression (Fig. 5D). PDE1A contributed to nearly 60–70% of total hydrolyzing PDE activity in colonic muscle cells isolated from the colons of TNBS-treated mice or control muscle strips treated for 48 hours with IL-1β or TNF-α compared with ∼16% in muscle cells from control mice (Fig. 7B). Together, these results indicate that PDE1A induction during inflammation contributes to the inhibition of cGMP formation. SNAP-stimulated PDE5 activity, in contrast, was not significantly different in colonic muscle cells of control (3986 ± 452 cpm/mg protein) and TNBS-treated mice (4145 ± 567 cpm/mg protein) or from muscle cells isolated from strips treated with IL-1β (4278 ± 510 cpm/mg protein) or TNF-α (4012 ± 602 cpm/mg protein) compared with control (3881 ± 595 cpm/mg protein).
Fig. 7.
Increase in PDE1A activity by inflammation. (A) Longitudinal muscle cells isolated from colons of control and TNBS-treated mice or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours were used to measure PDE1A activity as described in Materials and Methods. In some experiments, muscle strips were cultured with IL-1β (10 ng/ml) or TNF-α (1 nM) in the presence of NF-κB inhibitor MG132 (10 µM) for 48 hours. Values are the means ± S.E.M. of five experiments. **P < 0.01, significant increase in PDE1A activity compared with control. (B) Longitudinal muscle cells isolated from colons of control and TNBS-treated mice or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours were used to measure cGMP-hydrolyzing activity in the presence or absence of PDE1 inhibitor vinpocetine (50 µM). Total cGMP-hydrolyzing activity reflects the activity in the absence of vinpocetine. PDE1A activity was calculated as the percentage of total cGMP-hydrolyzing activity that was inhibited by vinpocetine. Values are the means ± S.E.M. of five experiments. **P < 0.01, significant increase in PDE1A activity compared with control.
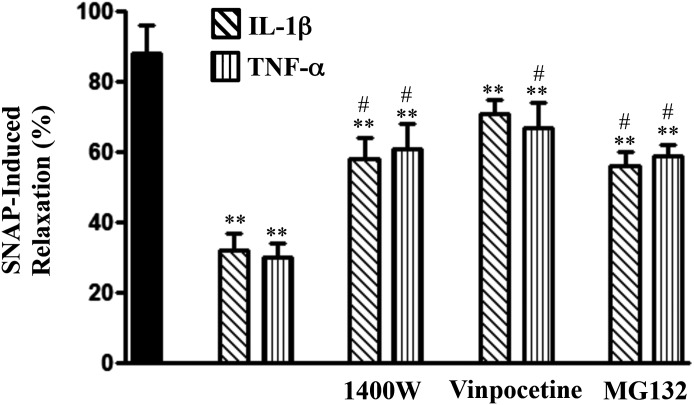
Inhibition of SNAP-Stimulated Relaxation in Colonic Longitudinal Muscle Cells Isolated from TNBS-Treated Mice or from Control Muscle Strips Treated with IL-1β or TNF-α.
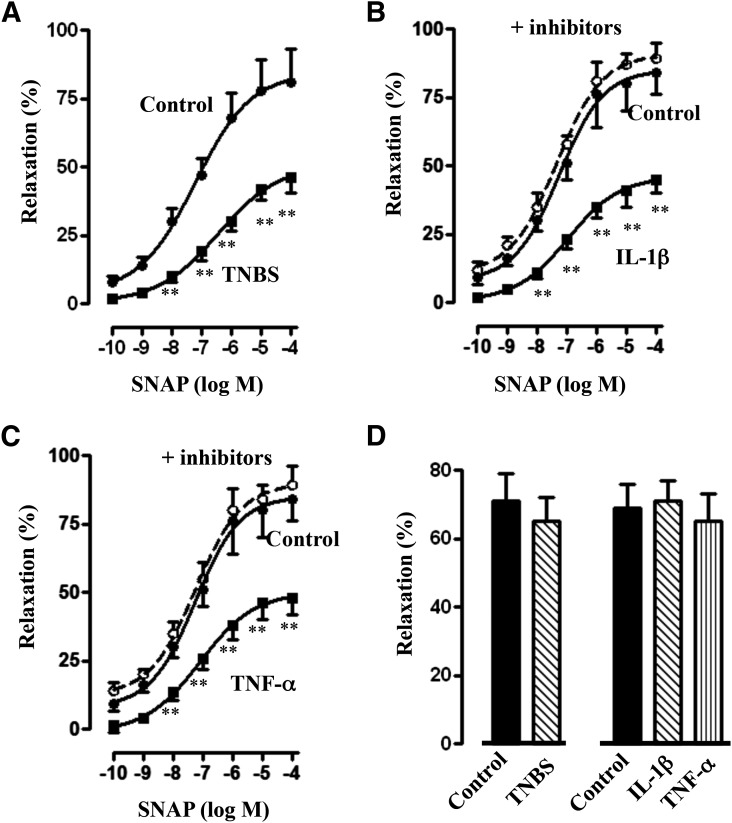
SNAP-stimulated relaxation, measured in the absence of IBMX, was inhibited in longitudinal muscle cells isolated from control muscle strips treated for 48 hours with IL-1β or TNF-α (Fig. 8). Maximal relaxation induced by 10 μM SNAP was inhibited by 60–65% (P < 0.05; n = 5–6). Inhibition of relaxation induced by treatment with IL-1β or TNF-α was partly reversed in the presence of 1400W, vinpocetine, or MG132 (Fig. 8). SNAP induced relaxation in colonic longitudinal muscle cells isolated from control and TNBS-treated mice. Relaxation, however, was attenuated and the concentration-response curve was shifted to the right in muscle cells isolated from TNBS-treated mice (Fig. 9A). Similarly, SNAP-induced relaxation was attenuated and the concentration-response curve was shifted to the right in muscle cells isolated from muscle strips treated for 48 hours with IL-1β (10 ng/ml) or TNF-α (1 nM) (Fig. 9, B and C). Inhibition of relaxation by IL-1β or TNF-α was blocked when the muscle strips were treated with IL-1β or TNF-α in the presence of both 1400W and vinpocetine (Fig. 9, B and C). The inhibition of relaxation and partial reversal by 1400W, vinpocetine, or MG132 and complete reversal by a combination of both 1400W and vinpocetine paralleled the changes in cGMP formation. Control studies showed that relaxation in longitudinal muscle cells isolated from the colons of TNBS-treated mice or from control muscle strips treated for 48 hours with IL-1β or TNF-α in response to a PDE-resistant analog of cGMP, 8-Bromo-cGMP, was not significantly different (Fig. 9D), suggesting that inhibition of SNAP-induced relaxation by cytokines or during inflammation was due to a decrease in cGMP levels, but not due to inhibition of the PKG effect on target proteins.
Fig. 8.
Suppression of SNAP-induced relaxation by proinflammatory cytokines and reversal of inhibition by blockade of iNOS, NF-κB, or PDE1 activity. Longitudinal muscle cells were isolated from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours. In some experiments, muscle strips were cultured with IL-1β or TNF-α in the presence of 1400W (10 µM), vinpocetine (50 µM), or MG132 (10 µM) for 48 hours. Cells were treated with 10 µM SNAP for 5 minutes followed by carbachol for 30 seconds to measure initial Ca2+-dependent contraction. Smooth muscle cell contraction was measured by scanning micrometry, and relaxation was expressed as the percent inhibition of carbachol-induced contraction. Values are the means ± S.E.M. of five to six experiments. **Significant inhibition in SNAP-induced relaxation by IL-1β or TNF-α compared with control SNAP-induced relaxation (solid bar). #P < 0.05, significant reversal of inhibition by 1400W, vinpocetine, or MG132.
Fig. 9.
Suppression of SNAP-induced relaxation by inflammation and complete reversal of inhibition by blockade of iNOS and PDE1 activity. Longitudinal muscle cells isolated from colons of control and TNBS-treated mice (A) or from muscle strips cultured in the presence of IL-1β (10 ng/ml) (B) or TNF-α (1 nM) (C) for 48 hours were treated with different concentrations of SNAP for 5 minutes and carbachol for 30 seconds to measure initial Ca2+-dependent contraction. In some experiments, muscle strips were cultured with IL-1β or TNF-α in the presence of both iNOS (1400W, 10 µM) and PDE1 (vinpocetine, 50 µM) inhibitors. Smooth muscle cell contraction was measured by scanning micrometry, and relaxation was expressed as the percent inhibition of carbachol-induced contraction. Values are the means ± S.E.M. of five to six experiments. **P < 0.01, significant inhibition of SNAP-induced relaxation. (D) Longitudinal muscle cells isolated from colons of control and TNBS-treated mice or from muscle strips cultured in the presence of IL-1β (10 ng/ml) or TNF-α (1 nM) for 48 hours were treated with PDE-resistant 8-Bromo-cGMP (10 µM) for 5 minutes and carbachol (1 µM) for 30 seconds to measure initial Ca2+-dependent contraction. Smooth muscle cell contraction was measured by scanning micrometry, and relaxation was expressed as the percent inhibition of carbachol-induced contraction. Values are the means ± S.E.M. of four experiments.
Discussion
Inhibitory transmitters induce relaxation of gastrointestinal smooth muscle by stimulating the production of cAMP and cGMP, leading to the activation of cAMP-dependent protein kinase and PKG respectively (Murthy, 2006). The levels of cAMP and cGMP are regulated by the combined activities of cyclases (AC5/6 and sGC) and PDEs (Soderling and Beavo, 2000; Francis et al., 2001; Ahmad et al., 2012). The present study characterized regulation of cGMP levels and muscle relaxation in colonic longitudinal smooth muscle during inflammation in vivo and in response to proinflammatory cytokines in vitro. We have demonstrated the expression and activities of sGC and cGMP-hydrolyzing PDE5 and PDE1A and smooth muscle relaxation in colonic longitudinal muscle cells isolated from TNBS-treated mice and from colonic muscle strips cultured for 48 hours with IL-1β or TNF-α. We showed that 1) cGMP levels in response to NO donor are regulated by the stimulation of sGC and PDE5 activity, and 2) during inflammation in vivo and in response to proinflammatory cytokines in vitro, cGMP formation and muscle relaxation are inhibited. The decrease in cGMP levels reflected the combined effect of iNOS-mediated S-nitrosylation of sGC and inhibition of its activity, and upregulation of PDE1A expression and increase in cGMP hydrolysis (Fig. 10). As shown in previous studies, relaxation in response to cAMP-elevating agents (e.g., forskolin) was also inhibited during inflammation in vivo and in response to proinflammatory cytokines in vitro (Mahavadi et al., 2014). Inhibition of relaxation in response to forskolin was due to inhibition of AC5/6 activity via iNOS-mediated S-nitrosylation, and phosphoinositide 3-kinase– and extracellular signal-regulated kinase 1/2–mediated phosphorylation, and stimulation of cAMP-specific PDE4D5 activity. The combined inhibition of AC5/6 activity and stimulation of PDE4D5 decreased cAMP formation and smooth muscle relaxation.
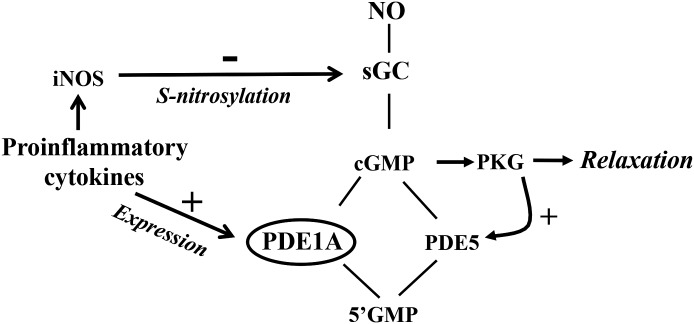
Fig. 10.
Schematic diagram demonstrating the effects of proinflammatory cytokines on pathways that regulate cGMP levels in colonic longitudinal smooth muscle cells. In longitudinal smooth muscle, cGMP levels are regulated by the activities of sGC and PDE5; the activity of the latter is stimulated via PKG-mediated phosphorylation. Proinflammatory cytokines inhibited sGC activity via iNOS-mediated S-nitrosylation, induced PDE1A expression via NF-κB, and stimulated cGMP hydrolysis, leading to suppression of cGMP formation and a decrease in muscle relaxation.
Proinflammatory cytokines acting via transcription factors and regulatory kinases induce changes in the expression and activity of various signaling molecules that regulate smooth muscle contraction and relaxation (Akiho et al., 2002; Shi et al., 2003; Cao et al., 2004; Jin et al., 2004; Salinthone et al., 2004; Singer et al., 2004; Shi and Sarna, 2005; Zhao et al., 2005, 2006; Khan and Collins, 2006; Ohama et al., 2007; Ihara et al., 2012; Shea-Donohue et al., 2012; Yang et al., 2013). Previous studies have identified that signaling pathways that mediate initial and sustained contraction in intestinal longitudinal muscle are distinct from those in circular muscle (Grider and Makhlouf, 1988; Murthy, 2006). Exposure to proinflammatory cytokines leads to hypercontractility of longitudinal and hypocontractility of circular muscle, and the differential effects of cytokines are due to differences in the signaling targets.
Several studies have examined the changes in the signaling mechanisms that lead to hypocontraction of circular muscle and hypercontraction of longitudinal muscle (Martinolle et al., 1997; Akiho et al., 2002; Ohama et al., 2003, 2007; Shi et al., 2003; Cao et al., 2004; Jin et al., 2004; Shi and Sarna, 2005; Zhao et al., 2005, 2006; Khan and Collins, 2006). Our previous studies have shown that exposure of circular muscle to proinflammatory cytokines induces upregulation of RGS4 expression via NF-κB and downregulation of CPI-17 expression, an endogenous inhibitor of MLCP, which leads to a decrease in contraction, whereas exposure of longitudinal muscle induces an increase in myosin light-chain kinase activity via NF-κB–dependent activation of the 5′-adenosine monophosphate–activated kinase/cAMP-dependent protein kinase pathway and upregulation of LARG (leukemia-associated RhoGEF) expression and the RhoA/Rho kinase pathway via jun kinase, which leads to an increase in contraction (Hu et al., 2007, 2008, 2009; Alshboul et al., 2014; Nalli et al., 2014). The disparate changes in the signaling targets induced by cytokines in circular and longitudinal muscle disrupt the functional coordination of the two muscle layers.
The present study also provides evidence for specific changes in the activity of sGC and cGMP-hydrolyzing PDEs that lead to a decrease in cGMP levels and muscle relaxation. In normal muscle, cGMP levels are regulated by the balance in the activity of sGC and cGMP-specific PDE5. Exposure of longitudinal muscle to inflammation in vivo or proinflammatory cytokines in vitro caused S-nitrosylation of sGC. The resultant decrease in sGC activity caused a decrease in cGMP formation and smooth muscle relaxation. An iNOS inhibitor, 1400W, completely reversed the effect of cytokines on S-nitrosylation of sGC and its activity, suggesting that the effect is mediated by iNOS. In contrast, inhibition of cGMP levels and muscle relaxation, measured by the absence of IBMX, were partly reversed by 1400W or PDE1 inhibitor vinpocetine and completely reversed in the presence of both inhibitors. Control studies demonstrated that relaxation in response to PDE-resistant cGMP analog in muscle cells isolated from the colon of TNBS-treated mice or from control muscle strips treated for 48 hours with IL-1β or TNF-α was similar compared with control. These results suggest that the upregulation of PDE1A activity is responsible, in part, to the decreased relaxation in response to cytokines. Exposure of longitudinal muscle to inflammation in vivo or proinflammatory cytokines in vitro caused upregulation of PDE1A, but not PDE1B and PDE1C expression. The contribution of PDE1A to total cGMP-hydrolyzing activity was increased during inflammation. PDE5 expression and activity were not altered in TNBS-treated mice and muscle strips exposed to either cytokine. PDE1A hydrolyzes cGMP with much higher affinity than cAMP, and the activity of PDE1 family members, including PDE1A, is stimulated by Ca2+/calmodulin, albeit with different affinities. PDE1A has higher affinity for calmodulin and is stimulated at much lower concentrations of Ca2+ than the other isozymes (Sharma et al., 2006). Induction of PDE1A activity leading to a decrease in cGMP levels in response to cytokines could also contribute to the hypercontractile response of longitudinal muscle during inflammation. The cytokine-activated signal transduction pathway leading to PDE1A expression is characterized by activation of NF-κB. Inhibition of the NF-κB pathway suppressed cytokine-mediated upregulation of PDE1A. Previous studies have shown that proinflammatory cytokines (IL-1β or TNF-α) cause activation of NF-κB via the canonical pathway involving IκBα kinase–mediated phosphorylation of IκBα, leading to degradation of IκBα and nuclear translocation of p65/p50 subunits.
In conclusion, our results demonstrate that, during inflammation in colonic longitudinal muscle, cGMP levels and smooth muscle relaxation are inhibited by the combined effects of iNOS-mediated S-nitrosylation of sGC and NF-κB–dependent PDE1A expression. The effect of TNBS-induced inflammation on sGC nitrosylation and PDE1A expression closely mimicked those elicited by exposure to IL-1β or TNF-α.
Abbreviations
- 1400W
N-([3-(aminomethyl)phenyl]methyl)ethanimidamide dihydrochloride
- AC5/6
adenylyl cyclase 5/6
- IBMX
3-isobutyl-1-methylxanthine
- IL-1β
interleukin-1β
- iNOS
inducible nitric oxide synthase
- Mes
2-[N-morpholino] ethanesulfonic acid
- MG132
benzyl N-[(2S)-4-methyl-1-[[(2S)-4-methyl-1-[[(2S)-4-methyl-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]amino]-1-oxopentan-2-yl]carbamate
- MLCP
myosin light-chain phosphatase
- NF-κB
nuclear factor κB
- NO
nitric oxide
- PCR
polymerase chain reaction
- PDE
phosphodiesterase
- PKG
cGMP-dependent protein kinase
- Rp-cGMPS
guanosine-3′,5′-cyclic monophosphorothioate, Rp-isomer
- sGC
soluble guanylyl cyclase
- SNAP
S-nitroso-N-acetylpenicillamine
- TNBS
2,4,6-trinitrobenzene sulfonic acid
- TNF-α
tumor necrosis factor α
Authorship Contributions
Participated in research design: Grider, Murthy.
Conducted experiments: Rajagopal, Mahavadi, Nalli, Kumar, Hu, Bhattacharya.
Contributed new reagents or analytic tools: Grider.
Performed data analysis: Rajagopal, Murthy.
Wrote or contributed to the writing of the manuscript: Rajagopal, Hu, Grider, Murthy.
Footnotes
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK15564 (to K.S.M.)]. The authors declare no conflicts of interest.
References
- Ahmad F, Degerman E, Manganiello VC. (2012) Cyclic nucleotide phosphodiesterase 3 signaling complexes. Horm Metab Res 44:776–785. [DOI] [PubMed] [Google Scholar]
- Akiho H, Blennerhassett P, Deng Y, Collins SM. (2002) Role of IL-4, IL-13, and STAT6 in inflammation-induced hypercontractility of murine smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 282:G226–G232. [DOI] [PubMed] [Google Scholar]
- Alkahtani R, Mahavadi S, Al-Shboul O, Alsharari S, Grider JR, Murthy KS. (2013) Changes in the expression of smooth muscle contractile proteins in TNBS- and DSS-induced colitis in mice. Inflammation 36:1304–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shboul O, Mahavadi S, Sriwai W, Grider JR, Murthy KS. (2013) Differential expression of multidrug resistance protein 5 and phosphodiesterase 5 and regulation of cGMP levels in phasic and tonic smooth muscle. Am J Physiol Gastrointest Liver Physiol 305:G314–G324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shboul O, Nalli AD, Kumar DP, Zhou R, Mahavadi S, Kuemmerle JF, Grider JR, Murthy KS. (2014) Jun kinase-induced overexpression of leukemia-associated Rho GEF (LARG) mediates sustained hypercontraction of longitudinal smooth muscle in inflammation. Am J Physiol Cell Physiol 306:C1129–C1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Vrees MD, Potenti FM, Harnett KM, Fiocchi C, Pricolo VE. (2004) Interleukin 1β-induced production of H2O2 contributes to reduced sigmoid colonic circular smooth muscle contractility in ulcerative colitis. J Pharmacol Exp Ther 311:60–70. [DOI] [PubMed] [Google Scholar]
- Corbin JD, Zoraghi R, Francis SH. (2009) Allosteric-site and catalytic-site ligand effects on PDE5 functions are associated with distinct changes in physical form of the enzyme. Cell Signal 21:1768–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy MA, Rattan S. (2011) Role of rho kinase in the functional and dysfunctional tonic smooth muscles. Trends Pharmacol Sci 32:384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. (2012) Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem 81:533–559. [DOI] [PubMed] [Google Scholar]
- Francis SH, Busch JL, Corbin JD, Sibley D. (2010) cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev 62:525–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SH, Turko IV, Corbin JD. (2001) Cyclic nucleotide phosphodiesterases: relating structure and function. Prog Nucleic Acid Res Mol Biol 65:1–52. [DOI] [PubMed] [Google Scholar]
- Friebe A, Wedel B, Harteneck C, Foerster J, Schultz G, Koesling D. (1997) Functions of conserved cysteines of soluble guanylyl cyclase. Biochemistry 36:1194–1198. [DOI] [PubMed] [Google Scholar]
- Grider JR, Makhlouf GM. (1988) Contraction mediated by Ca++ release in circular and Ca++ influx in longitudinal intestinal muscle cells. J Pharmacol Exp Ther 244:432–437. [PubMed] [Google Scholar]
- Hazelgrove KB, Flynn RS, Qiao LY, Grider JR, Kuemmerle JF. (2009) Endogenous IGF-I and α v β3 integrin ligands regulate increased smooth muscle growth in TNBS-induced colitis. Am J Physiol Gastrointest Liver Physiol 296:G1230–G1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Li F, Mahavadi S, Murthy KS. (2008) Interleukin-1beta up-regulates RGS4 through the canonical IKK2/IkappaBalpha/NF-kappaB pathway in rabbit colonic smooth muscle. Biochem J 412:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Li F, Mahavadi S, Murthy KS. (2009) Upregulation of RGS4 expression by IL-1β in colonic smooth muscle is enhanced by ERK1/2 and p38 MAPK and inhibited by the PI3K/Akt/GSK3β pathway. Am J Physiol Cell Physiol 296:C1310–C1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Mahavadi S, Li F, Murthy KS. (2007) Upregulation of RGS4 and downregulation of CPI-17 mediate inhibition of colonic muscle contraction by interleukin-1β. Am J Physiol Cell Physiol 293:C1991–C2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara E, Chappellaz M, Turner SR, MacDonald JA. (2012) The contribution of protein kinase C and CPI-17 signaling pathways to hypercontractility in murine experimental colitis. Neurogastroenterol Motil 24:e15–e26. [DOI] [PubMed] [Google Scholar]
- Jin X, Malykhina AP, Lupu F, Akbarali HI. (2004) Altered gene expression and increased bursting activity of colonic smooth muscle ATP-sensitive K+ channels in experimental colitis. Am J Physiol Gastrointest Liver Physiol 287:G274–G285. [DOI] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. (2001) Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem 276:4527–4530. [DOI] [PubMed] [Google Scholar]
- Khan WI, Collins SM. (2006) Gut motor function: immunological control in enteric infection and inflammation. Clin Exp Immunol 143:389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumenacker JS, Hanafy KA, Murad F. (2004) Regulation of nitric oxide and soluble guanylyl cyclase. Brain Res Bull 62:505–515. [DOI] [PubMed] [Google Scholar]
- Krumenacker JS, Hyder SM, Murad F. (2001) Estradiol rapidly inhibits soluble guanylyl cyclase expression in rat uterus. Proc Natl Acad Sci USA 98:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle JF. (1998) Synergistic regulation of NOS II expression by IL-1 beta and TNF-alpha in cultured rat colonic smooth muscle cells. Am J Physiol 274:G178–G185. [DOI] [PubMed] [Google Scholar]
- Mahavadi S, Bhattacharya S, Kumar DP, Clay C, Ross G, Akbarali HI, Grider JR, Murthy KS. (2013) Increased PDE5 activity and decreased Rho kinase and PKC activities in colonic muscle from caveolin-1-/- mice impair the peristaltic reflex and propulsion. Am J Physiol Gastrointest Liver Physiol 305:G964–G974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahavadi S, Nalli AD, Kumar DP, Hu W, Kuemmerle JF, Grider JR, Murthy KS. (2014) Cytokine-induced iNOS and ERK1/2 inhibit adenylyl cyclase type 5/6 activity and stimulate phosphodiesterase 4D5 activity in intestinal longitudinal smooth muscle. Am J Physiol Cell Physiol 307:C402–C411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinolle JP, Garcia-Villar R, Fioramonti J, Bueno L. (1997) Altered contractility of circular and longitudinal muscle in TNBS-inflamed guinea pig ileum. Am J Physiol 272:G1258–G1267. [DOI] [PubMed] [Google Scholar]
- Mayer B, Kleschyov AL, Stessel H, Russwurm M, Münzel T, Koesling D, Schmidt K. (2009) Inactivation of soluble guanylate cyclase by stoichiometric S-nitrosation. Mol Pharmacol 75:886–891. [DOI] [PubMed] [Google Scholar]
- Mayer B, Pfeiffer S, Schrammel A, Koesling D, Schmidt K, Brunner F. (1998) A new pathway of nitric oxide/cyclic GMP signaling involving S-nitrosoglutathione. J Biol Chem 273:3264–3270. [DOI] [PubMed] [Google Scholar]
- Murthy KS. (2001) Activation of phosphodiesterase 5 and inhibition of guanylate cyclase by cGMP-dependent protein kinase in smooth muscle. Biochem J 360:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KS. (2006) Signaling for contraction and relaxation in smooth muscle of the gut. Annu Rev Physiol 68:345–374. [DOI] [PubMed] [Google Scholar]
- Murthy KS, Makhlouf GM. (1998) Differential regulation of phospholipase A2 (PLA2)-dependent Ca2+ signaling in smooth muscle by cAMP- and cGMP-dependent protein kinases. Inhibitory phosphorylation of PLA2 by cyclic nucleotide-dependent protein kinases. J Biol Chem 273:34519–34526. [DOI] [PubMed] [Google Scholar]
- Murthy KS, Zhou H, Grider JR, Makhlouf GM. (2003) Inhibition of sustained smooth muscle contraction by PKA and PKG preferentially mediated by phosphorylation of RhoA. Am J Physiol Gastrointest Liver Physiol 284:G1006–G1016. [DOI] [PubMed] [Google Scholar]
- Murthy KS, Zhou H, Makhlouf GM. (2002) PKA-dependent activation of PDE3A and PDE4 and inhibition of adenylyl cyclase V/VI in smooth muscle. Am J Physiol Cell Physiol 282:C508–C517. [DOI] [PubMed] [Google Scholar]
- Nalli AD, Kumar DP, Mahavadi S, Al-Shboul O, Alkahtani R, Kuemmerle JF, Grider JR, Murthy KS. (2014) Hypercontractility of intestinal longitudinal smooth muscle induced by cytokines is mediated by the nuclear factor-κB/AMP-activated kinase/myosin light chain kinase pathway. J Pharmacol Exp Ther 350:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T, Hori M, Ozaki H. (2007) Mechanism of abnormal intestinal motility in inflammatory bowel disease: how smooth muscle contraction is reduced? J Smooth Muscle Res 43:43–54. [DOI] [PubMed] [Google Scholar]
- Ohama T, Hori M, Sato K, Ozaki H, Karaki H. (2003) Chronic treatment with interleukin-1beta attenuates contractions by decreasing the activities of CPI-17 and MYPT-1 in intestinal smooth muscle. J Biol Chem 278:48794–48804. [DOI] [PubMed] [Google Scholar]
- Pyriochou A, Papapetropoulos A. (2005) Soluble guanylyl cyclase: more secrets revealed. Cell Signal 17:407–413. [DOI] [PubMed] [Google Scholar]
- Rybalkin SD, Yan C, Bornfeldt KE, Beavo JA. (2003) Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circ Res 93:280–291. [DOI] [PubMed] [Google Scholar]
- Salinthone S, Singer CA, Gerthoffer WT. (2004) Inflammatory gene expression by human colonic smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 287:G627–G637. [DOI] [PubMed] [Google Scholar]
- Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. (2007) Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc Natl Acad Sci USA 104:12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed N, Kim DD, Fioramonti X, Iwahashi T, Durán WN, Beuve A. (2008) Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circ Res 103:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RK, Das SB, Lakshmikuttyamma A, Selvakumar P, Shrivastav A. (2006) Regulation of calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1): review. review Int J Mol Med 18:95–105. [PubMed] [Google Scholar]
- Shea-Donohue T, Notari L, Sun R, Zhao A. (2012) Mechanisms of smooth muscle responses to inflammation. Neurogastroenterol Motil 24:802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XZ, Lindholm PF, Sarna SK. (2003) NF-κ B activation by oxidative stress and inflammation suppresses contractility in colonic circular smooth muscle cells. Gastroenterology 124:1369–1380. [DOI] [PubMed] [Google Scholar]
- Shi XZ, Sarna SK. (2005) Transcriptional regulation of inflammatory mediators secreted by human colonic circular smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 289:G274–G284. [DOI] [PubMed] [Google Scholar]
- Singer CA, Salinthone S, Baker KJ, Gerthoffer WT. (2004) Synthesis of immune modulators by smooth muscles. BioEssays 26:646–655. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Beavo JA. (2000) Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol 12:174–179. [DOI] [PubMed] [Google Scholar]
- Somlyo AV, Khromov AS, Webb MR, Ferenczi MA, Trentham DR, He ZH, Sheng S, Shao Z, Somlyo AP. (2004) Smooth muscle myosin: regulation and properties. Philos Trans R Soc Lond B Biol Sci 359:1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. (2014) Regulation of endothelial nitric oxide synthase activity by protein-protein interaction. Curr Pharm Des 20:3514–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt TA, Naftilan AJ, Francis SH, Corbin JD. (1998) ANF elicits phosphorylation of the cGMP phosphodiesterase in vascular smooth muscle cells. Am J Physiol 274:H448–H455. [DOI] [PubMed] [Google Scholar]
- Xia T, Dimitropoulou C, Zeng J, Antonova GN, Snead C, Venema RC, Fulton D, Qian S, Patterson C, Papapetropoulos A, et al. (2007) Chaperone-dependent E3 ligase CHIP ubiquitinates and mediates proteasomal degradation of soluble guanylyl cyclase. Am J Physiol Heart Circ Physiol 293:H3080–H3087. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sun R, Grinchuk V, Fernández-Blanco JA, Notari L, Bohl JA, McLean LP, Ramalingam TR, Wynn TA, Urban JF, Jr, et al. (2013) IL-33-induced alterations in murine intestinal function and cytokine responses are MyD88, STAT6, and IL-13 dependent. Am J Physiol Gastrointest Liver Physiol 304:G381–G389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Morimoto M, Dawson H, Elfrey JE, Madden KB, Gause WC, Min B, Finkelman FD, Urban JF, Jr, Shea-Donohue T. (2005) Immune regulation of protease-activated receptor-1 expression in murine small intestine during Nippostrongylus brasiliensis infection. J Immunol 175:2563–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Urban JF, Jr, Morimoto M, Elfrey JE, Madden KB, Finkelman FD, Shea-Donohue T. (2006) Contribution of 5-HT2A receptor in nematode infection-induced murine intestinal smooth muscle hypercontractility. Gastroenterology 131:568–578. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sayed N, Pyriochou A, Roussos C, Fulton D, Beuve A, Papapetropoulos A. (2008) Protein kinase G phosphorylates soluble guanylyl cyclase on serine 64 and inhibits its activity. Arterioscler Thromb Vasc Biol 28:1803–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]