ABSTRACT
Bacterial cell surface glycans, such as capsular polysaccharides and lipopolysaccharides (LPS), influence host recognition and are considered key virulence determinants. The periodontal pathogen Porphyromonas gingivalis is known to display at least three different types of surface glycans: O-LPS, A-LPS, and K-antigen capsule. We have shown that PG0121 (in strain W83) encodes a DNABII histone-like protein and that this gene is transcriptionally linked to the K-antigen capsule synthesis genes, generating a large ∼19.4-kb transcript (PG0104-PG0121). Furthermore, production of capsule is deficient in a PG0121 mutant strain. In this study, we report on the identification of an antisense RNA (asRNA) molecule located within a 77-bp inverted repeat (77bpIR) element located near the 5′ end of the locus. We show that overexpression of this asRNA decreases the amount of capsule produced, indicating that this asRNA can impact capsule synthesis in trans. We also demonstrate that deletion of the 77bpIR element and thereby synthesis of the large 19.4-kb transcript also diminishes, but does not eliminate, capsule synthesis. Surprisingly, LPS structures were also altered by deletion of the 77bpIR element, and reactivity to monoclonal antibodies specific to both O-LPS and A-LPS was eliminated. Additionally, reduced reactivity to these antibodies was also observed in a PG0106 mutant, indicating that this putative glycosyltransferase, which is required for capsule synthesis, is also involved in LPS synthesis in strain W83. We discuss our finding in the context of how DNABII proteins, an antisense RNA molecule, and the 77bpIR element may modulate expression of surface polysaccharides in P. gingivalis.
IMPORTANCE The periodontal pathogen Porphyromonas gingivalis displays at least three different types of cell surface glycans: O-LPS, A-LPS, and K-antigen capsule. We have shown using Northern analysis that the K-antigen capsule locus encodes a large transcript (∼19.4 kb), encompassing a 77-bp inverted repeat (77bpIR) element near the 5′ end. Here, we report on the identification of an antisense RNA (asRNA) encoded within the 77bpIR. We show that overexpression of this asRNA or deletion of the element decreases the amount of capsule. LPS structures were also altered by deletion of the 77bpIR, and reactivity to monoclonal antibodies to both O-LPS and A-LPS was eliminated. Our data indicate that the 77bpIR element is involved in modulating both LPS and capsule synthesis in P. gingivalis.
INTRODUCTION
Encapsulation is a well-known mechanism that protects pathogenic bacteria from clearance by host immune defenses (1). This reduction in clearance can lead to persistent survival and thereby long-term interplay between the bacterium and host. Capsules not only reduce the ability of the host effectors to gain access to the bacterial cell but also mask the cell surface and thereby modulate the host's response to the bacterium. This model is supported by studies showing that encapsulated bacteria have an advantage in immune evasion (2). Synthesis of a capsule is considered to be a key virulence determinant of Porphyromonas gingivalis (3). Strains that produce K-antigen capsule are more resistant to phagocytosis (4) and cause a spreading type of infection in a murine lesion model (5). In contrast, nonencapsulated strains adhere more to cultured primary gingival epithelial cells and cause a severe localized abscess (6). Importantly, a capsule null mutant strain was shown to be a more potent inducer of cytokine synthesis by human gingival fibroblasts than the corresponding parent strain, indicating a role for capsule in cloaking P. gingivalis against innate immune responses (3). Although synthesis of capsule by P. gingivalis is an important virulence determinant, the regulatory mechanisms that control its synthesis have not been determined.
Lipopolysaccharides (LPS) are surface glycolipids that are tightly associated with the outer leaflet of Gram-negative bacteria and consist of a complex glycan structure covalently bound to a lipid anchor (lipid A). A conserved core oligosaccharide is linked to lipid A via 3-deoxy-d-manno-octulosonic acid, and in the case of P. gingivalis this core has been shown to consist of mannose, allosamine, glycerol, and phosphoethanolamine (7). Attached to this core is a high-molecular-weight polysaccharide of one of two clearly distinguishable types, generating two classes of LPS molecules (8, 9). The O-LPS consists of a repeating unit of four sugars, i.e., rhamnose, glucose, galactose, and either glucosamine or galactosamine (10, 11), while a novel anionic polysaccharide (APS) component, which is made up of a phosphorylated branched mannan, is attached to the core in the A-LPS molecule (9). Interestingly, A-LPS is immunologically connected to the posttranscriptional modification of Arg-gingipains (RgpA) since a monoclonal antibody (MAb 1B5) raised to RgpA cross-reacts with A-LPS (8, 12). Deletion mutants in porR (PG1138) and wbpB (PG2119) lack A-LPS and have less gingipain activity, indicating an involvement of these loci in A-LPS biosynthesis (13, 14). The mutants are also more susceptible to killing by the complement system, indicating a role of A-LPS synthesis in the serum resistance of P. gingivalis (14, 15). Both K-antigen capsule and LPS account for the serotype specificity of a particular strain. To date, three different O-antigen serotypes and at least six K-antigen serotypes of P. gingivalis have been identified (16, 17).
Sequence analysis of the P. gingivalis strain W83 genome indicates multiple polysaccharide synthesis loci (18); however, which genes synthesize the different surface polysaccharides is not clear. Genes in the PG0104-PG0121 locus have been shown to be required for K1 capsule synthesis (19, 20). Yet PG0106 (a putative UDP-phosphate alpha-N-acetylglucosaminyltransferase) mutant in strain 33277 is also deficient in LPS synthesis (21), indicating that PG0106 and possibly other genes in this locus are involved in synthesizing both K antigen and LPS molecules. Unlike the K-antigen locus, the coding sequence of another polysaccharide synthesis locus (PG1135 to PG1142) is highly conserved in various P. gingivalis strains (19). As mentioned above, PG1138 is essential for A-LPS biosynthesis and therefore contributes to resistance to complement killing, which may explain the high conservation in various P. gingivalis strains.
To date, only two different regulatory mechanisms have been identified that control synthesis of surface polysaccharides in P. gingivalis. One is a tyrosine phosphatase (Ltp1) encoded by PG1641, which controls expression of a number of genes encoding proteins involved in the synthesis of surface polysaccharides (22, 23). In the ltp1 deletion mutant, the expression of PG0106 and PG0116, which are located in the K-antigen locus, is downregulated, while porR (PG1138), porS (PG1137), wbbL (PG1140), a putative rhamnosyltransferase, PG0436, a predicted capsular polysaccharide transport protein, and rfbD (PG1561), a putative dTDP-4-dehydrorhamnose-3,5-epimerase, were found to be upregulated (22), indicating an underlying mechanism of repression. Just recently, Ltp1 was shown to inactivate (dephosphorylate) the tyrosine kinase Ptk1, which in turn was shown to control levels of exopolysaccharide production in both strain 33277 and W83 (24), showing that this tyrosine kinase modulates production and export of extracellular polysaccharide in P. gingivalis. The second regulatory mechanism that has been identified is the DNABII protein HU β-subunit (PG0121); specifically, genes in both the K-antigen synthesis locus and the A-LPS locus are downregulated in a PG0121 mutant (25, 26).
Previously, we showed by Northern analysis that the K-antigen locus encodes a large polycistronic message (PG0104 to PG0121) encompassing a 77-bp inverted repeat (77bpIR) region (25, 26), and we determined that the histone-like protein HU (PG0121), which is encoded at the 3′ end of the transcript, binds to the 77-bp sequence (27). Our working model has been that this interaction is involved in regulating transcription and/or RNA stability. In this study, we further characterized this 77bpIR element. We report on the identification of a 550-nucleotide (nt) antisense RNA (asRNA) transcript encoded within the 77bpIR element, demonstrating that transcription in the 77bpIR region is bidirectional. Furthermore, we show that overexpression of this asRNA in trans from a plasmid or deletion of the 77bpIR element alters the synthesis of surface polysaccharides.
MATERIALS AND METHODS
Strains, media, and chemicals.
Bacterial strains and plasmids used in this study are given in Table 1. P. gingivalis strains were maintained in a Coy anaerobic chamber in an atmosphere containing 5% hydrogen, 10% carbon dioxide, and 85% nitrogen at 37°C on Trypticase soy agar supplemented with 5% defibrinated sheep blood (Northeast Laboratory, Waterville, ME), 5 μg/ml hemin, and 1 μg/ml menadione (BAPHK). For liquid culture, the strains were grown in Trypticase soy broth (TSBHK) also supplemented with hemin and menadione. Recombinant strains were selected using erythromycin (Erm) at 10 μg/ml or tetracycline (Tc) at 1 μg/ml, as appropriate. Escherichia coli strains were maintained in Luria broth (LB) or on LB agar plates. Recombinant strains were grown on LB plates with ampicillin (Amp) at 100 μg/ml and erythromycin (Erm) at 200 μg/ml, as appropriate. Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich. Plasmids were constructed in E. coli DH5α and then transformed into P. gingivalis by electroporation (see the protocols below).
TABLE 1.
Strains and plasmids used in this study
| Strain designation, strain characteristics, or plasmid | Source or reference |
|---|---|
| P. gingivalis | |
| W83 | ATCC |
| ΔPG0106::Erm (Emr) in strain W83 | 20 |
| ΔPG0121::Erm (Emr) in strain W83 | 25 |
| ΔPG0104::Erm (Emr) in strain W83 | 25 |
| Δ77bpIR::Erm (Emr) in strain W83 | This study |
| W83-Tc (Tcr) control in strain W83 | This study |
| Δ77bpIR-C (Tcr) complementation | This study |
| Plasmids | |
| pHS17 | 44 |
| pT-COW (Cbr Tcr) | 45 |
| pGEM-T-easy | Promega |
| pTloop1 | This study |
Antisera.
The serotype-specific rabbit antiserum to P. gingivalis W83 and a rabbit polyclonal antibody to PG0121 have been described previously (25, 26). The A-LPS-specific monoclonal antibody 1B5 was obtained from Michael Curtis, Queen Mary University of London, London, England, and the O-LPS-specific monoclonal antibody 7F12 was obtained from Richard Darveau, University of Washington, Seattle, WA. Antiserum to FimA, the fimbria subunit in P. gingivalis, was graciously provided by Ashu Sharma at the University of Buffalo, Buffalo, NY.
Generation of cassettes for allelic exchange.
To investigate the function of the 77bpIR region, we created a deletion mutant. The cassette for allelic exchange to create this knockout strain was generated by overlap PCR using the 77bpIR primers listed in Table 2, as previously described (20). The sequence of the final PCR product was confirmed using the F3_77bpIR and R3_77bpIR primers. The PCR product was purified and then transformed as a linear fragment into P. gingivalis strain W83 to create the DEL77bpIR::Erm insertion-deletion mutant, designated here Δ77bpIR.
TABLE 2.
Primers used in this study
| Primer function and name | Sequence (5′–3′) |
|---|---|
| 77bpIR mutant construction, verification, and complementation | |
| ErmF | CCGATAGCTTCCGCTATTGC |
| ErmR | GAAGCTGTCAGTAGTATACC |
| Ermchk1 | CGTAAATGTTCAACCAAAGCTGTG |
| Ermchk2 | CTCAAGTCTCGATTCAGCAATTGC |
| R2-77bpIR | TGTAGATAAATTATTAGGTATACTACTGACAGCTTCTGGAATTATTACATTCTTTGTGCA |
| F2-77bpIR | ACCGATGAGCAAAAAAGCAATAGCGGAAGCGATCGGATCATTCCACTGGGATTTAGGA |
| F3-77bpIR | GCTCCGCGATGCCCTCAAA |
| R3-77bpIR | CCC AAG CAA CCA ATA ACAGACACA |
| 77bpIR complementation strain construction | |
| F3-77bpIR | GCTCCGCGATGCCCTCAAA |
| R1-77bpIR | TAACATCCCCAGAAGCACCAGACT |
| TetQ-F | CGTTCCATTGGCCCTCAAAC |
| TetQ-R | CTCCTGCCATTCATAGAGGC |
| Knock-in-R1 | GGAGCACAAGTGCTGTTGAC |
| pTloop1 plasmids | |
| Loop1Hind-Forw | CAAGAAGCTTTCCATATGTCAGAGATTCAAATCATAGAG |
| Loop1Eag-Rev | CAAGCGGCCGTTAATGGGACCTTTGCACAATAAGCA |
| Loop2Ale-Forw | CATTCACTGACGTGTGACAATCAAATCCTTTCAACG |
| Loop2Eag-Rev | TGATCGGCCGTCTGCACTGGCAATGTAACC |
| 5′/3′ RACE | |
| First-round PCR | |
| sORF + 140Rev | CGGATTTCCACAAGTGGAAAGATAG |
| sORF + 170Fwd | ACCAAATAGTCGCCTGACACA |
| Second-round PCR | |
| sORF + 130Rev | GTGGAAAGATAGCTTTCCACCG |
| sORF + 210Fwd | AAAGTGCCTGCTTATTGTGCAAAG |
| Third-round PCR | |
| sORF + 28Rev | ACATCCAACGACATGACCGT |
| 0106p1-Rev | GAGCGTAGCAATCGTTAGATC |
| qRT-PCR | |
| PG0108-Forw | CATGTGTAGAGGCTGCAAC |
| PG0108-Rev | GACGCTCTCGATAGATTAGATCAG |
| PG0113-Forw | CGTCCAGGAGATAGACTGATTGTTAC |
| PG0113-Rev | TGGTGGATAATAAGGCATCATAATCA |
| PG0118-Forw | GCGATGAATTGACAGAACAGCAT |
| PG0118-Rev | TTGCGTAATTTGGCTCATAATAACTG |
| PG0121-F2 | TGTAGCTGAAAAGGCCAACC |
| PG0121-R2 | GCACGCTCGCTCACTGAGAA |
| PG1138-F1 | GCTGCGAAAAAGTTCTGTCC |
| PG1138-R1 | GTATATCCCCCTCCCATTGC |
| PG_1141-Forw | GGCAGCAGATTATGCCATCA |
| PG_1141-Rev | GAGGCAAATCAGTGGCTACAA |
| 16S-Forw | TGTTACAATGGGAGGGACAAAGGG |
| 16S-Rev | TTACTAGCGAATCCAGCTTCACGG |
| asRNA RT-PCR | |
| asRNA-Forw | GATTGGAGGAGCACAAGTGCTGTT |
| asRNA-Rev | AGTGGAAAGATAGCTTTCCACCGG |
Construction of the Δ77bpIR knock-in complementation strain Δ77bpIR-C.
To confirm the role of the 77bpIR region in surface glycan regulation, a 2.5-kb fragment harboring the 77bpIR region was PCR amplified using the F3-77bpIR and R1-77bpIR primers and P. gingivalis W83 genomic DNA as a template. The DNA fragment was then TA cloned into the pGEM-T Easy vector (Promega), generating plasmid pGEM-hairpin. A fragment of approximately 2.5 kb containing the tetQ allele was ligated into a unique BsmBI restriction site located 400 bp upstream of the 77bpIR region in the pGEM-hairpin plasmid. The direction of the insertion was confirmed by PCR, and a plasmid harboring the tetQ allele in the antisense direction was chosen to generate a knock-in strain construct. Finally, a region of about 5.1 kb was amplified by PCR and electroporated into the Δ77bpIR strain, and a clone was selected for tetracycline resistance and erythromycin sensitivity, generating the complemented strain Δ77bpIR-C (Fig. 1 shows the tetQ insertion). All plasmids and strains were confirmed by sequencing at each step of construction. As a control, a 4.0-kb region including the tetQ allele insertion was amplified and electroporated into the wild-type strain to yield strain W83-Tc. Insertion of the tetQ allele in this region of the wild-type strain had no effect on the black pigmentation phenotype.
FIG 1.
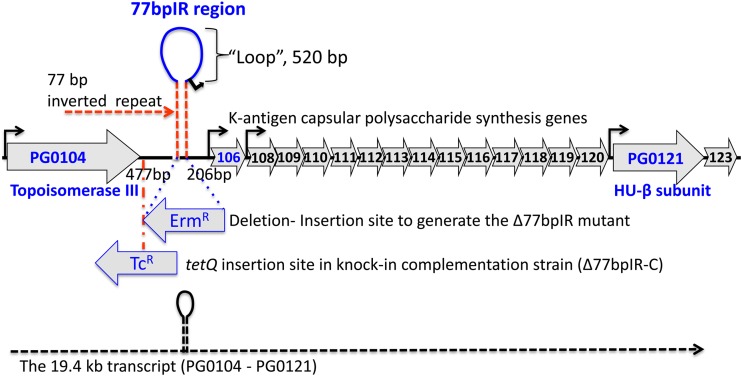
Schematic representation of the K-antigen capsule locus in P. gingivalis strain W83. The region is predicted to encode 17 open reading frames (PG0104 to PG0123). Shown is the 77-bp inverted repeat element (77bpIR) located 206 bp 5′ of the start codon of PG0106 and 477 bp 3′ of the stop codon of PG0104 (topoisomerase III). The element is predicted to form a large (667-bp) stem-loop structure, and the protein (HU-β subunit) encoded by PG0121 near the 3′ end of the locus has been shown to bind the inverted repeat. Also shown is the insertion site and antisense orientation of the erythromycin resistance cassette (Ermr) used to generate the inverted repeat element mutant strain (Δ77bpIR), as well as the insertion site and orientation of the tetracycline resistance cassette (Tcr) in the knock-in complementation strain (Δ77bpIR-C). Promoter regions are identified with black arrows, and the dashed line at the bottom of the figure represents the large 19.4-kb operon transcript, PG0104-PG0121, which encodes the 77bpIR element (shown as a stem-loop) near the 5′ end of the transcript.
Electroporation of P. gingivalis.
Electroporation was performed as previously described (25).
Isolation of total RNA.
Total RNA was isolated from 1 ml of the cells grown in 20 ml of TSBHK to an optical density at 600 nm (OD600) of 0.7 using a Direct-zol RNA MiniPrep kit (Zymo Research) according to the manufacturer's protocol. Contaminating genomic DNA was removed by treatment with Turbo DNA-free (Life Technology). The remainder of each culture was used for immunoblot analysis of surface polysaccharides.
qRT-PCR.
Total RNA (1 μg) was converted to cDNA using iScript reverse transcription supermix (Bio-Rad). cDNAs were diluted 100 times and used for quantitative reverse transcription-PCR (qRT-PCR). qRT-PCR was performed on an iCycler using iQ SYBR green supermix (Bio-Rad). Transcript levels were determined using the standard curve method and normalized to the copy number of 16S rRNA.
Gingipain activity assay.
Arginine- and lysine-specific gingipain activities were assayed using N-α-benzoyl-l-arginine-p-nitroanilide and N-α-acetyl-l-lysine-p-nitroanilide as colorimetric substrates as described previously (28). Briefly, 1 ml of bacterial culture at an OD600 of ∼0.7 was centrifuged to pellet bacteria, and the supernatant was removed and saved for analysis. The pellet was then suspended in fresh TSBHK medium to a volume of 1 ml. Both pellet and supernatant fractions were diluted 1/10 in assay buffer (200 mM Tris, 5 mM CaCl2, 150 mM NaCl, and 10 mM l-cysteine at pH 7.6) and serially diluted in the same buffer in a microtiter plate in a total volume of 50 μl. After 10 min of incubation at 37°C to equilibrate temperature, an equal volume of the appropriate substrate (100 μM) was added, and the reaction was allowed to proceed at 37°C for 2 h. Activity was measured as an increase in OD405.
Electron microscopy.
Bacterial cell wall structure was examined by electron microscopy after cells were stained with ruthenium red, as previously described (26). Strains were streaked to BAPHK agar plates and harvested after 24 h of growth. The Electron Microscopy and Bio-Imaging core at the University of Florida Interdisciplinary Center for Biotechnology Research performed the staining and imaging of cells.
Western blotting.
For Western blotting, surface polysaccharides were extracted by suspending equal amounts of bacteria (normalized by the OD600 of 1.4) in deionized water, followed by autoclaving for 30 min at 120°C. After cooling, samples were centrifuged to pellet insoluble material, and the supernatants were separated by SDS-PAGE and transferred to nitrocellulose membranes overnight at 20 V, as described previously (29). For analysis of protein antigens, cultures were centrifuged to pellet bacteria and suspended in cold phosphate-buffered saline (PBS) containing a protease inhibitor cocktail and 5 mM tosyllysine chloromethyl ketone (TLCK) to minimize degradation of proteins and analyzed as above. Antibody incubations were performed in Tris-buffered saline, pH 7.5, containing 0.1% Tween 20 and 5% nonfat dry milk. Species and isotype-appropriate horseradish peroxidase-linked secondary antibodies were used in all assays, and detection was done with a chemiluminescent substrate (Pierce).
For detection of K-antigen capsule, K-antigen antiserum was used in conjunction with an anti-IgM secondary antibody. As previously described (25), the K-antigen antiserum was generated with W83 whole cells; therefore, the use of an anti-IgG secondary antibody was found to be nonspecific. Surface carbohydrates as immunogens do not always induce switching from the isotype IgM to the isotype IgG; therefore, we tested an anti-IgM secondary antibody for the required specificity. Indeed, blots of wild-type bacteria with anti-IgM secondary showed a high-molecular-weight smear characteristic of capsule. Furthermore, the PG0106 capsule-deficient strain showed no staining; hence, the use of anti-IgM secondary antibody provided the specificity necessary for the detection of capsule. The anti-IgM secondary antibody was also employed in enzyme-linked immunosorbent assays (ELISAs) for K-antigen capsule (see below).
LPS analysis.
P. gingivalis cells harvested in the absence of proteinase inhibitors were digested with proteinase K in the presence of SDS at 60°C as described previously by Hitchcock and Brown (30), subjected to SDS-PAGE using 12% polyacrylamide gels, and stained with ammoniacal silver (31) to visualize LPS.
ELISA for quantification of K-antigen capsule.
An ELISA was developed to compare the quantities of capsule present in the mutant and parent strains using anti-IgM secondary antibody for specificity (see rationale for the use of anti-IgM in the discussion above of the Western blotting method). Briefly, microtiter plates (Microlon; Greiner Bio-One) were coated overnight at 4°C with autoclaved P. gingivalis extracts (as above for blots) diluted 1/1,000 in 50 mM carbonate buffer, pH 9.6, and further diluted serially 2-fold on the plate. Antigen-coated plates were blocked with 5% nonfat dry milk in PBS. After plates were washed with PBS containing 0.1% Tween 20, they were reacted with anti-W83 rabbit serum at a 1/2,000 final dilution in PBS containing 0.1% bovine serum albumin and 0.1% Tween 20 for 1 h at room temperature. After plates were washed as above, plate-bound antibody was probed with a 1/5,000 dilution of goat anti-rabbit IgM–horseradish peroxidase conjugate (Southern Biotech) and detected with a colorimetric substrate (3,3′,5,5′-tetramethylbenzidine [TMB]; Sigma).
Growth rate analysis.
P. gingivalis strains were grown on BAPHK agar plates from frozen stocks and incubated anaerobically for 24 h at 37°C. Three biological replicates were inoculated into 5 ml of TSBHK broth and incubated anaerobically for another 24 h. Each culture was then subcultured as three technical replicates of 5 ml each with a 1:50 dilution in 15-ml optically clear test tubes and incubated anaerobically for another 30 h. To monitor growth, the test tubes were inserted into a spectrophotometer (Biowave cell density meter CO8000; BioExpress), and OD600 readings were acquired every hour for 30 h. All inoculations and OD measurements were performed in the anaerobic chamber. Finally, nine OD readings at each time point were averaged. Error bars show standard deviations.
5′/3′ RACE.
Rapid amplification of cDNA ends (RACE) was carried out as previously described, with minor modification (32, 33). Specifically, three rounds of nested PCR were used to amplify the fragment of interest after a self-ligated cDNA was obtained, whereas the original protocol (33) used one round of PCR amplification. Primers for nested PCR were designed to specifically amplify the region of interest. For this purpose, regions in the P. gingivalis genome with similar 77bpIR element sequences were aligned using the ClustalW web site. Primers were designed as a 100% match only to the sequence in between PG0104 and PG0106. These primers (listed in Table 2) have a one- or two-base mismatch in the 3′ end of each of the primers to other 77bpIR regions, avoiding amplification of other similar regions in the genome. After a third round of nested PCR, the amplified fragments were cloned into pCR2.1 TOPO vector. Sequencing was performed by the University of Florida sequencing core facility.
Generation of plasmid pTloop.
As shown in Fig. 1, there are 520 bp between the 77-bp inverted repeats, which we refer to as the loop region. To test if the deletion could be complemented in trans and to explore the function of the loop in regulating synthesis of surface polysaccharides, we generated plasmid pTloop1 using the P. gingivalis low-copy-number plasmid pT-COW. Plasmid pTloop1 carries the DNA between the 77-bp inverted repeats. The insert was generated with the primer set Loop1Hind-Forw and Loop1Eag-Rev (Table 1). Plasmid pTloop1 was transformed into both the Δ77bpIR mutant strain and the parent strain W83. The same strains harboring plasmid pT-COW were also generated for control purposes. Tetracycline resistance (1 μg/ml) was used for selection.
Reverse transcription-PCR.
Total RNA was isolated from 3-ml cultures of P. gingivalis (strain W83 harboring plasmid pT-COW or pTloop1) grown to an OD550 of 0.7 or 1.1 in TSBHK with Tc (1 μg/ml) using a Direct-zol RNA MiniPrep kit (Zymo Research), according to the manufacturer's protocol. To remove any remaining DNA contamination, the RNA preparation was digested with Turbo DNase (Ambion). Total RNA (2 μg) was converted to cDNA using iScript Reverse Transcription Supermix (Bio-Rad). PCR was performed using Phusion polymerase (New England BioLabs) with primers specific for the antisense transcript encoded within the loop as well as for 16S rRNA as a control (Table 2). A negative control without reverse transcriptase and a genomic DNA positive control were also included.
RESULTS
Genomic organization of the K-antigen capsule locus.
The complete genome sequence of P. gingivalis strain W83 has been reported previously (18). The annotation in the Comprehensive Microbial Resource (CMR) database predicted that genes PG0106 through PG0120 were part of an operon, and our previous studies confirmed this annotation (20, 26). As previously reported (25), a large (77-bp) inverted repeat (77bpIR) region is located 5′ to PG0106. This element and the genes flanking the 77bpIR region are shown in Fig. 1. As indicated, one side of the inverted repeat begins 206 bp upstream of the PG0106 start codon, and its complement is located 477 bp downstream of the stop codon for PG0104. The loop itself consists of 520 bp. Recently, we demonstrated that the HU protein (PG0121), which is encoded at the 3′ end of the locus, binds to the 77bpIR element with high affinity in vitro, and we biochemically characterized the interaction (25, 27).
Deletion of the 77bpIR element confers altered colony morphology, growth rate, and presentation of surface polysaccharides.
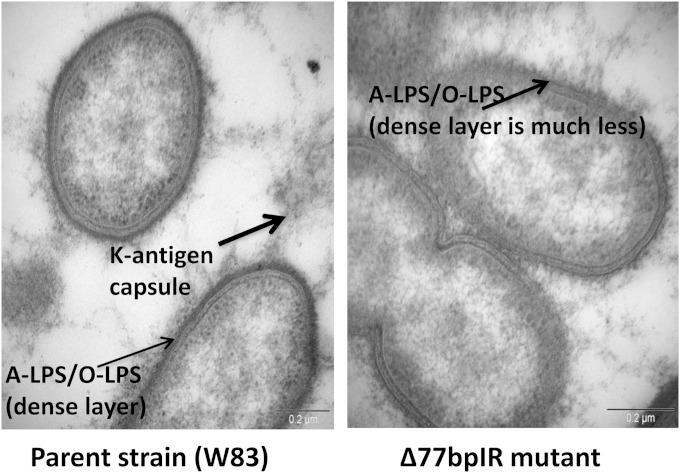
To generate strain Δ77bpIR, the entire 77bpIR region was replaced with an erythromycin resistance cassette, as shown in Fig. 1. The mutant strain formed nonpigmented colonies when grown on blood agar, as has been observed for multiple surface glycan mutants in P. gingivalis (see Discussion), consistent with an altered cell wall structure. Interestingly, over time (3 to 4 days) the colonies become pigmented, especially in the heavy streak zones. Importantly, the complemented strain (Δ77bpIR-C) that was generated by reconstructing the 77bpIR region on the chromosome of the Δ77bpIR mutant (as described in Materials and Methods) formed pigmented colonies indistinguishable from those of the parent strain (data not shown). In addition, as shown in Fig. 2, electron microscopy using ruthenium red staining for carbohydrates confirmed an altered expression of surface polysaccharides in the 77bpIR mutant strain; specifically, there was a decrease in the overall amount of cell surface-associated polysaccharides. In particular, the dense layer adjacent to the cell surface was absent in the Δ77bpIR mutant.
FIG 2.
Transmission electron micrographs of the parent strain W83 and the corresponding Δ77bpIR mutant. The cells were stained with ruthenium red to detect surface polysaccharides, as described in Materials and Methods. The arrows indicate the different types of polysaccharides (LPS or K-antigen capsule) on the surface of the cells. The mutant strain shows a significant decrease in the heavily stained dense layer of polysaccharide (A-LPS or O-LPS) that surrounds the parent strain.
Growth rate.
Growth of the 77bpIR mutant strain was compared to that of the parent strain W83 in TSBHK. Deletion of the 77bpIR element resulted in a modest reduction in growth rate, which may be explained by a decrease in gingipain activity (data not shown).
The Δ77bpIR mutant has reduced K-antigen capsule expression.
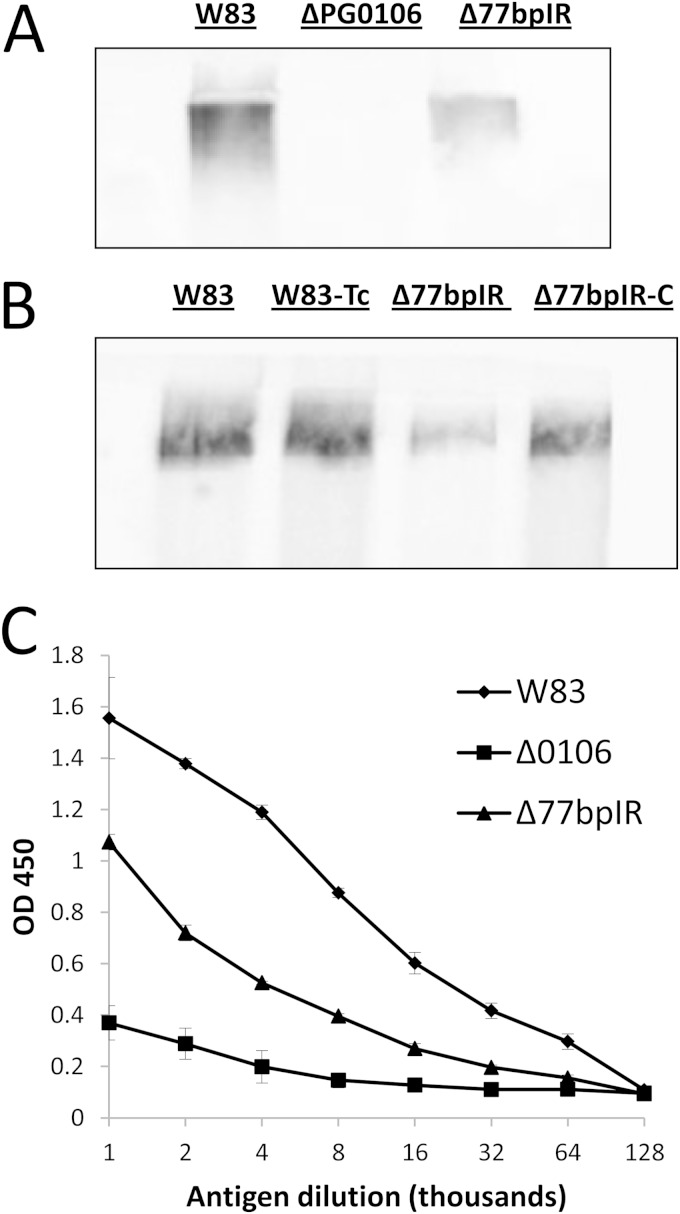
Since our working model has been that the 77bpIR element is involved in regulating transcription and/or stability of the K-antigen capsule operon transcript and thereby the amount of capsule produced, we examined the Δ77bpIR mutant for the presence of capsule by immunoblotting with a K1-specific antiserum. As seen in Fig. 3A the antiserum reacted with an indistinct very-high-molecular-weight band in extracts from wild-type W83 but not the PG0106 mutant, which we previously determined lacks K antigen (20). The Δ77bpIR mutant strain, on the other hand, showed a lower level of staining than the parent strain, consistent with reduced K-antigen expression, as would be predicted if the 77bpIR element enhanced transcription or RNA stability of the K-antigen operon. As can be seen in Fig. 3B, the complemented strain (Δ77bpIR-C) has a level of staining comparable to that of the wild type, showing that deletion of the 77bpIR element is responsible for the K-antigen defect. The same extracts were also tested in an ELISA format (Fig. 3C). Consistent with the blot, the Δ77bpIR mutant strain showed reduced K-antigen content. These results support the hypothesis that the 77bpIR element is involved in enhancing K-antigen capsule synthesis, yet K-antigen synthesis is not dependent on this element but proceeds at a basal level in its absence.
FIG 3.
Quantification of K-antigen capsule by immunoblotting and ELISA, using anti-W83 whole-cell antiserum in combination with an anti-IgM secondary antibody. (A) Immunoblotting of the mutant strain Δ77bpIR shows reduced reactivity to K1-specific antiserum compared to the parent strain W83. The K-antigen null strain (ΔPG0106) was used as a negative control. (B) The complemented strain (Δ77bpIR-C) exhibited wild-type levels of K-antigen capsule. (C). Capsule contents of the parent strain W83, Δ77bpIR, and ΔPG0106 (Δ0106) were quantitated by ELISA. The Δ77bpIR mutant has reduced amounts of K-antigen capsule compared to the parent strain. Error bars represent standard deviations of technical replicates.
The Δ77bpIR mutant strain has an altered LPS profile.
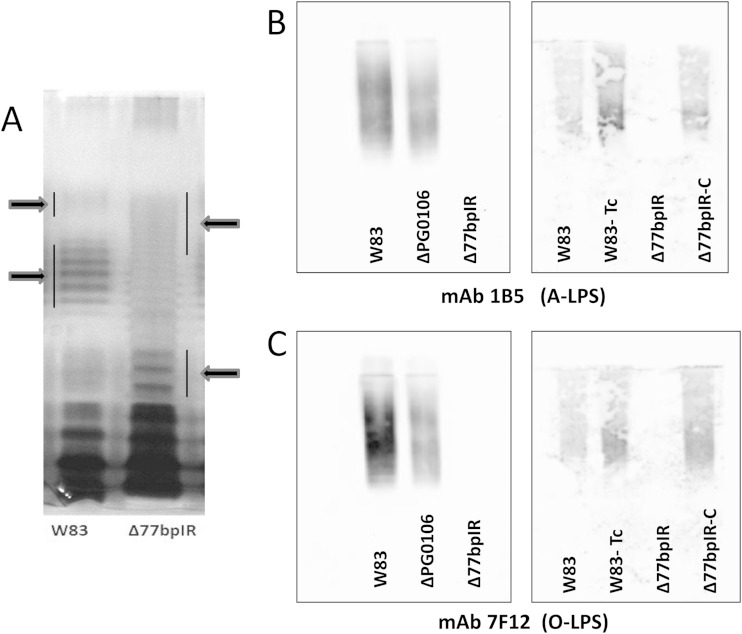
We previously reported that deletion of the HU protein PG0121, which is encoded at the 3′ end of the capsule locus, has altered polysaccharide content and that a variety of loci are differentially expressed in the PG0121 deletion mutant, including genes in known polysaccharide synthesis loci, including the APS synthesis locus PG1135-PG1142 (25, 26). It has also been reported that deletion of PG0106 in P. gingivalis strain 33277, the first gene in the K-antigen operon, yields a strain with an altered LPS profile on silver-stained gels of purified LPS (21). To examine the possibility that the structure of the LPS molecules is also modulated by the 77bpIR element, proteinase K-digested whole-cell lysates were compared for LPS profile by silver ammonia-stained SDS-PAGE gels. As can be seen in Fig. 4A, the Δ77bpIR mutant displayed a strikingly different pattern from that of the parent strain with an overall shift to lower molecular weight and a reduction in the intensity of the high-molecular-weight clusters. An analogous change in staining pattern has been observed for other P. gingivalis mutants known to affect cell wall structure, notably the PorR locus (PG1138 to PG1142) and the VimA locus (PG0880 to PG0885) (34); however, the nature of the alterations in LPS structure have not been elucidated.
FIG 4.
LPS laddering pattern on silver-stained gels and quantification of A-LPS and O-LPS by immunoblotting. (A) Proteinase K-digested bacteria from the parent strain W83 and the Δ77bpIR mutant were separated by SDS-PAGE and stained with ammoniacal silver to visualize LPS. The Δ77bpIR mutant displayed an altered banding pattern with an overall shift to lower molecular weights. (B and C) Autoclaved extracts of W83, Δ77bpIR, and ΔPG0106 were subjected to SDS-PAGE and Western blotting, as described in Materials and Methods. Membranes were probed with antibodies reactive to A-LPS and O-LPS. The ΔPG0106 mutant has reduced reactivity to antibodies to A-LPS and O-LPS. The Δ77bpIR mutant did not react to antibody to A-LPS or O-LPS. Reactivity to both A-LPS and O-LPS was restored to wild-type levels in the complemented strain Δ77bpIR-C.
The Δ77bpIR mutant strain has reduced expression of both O-LPS and A-LPS.
To further characterize the LPS of the Δ77bpIR mutant strain, monoclonal antibodies specific for either the A-LPS (1B5) or the O-LPS (7F12) were used in Western analysis. Immunoblotting (Fig. 4B and C) revealed that autoclaved extracts of the 77bpIR mutant were essentially devoid of polysaccharides reacting to either of these antibodies, indicating either that both of these types of LPS are no longer being synthesized or that the basic structures are being synthesized in a modified form such that they no longer contain the relevant epitopes recognized by the monoclonal antibody. Preliminary analysis of sugar composition of LPS purified from each strain and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry of lipid A did not identify significant differences between the strains (data not shown), consistent with the change in antibody reactivity being due to changes in LPS modification rather than loss of complete LPS species. Recently, a dramatic shift in LPS molecular weight in a wbpB mutant strain using an anti-O-LPS monoclonal was reported (13). This result is consistent with the idea that P. gingivalis is able to make multiple LPS types that might vary in antibody reactivity. It was also observed that the ΔPG0106 strain W83 mutant, defective in the first gene of the K-antigen locus, displayed decreased reactivity to the LPS antibodies (Fig. 4B and C), indicating that genes in the K-antigen synthesis locus may also be involved in LPS synthesis.
Deletion of HU (PG0121) affects the synthesis of LPS.
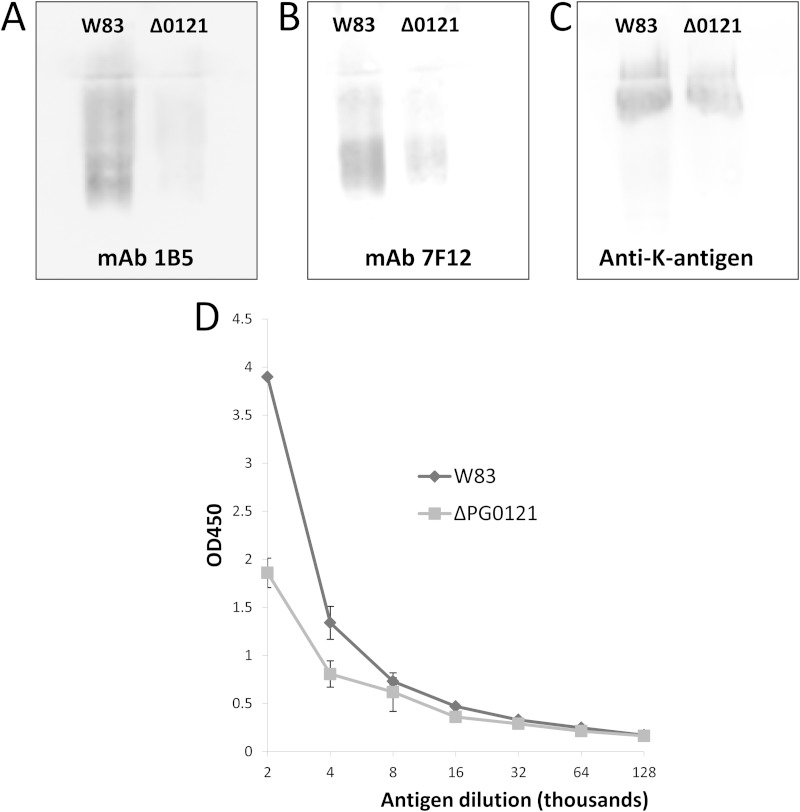
Since deletion of the 77bpIR region, which we have shown to interact with the HU protein PG0121, leads to significant alterations in LPS, we examined the LPS of strain ΔPG0121 using the LPS antibodies. As seen in Fig. 5A and B, although the LPS banding pattern was the same as that of the parent strain (data not shown), reactivity of the PG0121 deletion strain to both LPS antibodies was present but was greatly reduced, indicating that the synthesis of LPS, as well as the amount of K-antigen, is modulated by HU (PG0121).
FIG 5.
Quantification of surface polysaccharides in the HU protein (PG0121) mutant by immunoblotting and ELISA. Shown are immunoblots determining the presence of LPS and K-antigen capsule using antibodies for A-LPS (1B5) (A), O-LPS (7F12) (B), and capsule (anti-serotype K1) (C). The HU protein mutant (Δ0121) has reduced reactivity to both types of LPS and K-antigen antibodies. (D) K-antigen capsule contents of the parent strain W83 and the ΔPG0121 mutant were quantitated by ELISA. The ΔPG0121 mutant has reduced amounts of K-antigen capsule compared to the parent strain. Error bars represent standard deviations of technical replicates.
The Δ77bpIR mutant strain has an altered expression profile of genes involved in the synthesis of surface glycans.
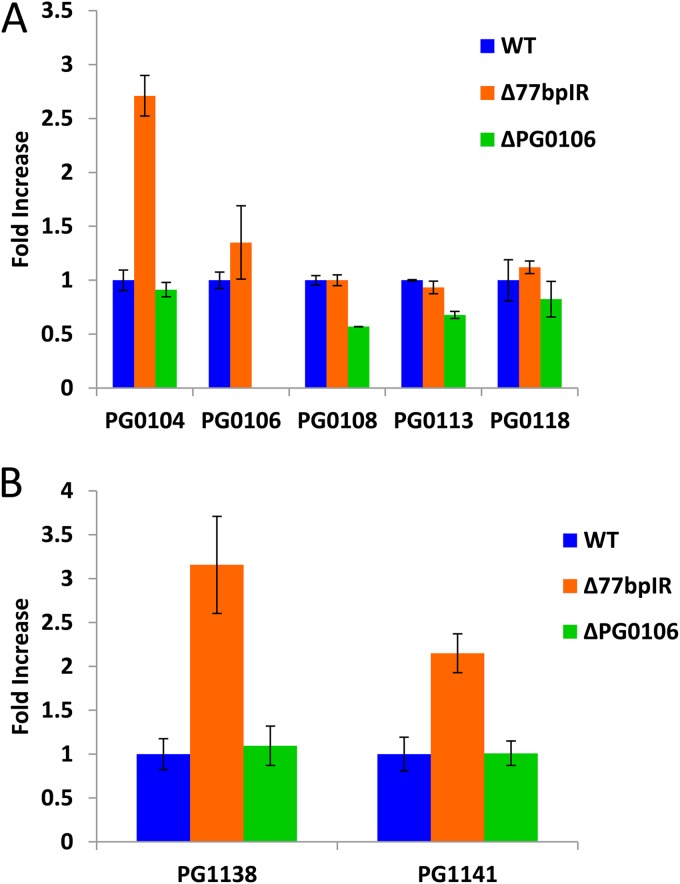
Since the 77bpIR element is encoded within the 5′ end of the large 19.4-kb transcript (Fig. 1), deletion of this element abolishes the synthesis of this transcript. In order to evaluate the effect of the 77bpIR deletion on transcription of genes within this locus, we performed quantitative RT-PCR (qRT-PCR) analysis. As shown in Fig. 6A, six of the genes located in this locus (PG0104, PG0106, PG0108, PG0113, PG0118, and PG0121) were examined. The genes flanking the 77bpIR (PG0104 and PG0106) were upregulated in the mutant, yet the transcript levels of downstream genes, including PG0108, PG0113, and PG0118, were not altered compared to those of the parent strain W83. When genes from the locus reported to be associated with A-LPS synthesis were examined, an anticipated reduction in expression was not seen. As shown by the data in Fig. 6B, two genes, PG1138 and PG1141, known to be associated with A-LPS synthesis were found to be upregulated at the transcript level 3- and 2-fold, respectively. Since deletion of these genes is associated with loss of A-LPS and altered LPS structure (35), it is surprising that their upregulation is seemingly associated with the same phenotype, i.e., a loss of A-LPS and an altered LPS structure.
FIG 6.
Quantitative PCR analysis of genes involved in synthesis of surface polysaccharides. The parent strain W83, strain Δ77bpIR, and strain ΔPG0106 were grown to mid-log phase, and transcription of select genes was evaluated by qRT-PCR. (A) Transcription of K-antigen capsule synthesis genes (PG0108, PG0113, and PG0118) is not significantly altered by deletion of the 77bpIR element; however, PG0104, located upstream from the 77bpIR element, and PG0106, just down stream from the element, showed an increase in expression levels. Expression of the K-antigen capsule synthesis gene is downregulated in the ΔPG0106 K-antigen null mutant. (B) Genes reported to be associated with LPS synthesis are upregulated in the Δ77bpIR mutant. The cDNA samples analyzed for K-antigen capsule synthesis genes were further analyzed for changes in expression in genes associated with synthesis of LPS. Both PG1138 and PG1142, located in the PorR locus, have been reported to affect the presence of A-LPS. Error bars represent standard deviations of technical replicates.
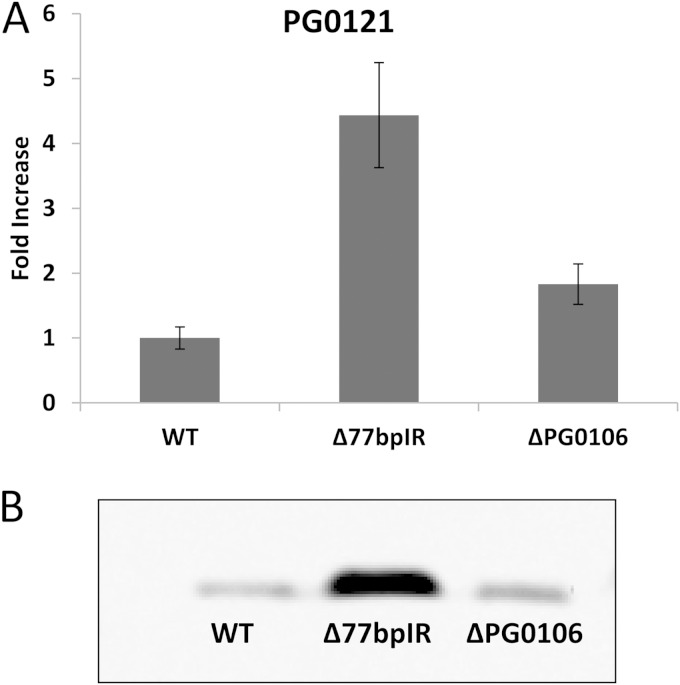
The Δ77bpIR mutant strain has increased expression of PG0121.
We also examined differential expression of PG0121 in the Δ77bpIR mutant. Previously, we showed that PG0121 is transcriptionally linked to the capsule synthesis genes yet is also transcribed separately from its own promoter as a monocistronic message. We hypothesized that synthesis of the large 19.4-kb transcript interferes with transcription from the promoter element just upstream of PG0121. As seen in Fig. 7A, qRT-PCR analysis determined that expression of PG0121 increased approximately 5-fold in the Δ77bpIR mutant, and this increase in expression level was also observed at the protein level, as indicated by immunoblotting using an anti-PG0121 serum (Fig. 7B). Thus, we can conclude that the 77bpIR element represses expression of HU (PG0121).
FIG 7.
Quantitative PCR and Western analysis of the HU protein (PG0121). (A) Transcriptional analysis of PG0121 expression determined on the same cDNA samples used for data presented in Fig. 6. PG0121 was found to be upregulated approximately 5-fold in the Δ77bpIR mutant compared to the level in the parent strain. Error bars represent standard deviations of technical replicates. (B) Western blot of the bacteria from the same cultures probed with anti-PG0121 serum showing that PG0121 is also upregulated at the protein level. Transcription and translation of PG0121 are upregulated in the Δ77bpIR mutant.
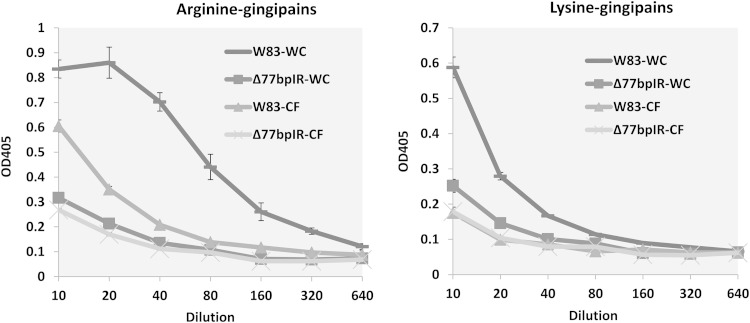
The Δ77bpIR mutant strain has reduced gingipain activity.
P. gingivalis mutants that no longer express A-LPS are known to have reduced cell-associated gingipain activity since A-LPS plays a role in anchoring the proteinases to the cell wall (35). We tested both washed cells and cell-free spent medium of both the parent strain and the mutant for arginine-specific (Rgp) and lysine-specific (Kgp) activities. As has been reported for strains with mutations in the PorR and VimA loci, cell-associated Rgp and Kgp activities were greatly reduced in the Δ77bpIR mutant (Fig. 8). In contrast to these other strains, however, gingipain activity was not preferentially found in the medium. Our data do not distinguish, however, whether the low activity in the medium was due to lack of secretion of the gingipains or whether the proteins are secreted in an inactive form. In addition, a W83 strain with a mutation in VimA has been reported to express a fimbrial protein with reactivity to anti-FimA antibody that is not present in wild-type P. gingivalis W83 (36). We were unable to detect an anti-FimA reactive band in either the parent strain or the Δ77bpIR mutant (data not shown), indicating that the 77bpIR region does not modulate expression of this protein.
FIG 8.
Arginine- and lysine-specific gingipain activity in whole cells or cell-free supernatants. P. gingivalis washed cells (WC) or the corresponding spent growth medium (CF) was assayed for either arginine-specific or lysine-specific gingipain activity using synthetic chromogenic substrates, as described in Materials and Methods. Cultures were adjusted to the same optical densities, and pelleted bacteria were resuspended to a volume equal to that of the adjusted culture to allow a direct assessment of the amount of secreted versus cell-associated activity. Gingipain activity in the Δ77bpIR mutant is reduced compared to that in the parent strain W83. Error bars represent standard deviations of technical replicates.
Validation of an antisense RNA encoded in the 77bpIR element and the effect of overexpression of this asRNA on polysaccharide synthesis.
Using 5′/3′ RACE analysis, we identified an asRNA encoded between the 77-bp inverted repeats. As shown in Fig. 9, this RNA molecule is 550 nucleotides in length and has an internal 32-bp inverted repeat. To explore the function of the loop region between the 77-bp inverted repeats, we generated a P. gingivalis pT-COW plasmid harboring the DNA (520 bp) between the repeats. This plasmid, designated pTloop1 was transformed into the Δ77bpIR mutant strain as well as the parent strain W83 (to generate an asRNA-overexpressing strain). As shown in Fig. 10A, strain W83 harboring pTloop1 overexpresses the asRNA, and the strain is defective in K-antigen capsule (Fig. 10B) compared to the control parent strain harboring plasmid pT-COW, indicating that the asRNA is a trans-acting molecule. In addition, overexpression of the asRNA also resulted in reduced reactivity to O-LPS (data not shown). Importantly, the phenotype of the Δ77bpIR mutant was complemented when the 77bpIR mutation was restored on the chromosome; however, the 77bpIR element did not complement when provided in trans on a plasmid (data not shown), indicating that the 77bpIR is, at least in part, also a cis-acting element.
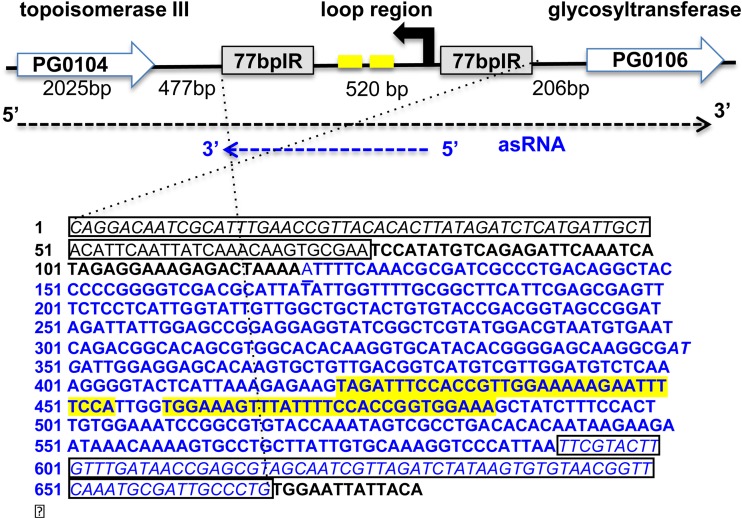
FIG 9.
Schematic of the region from PG0104 to PG0106 and the results from 5′/3′ RACE analysis of the asRNA transcript. Shown is the antisense DNA sequence of the 77bpIR region. The 77-bp inverted repeats flanking the region are outlined with a box. Blue font maps the identified asRNA coding region (550 nt). A 32-bp inverted repeat within the asRNA is indicated in yellow. The 5′ end of the asRNA transcript begins within the loop region (44 bp from the 77bpIR), and the 3′ end is at the end of the 3′ 77-bp inverted repeat.
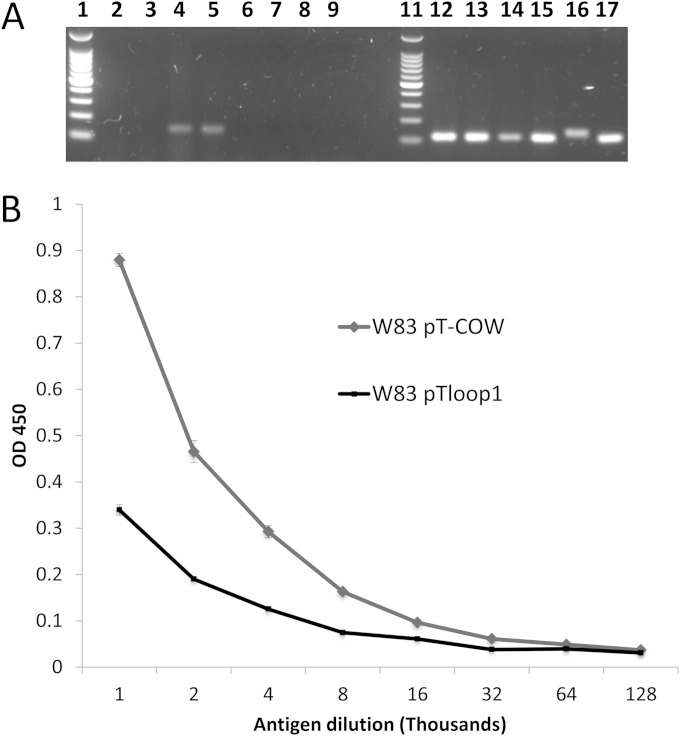
FIG 10.
Reverse transcription to validate overexpression of asRNA in strain W83 harboring plasmid pTloop1 and ELISA showing reduced synthesis of K antigen when asRNA is overexpressed. (A) For comparison, total RNA was extracted from strain W83 harboring pT-COW (control) and W83 harboring plasmid pTloop1 grown to either the late exponential phase (OD550 of 0.7) of growth or stationary phase (OD550 of 1.1). Lane 1, 100-bp molecular weight marker; lane 2, control strain, stationary phase, asRNA; lane 3, control strain, late exponential phase, asRNA; lane 4, pTloop1 strain, stationary phase, asRNA; (5) pTloop1 strain, late exponential phase, asRNA; lanes 6 to 9, no-RT controls of samples in lanes 2 to 5; lane 11, 100-bp molecular weight marker; lane 12, control strain, stationary phase, 16S; lane 13, control strain, late exponential phase, 16S; lane 14, pTloop1 strain, stationary phase, 16S; lane 15, pTloop1 strain, late exponential phase, 16S; lane 16, genomic DNA, asRNA; lane 17, genomic DNA, 16S. Transcriptional analysis verified that strain W83 harboring plasmid pTloop1 overexpresses the asRNA encoded in the loop region during both late exponential and stationary phases of growth. (B) K-antigen capsule content of strain W83 with plasmid pT-COW or pTloop1 was quantitated by ELISA. The strain overexpressing the asRNA has reduced amounts of K-antigen capsule compared to the control strain. Error bars represent standard deviations of technical replicates.
DISCUSSION
Cell surface components, such as capsule, O-LPS, and A-LPS, as well as the cysteine proteases known as gingipains, are important determinants of P. gingivalis virulence. In this article, we show that deletion of a 77bpIR element at the 5′ end of the K-antigen capsule synthesis locus alters the presentation of all three cell surface glycans and reduces Arg- and Lys-gingipain activity. We also show that deletion of the element cannot be complemented in trans, yet the wild-type phenotype was reestablished when the element was restored on the chromosome, indicating that the 77bpIR element is a cis-acting element. Although only a small hypothetical open reading frame is predicted to be encoded within the 77bpIR element, given the size of the loop region (520 bp), we hypothesized that the loop may encode an asRNA and/or a small peptide. Using 5′/3′ RACE, we validated the synthesis of an antisense RNA molecule. This transcript initiates within the loop region, 44 bp from the 77-bp inverted repeat, and terminates with the other 77-bp inverted repeat encoded at its 3′ end. Furthermore, the molecule has an internal 32-nt inverted repeat (separated by only 4 nucleotides), indicating that this asRNA has secondary structure, which may provide stability. Importantly, we show that this asRNA (even without the 77 bp encoded at the 3′ end) can alter expression of surface polysaccharides when they are provided in trans from a plasmid. Thus, the 77bpIR element encodes both cis- and trans-acting functions. Additional studies are required to determine if the antisense transcript is a bona fide asRNA and/or encodes a peptide that is involved in signaling; however, given the antisense orientation of the transcript and the predicted secondary structure, as well as a predicted coding region for a small peptide, our current hypothesis is that this transcript is an asRNA that may also encode a peptide.
In previous studies, we determined that expression of a histone-like DNA binding protein (HU β-subunit, PG0121) was required for wild-type transcript levels of the K-antigen synthesis genes located in the PG0104-PG0120 locus (25), showing that PG0121 enhances expression at the level of transcription and that this results in a concomitant increase in the amount of capsule produced. We also determined that PG0121 preferentially binds to the 77bpIR sequence (27). Our working model has been that the binding of the HU protein PG0121 to the mRNA secondary structures could alter the RNA sufficiently to allow transcriptional readthrough and the subsequent translation through a mechanism known as antitermination (37, 38) and that it does so in combination with the topoisomerase (PG0104) encoded just upstream of the 77bpIR element. Hence, our current findings that deletion of the 77bpIR element does not result in lower transcript levels of the K-antigen synthesis genes were unexpected. To reconcile this finding, we postulate that there may be transcriptional interference of tandem promoters, whereby the activity of the upstream PG0104 promoter, which generates the large ∼19.4-kb transcript, interferes with the downstream promoters in the locus. When the 77bpIR element is deleted, the downstream promoters are more active. In support of this model, upregulation was observed for PG0106 and PG0121 in the Δ77bpIR mutant; however, additional expression studies using Northern analysis are required to determine if this hypothesis is correct. Given our finding that the 77bpIR is not required for capsule synthesis and our earlier findings that a ΔPG0106 mutant (a capsule null strain) can be complemented in trans, the most direct interpretation of our results is that the 77bpIR element is involved in enhancing K-antigen capsule synthesis. Yet capsule synthesis is not dependent on this element but proceeds at a basal level in its absence, and the promoters downstream of the 77bpIR element are sufficient for this synthesis.
An unexpected finding in our studies was that synthesis of LPS was also altered by deletion of the 77bpIR element. Interpretation of this result is complicated by uncertainty over which genes harbored in the P. gingivalis genome are required for LPS polysaccharide synthesis. Genes from two loci that have been shown to be involved in A-LPS synthesis, PG1138 to PG1142 (Fig. 6B) and PG0880 to PG0885 (data not shown), were examined in the Δ77bpIR strain, and expression was found to be unchanged or increased. It is unclear whether uncharacterized genes from other loci that are critical for LPS synthesis are downregulated, resulting in altered LPS synthesis in spite of the continued transcription from the known loci, or whether posttranscriptional control is important for LPS regulation, as is suggested for EPS synthesis. Since the 77bpIR region appears to regulate the protein levels of HU (PG0121), it is tempting to suggest that PG0121 also acts to alter transcription of LPS biosynthesis genes. We have determined that both the PG0121 mutant and the 77bpIR mutant produced altered, yet distinctly different, LPS profiles in spite of one having increased PG0121 levels while the other is deficient; this supports the hypothesis that the equilibrium of the histone-like protein subunits in the cell may modulate the synthesis of these surface glycans.
Another unexplored function of the 77bpIR-encoding transcript is it may provide a mechanism that allows for synthesis of an as yet unidentified surface glycan that may be preferentially expressed under only certain growth conditions. In support of this, our studies determined that deletion of PG0106, which is essential for K-antigen synthesis, also alters LPS synthesis, as had been reported previously for another strain (21). This effect is not due to a change in the transcription of PG0121 downstream in the operon (Fig. 7), and PG0106 is not required for LPS synthesis since the amount of LPS detected by immunoblotting was only decreased, not eliminated. A potential explanation of this finding is that more than one type of LPS molecule cross-reacts with the O-LPS antiserum. If all types are not synthesized, an overall decrease in the amount of LPS would be detected. This would indicate that PG0106 is important in the synthesis of capsule and a certain subtype of LPS and that the 77bpIR element functions to modulate the levels of PG0106. All together, the data indicate a potential link between capsule and LPS synthesis and transcription of the 77bpIR stem-loop-encoding transcript(s). Moreover, we postulate that the large 77bpIR stem-loop structure formed at the 5′ end of the mRNA likely plays a role in RNA stability and that this stability may be critical under certain conditions, such as those imposed during persistence as a human commensal.
Transcriptome studies have discovered that there is pervasive antisense transcription in bacteria (39, 40). Antisense RNA molecules are by far the most abundant transcripts in the bacterial cell and function to modulate transcription, RNA stability, and/or translation, and they can be both cis- and trans-acting molecules (reviewed in references 41 and 42). The sizes of asRNAs have a wide range (∼100 to 7,000 nucleotides), and the abundance of these molecules can vary from barely detectable to high levels. Moreover, depending on function, they can be transcribed at the same time or under opposing conditions as the sense strand (41). Furthermore, these transcripts can have dual functions, acting as antisense RNAs and as peptide-encoding mRNAs (43). Although we have only just begun to investigate the asRNA in the loop region, since overexpression results in less capsule synthesis, the function of this antisense transcript may be to target the large transcript for degradation, a transcript that would otherwise be stable, given the large stem-loop structure at the 5′ end. Discovering that the asRNA can impact synthesis of surface polysaccharides in trans complicates the interpretation of our findings. As a cis-acting element, the altered phenotype could be attributed to the K-antigen locus; yet since there are additional highly similar 77bpIR elements encoded on the chromosome and since one in particular is located in the VimA locus, this direct interpretation may not be correct. Experiments to determine how transcription of the asRNA alters synthesis of surface glycans are ongoing in our laboratories.
In regard to regulation, another potential layer of posttranscriptional regulation may involve activation of structural enzymes via phosphorylation. As indicated earlier, the tyrosine kinase Ptk1 regulates the levels of exopolysaccharide production in both strain 33277 and W83 (24), and this kinase can phosphorylate a UDP-acetyl-mannosamine dehydrogenase encoded in the K-antigen capsule locus (PG0108). Although it has yet to be determined if Ptk1 modulates production of K-antigen capsule, given the role of PG0108 in K-antigen synthesis, Ptk1 and its cognate Ltp1 tyrosine phosphatase are a potential mechanism of posttranscriptional control that may also modulate synthesis of capsular polysaccharide. In summary, we have found that the synthesis of P. gingivalis surface glycans, O-LPS, A-LPS, and K-antigen capsule, involves HU (PG0121) and a 77bpIR element that lies in the 5′ region of the K-antigen synthesis locus. Our studies combined with published data indicate that a multilayer regulatory system exists that controls and coordinates expression of surface glycans. Future studies will focus on defining these regulatory mechanisms, which appear to involve posttranscriptional regulation and coordinated expression of biosynthetic enzymes from multiple loci in the chromosome.
ACKNOWLEDGMENTS
This research was supported by NIH grants R01DE019117 and R21DE023924 from the National Institute of Dental and Craniofacial Research awarded to M.E.D. and N.G., respectively.
REFERENCES
- 1.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 2.Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MA. 2014. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev 38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunner J, Scheres N, El Idrissi NB, Deng DM, Laine ML, van Winkelhoff AJ, Crielaard W. 2010. The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol 10:5. doi: 10.1186/1471-2180-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, Curtis MA, Lewis JP. 2011. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun 79:4533–4542. doi: 10.1128/IAI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laine ML, van Winkelhoff AJ. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol 13:322–325. doi: 10.1111/j.1399-302X.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 6.Irshad M, van der Reijden WA, Crielaard W, Laine ML. 2012. In vitro invasion and survival of Porphyromonas gingivalis in gingival fibroblasts; role of the capsule. Arch Immunol Ther Exp (Warsz) 60:469–476. doi: 10.1007/s00005-012-0196-8. [DOI] [PubMed] [Google Scholar]
- 7.Rangarajan M, Aduse-Opoku J, Hashim A, Paramonov N, Curtis MA. 2013. Characterization of the alpha- and beta-mannosidases of Porphyromonas gingivalis. J Bacteriol 195:5297–5307. doi: 10.1128/JB.00898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 9.Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J Bacteriol 190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farquharson SI, Germaine GR, Gray GR. 2000. Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol Immunol 15:151–157. doi: 10.1034/j.1399-302x.2000.150302.x. [DOI] [PubMed] [Google Scholar]
- 11.Paramonov N, Bailey D, Rangarajan M, Hashim A, Kelly G, Curtis MA, Hounsell EF. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur J Biochem 268:4698–4707. doi: 10.1046/j.1432-1327.2001.02397.x. [DOI] [PubMed] [Google Scholar]
- 12.Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun 67:3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. 2014. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci Rep 4:5056. doi: 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slaney JM, Curtis MA. 2008. Mechanisms of evasion of complement by Porphyromonas gingivalis. Front Biosci 13:188–196. doi: 10.2741/2669. [DOI] [PubMed] [Google Scholar]
- 15.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. 2006. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect Immun 74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims TJ, Schifferle RE, Ali RW, Skaug N, Page RC. 2001. Immunoglobulin G response of periodontitis patients to Porphyromonas gingivalis capsular carbohydrate and lipopolysaccharide antigens. Oral Microbiol Immunol 16:193–201. doi: 10.1034/j.1399-302X.2001.160401.x. [DOI] [PubMed] [Google Scholar]
- 17.van Winkelhoff AJ, Appelmelk BJ, Kippuw N, de Graaff J. 1993. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol 8:259–265. doi: 10.1111/j.1399-302X.1993.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 18.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol 185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, Boutaga K, Laine ML, Van Winkelhoff AJ, Curtis MA. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun 74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey ME, Duncan MJ. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol 188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakao R, Senpuku H, Watanabe H. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun 74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, Demuth DR, Lamont RJ. 2008. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol 69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simionato MR, Tucker CM, Kuboniwa M, Lamont G, Demuth DR, Tribble GD, Lamont RJ. 2006. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun 74:6419–6428. doi: 10.1128/IAI.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright CJ, Xue P, Hirano T, Liu C, Whitmore SE, Hackett M, Lamont RJ. 2014. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen 3:383–394. doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberti-Segui C, Arndt A, Cugini C, Priyadarshini R, Davey ME. 2010. HU protein affects transcription of surface polysaccharide synthesis genes in Porphyromonas gingivalis. J Bacteriol 192:6217–6229. doi: 10.1128/JB.00106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priyadarshini R, Cugini C, Arndt A, Chen T, Tjokro NO, Goodman SD, Davey ME. 2013. The nucleoid-associated protein HUβ affects global gene expression in Porphyromonas gingivalis. Microbiology 159:219–229. doi: 10.1099/mic.0.061002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjokro NO, Rocco CJ, Priyadarshini R, Davey ME, Goodman SD. 2014. A biochemical analysis of the interaction of Porphyromonas gingivalis HU PG0121 protein with DNA. PLoS One 9:e93266. doi: 10.1371/journal.pone.0093266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veillard F, Potempa B, Poreba M, Drag M, Potempa J. 2012. Gingipain aminopeptidase activities in Porphyromonas gingivalis. Biol Chem 393:1471–1476. doi: 10.1515/hsz-2012-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. 1992. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. 1979. Biotechnology 24:145–149. [PubMed] [Google Scholar]
- 30.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- 32.Hirano T, Beck DA, Demuth DR, Hackett M, Lamont RJ. 2012. Deep sequencing of Porphyromonas gingivalis and comparative transcriptome analysis of a LuxS mutant. Front Cell Infect Microbiol 2:79. doi: 10.3389/fcimb.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol 63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 34.Aruni AW, Robles A, Fletcher HM. 2013. VimA mediates multiple functions that control virulence in Porphyromonas gingivalis. Mol Oral Microbiol 28:167–180. doi: 10.1111/omi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183–1191. [DOI] [PubMed] [Google Scholar]
- 36.Osbourne DO, Aruni W, Roy F, Perry C, Sandberg L, Muthiah A, Fletcher HM. 2010. Role of vimA in cell surface biogenesis in Porphyromonas gingivalis. Microbiology 156:2180–2193. doi: 10.1099/mic.0.038331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. 2009. A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J Bacteriol 191:7288–7295. doi: 10.1128/JB.00500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henkin TM, Yanofsky C. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repoila F, Majdalani N, Gottesman S. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 41.Georg J, Hess WR. 2011. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomason MK, Storz G. 2010. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. 2010. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol 76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- 44.Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun 63:1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl Environ Microbiol 62:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]