ABSTRACT
Xylulose 5-phosphate/fructose 6-phosphate phosphoketolase (Xfp), which catalyzes the conversion of xylulose 5-phosphate (X5P) or fructose 6-phosphate (F6P) to acetyl phosphate, plays a key role in carbohydrate metabolism in a number of bacteria. Recently, we demonstrated that the fungal Cryptococcus neoformans Xfp2 exhibits both substrate cooperativity for all substrates (X5P, F6P, and Pi) and allosteric regulation in the forms of inhibition by phosphoenolpyruvate (PEP), oxaloacetic acid (OAA), and ATP and activation by AMP (K. Glenn, C. Ingram-Smith, and K. S. Smith. Eukaryot Cell 13:657–663, 2014). Allosteric regulation has not been reported previously for the characterized bacterial Xfps. Here, we report the discovery of substrate cooperativity and allosteric regulation among bacterial Xfps, specifically the Lactobacillus plantarum Xfp. L. plantarum Xfp is an allosteric enzyme inhibited by PEP, OAA, and glyoxylate but unaffected by the presence of ATP or AMP. Glyoxylate is an additional inhibitor to those previously reported for C. neoformans Xfp2. As with C. neoformans Xfp2, PEP and OAA share the same or possess overlapping sites on L. plantarum Xfp. Glyoxylate, which had the lowest half-maximal inhibitory concentration of the three inhibitors, binds at a separate site. This study demonstrates that substrate cooperativity and allosteric regulation may be common properties among bacterial and eukaryotic Xfp enzymes, yet important differences exist between the enzymes in these two domains.
IMPORTANCE Xylulose 5-phosphate/fructose 6-phosphate phosphoketolase (Xfp) plays a key role in carbohydrate metabolism in a number of bacteria. Although we recently demonstrated that the fungal Cryptococcus Xfp is subject to substrate cooperativity and allosteric regulation, neither phenomenon has been reported for a bacterial Xfp. Here, we report that the Lactobacillus plantarum Xfp displays substrate cooperativity and is allosterically inhibited by phosphoenolpyruvate and oxaloacetate, as is the case for Cryptococcus Xfp. The bacterial enzyme is unaffected by the presence of AMP or ATP, which act as a potent activator and inhibitor of the fungal Xfp, respectively. Our results demonstrate that substrate cooperativity and allosteric regulation may be common properties among bacterial and eukaryotic Xfps, yet important differences exist between the enzymes in these two domains.
INTRODUCTION
Xylulose 5-phosphate (X5P)/fructose 6-phosphate (F6P) phosphoketolase (Xfp), a member of the thiamine pyrophosphate (TPP)-dependent enzyme family, catalyzes the production of acetyl phosphate from the breakdown of xylulose 5-phosphate (equation 1; EC 4.1.2.9) or fructose 6-phosphate (equation 2; EC 4.1.2.22). In lactic acid bacteria and bifidobacteria, Xfp partners with either acetate kinase (Ack) to generate acetate and ATP (equation 3) or phosphotransacetylase (Pta) to generate acetyl coenzyme A (acetyl-CoA) and Pi (equation 4) (1, 2). More recently, Xfp open reading frames (ORFs) have been discovered in euascomycete and basidiomycete fungi as well (3). In fungi, Xfp is believed to partner with Ack, since all fungi that have an Ack ORF have at least one, and in some cases two, Xfp ORFs but lack Pta (3).
| (1) |
| (2) |
| (3) |
| (4) |
Xfp has been biochemically and kinetically characterized from several bacterial species, including Lactobacillus plantarum (referred to by Yevenes and Frey as L. plantarum Xpk2) (2), Bifidobacterium spp. (1, 4), Lactococcus lactis (5), Leuconostoc mesenteroides (5), and Pseudomonas aeruginosa (5), and, more recently, one fungal species, Cryptococcus neoformans Xfp2 (6). The Bifidobacterium Xfp and the L. plantarum, L. lactis, L. mesenteroides, and P. aeruginosa Xfps displayed dual substrate specificity for both substrates X5P and F6P and followed Michaelis-Menten kinetics (1, 2, 4, 5). C. neoformans Xfp2 also displays dual substrate specificity but does not follow Michaelis-Menten kinetics (6). Instead, kinetic characterization of C. neoformans Xfp2 indicated the existence of both substrate cooperativity and allosteric regulation. C. neoformans Xfp2 was found to be inhibited by ATP, phosphoenolpyruvate (PEP), and oxaloacetic acid (OAA) and is activated by AMP (6). Substrate cooperativity and allosteric regulation have not been reported for any characterized bacterial Xfp (1, 2, 4, 5).
In this paper, we describe the characterization of L. plantarum Xfp, in which kinetic parameters were determined using the Hill equation, and the influence of potential allosteric effectors on L. plantarum Xfp activity was examined. L. plantarum Xfp was found to be an allosteric enzyme inhibited by PEP and OAA but unaffected by the presence of AMP or ATP. Additionally, glyoxylate was discovered to be an inhibitor of both C. neoformans Xfp2 and L. plantarum Xfp. Our results suggest that substrate cooperativity and allosteric regulation are common properties among bacterial and eukaryotic Xfp enzymes but are tailored to fit the metabolic pathways of the microbe.
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Sigma-Aldrich, VWR, Fisher Scientific, or Gold Biotechnology. The recombinant plasmid pET28b-xpk2 in Escherichia coli BL21(DE3) was kindly provided by Perry Frey (University of Wisconsin—Madison) for the production of recombinant L. plantarum Xfp (2).
Production and purification of recombinant L. plantarum Xfp.
BL21(DE3) containing the recombinant plasmid pET28b-xpk2 was grown in Luria-Bertani (LB) medium with 25 μg/ml kanamycin at 37°C to an absorbance of ∼0.8 at 600 nm. Recombinant L. plantarum Xfp production was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were allowed to grow overnight at room temperature and harvested by centrifugation.
Cells were suspended in buffer A (25 mM Tris, 150 mM sodium chloride, 20 mM imidazole, 1 mM dithiothreitol [DTT], and 10% glycerol [pH 7.4]) and lysed by two passages through a French pressure cell at approximately 130 MPa. Cell lysate was clarified by ultracentrifugation at 100,000 × g for 1.5 h. The supernatant was applied to a 5-ml His-Trap HP column (GE Healthcare) and subjected to column chromatography using an AKTA fast protein liquid chromatographer (GE Healthcare). After washing with at least seven column volumes of buffer A to remove any unbound protein, the column was subjected to a linear gradient of 20 to 500 mM imidazole to remove all column-bound protein. Fractions determined to contain L. plantarum Xfp by SDS-PAGE and by activity assays were pooled and dialyzed overnight against buffer containing 25 mM Tris, 1 mM DTT, and 10% glycerol (pH 7.0). Protein concentration was determined using a modified Bradford assay (7) with bovine serum albumin as the standard, and purified protein was stored at −80°C.
Production and purification of recombinant C. neoformans Xfp2.
The recombinant plasmid pET21b-XFP2 synthesized by GenScript was transformed into the E. coli expression strain RosettaBlue (Novagen). The recombinant strain was grown in LB medium supplemented with 1% dextrose to reduce basal-level transcription from the T7 RNA polymerase gene under the control of the L8-UV5 promoter in the DE3 prophage prior to the addition of IPTG, 50 μg/ml ampicillin, and 34 μg/ml chloramphenicol. At an absorbance of ∼0.8 at 600 nm, IPTG was added to a final concentration of 1 mM to induce production of the enzyme. Expression was allowed to proceed overnight at room temperature, and cells were harvested by centrifugation. Recombinant C. neoformans Xfp2 was purified as previously described (6).
Xfp assay.
Enzymatic activity was measured using the hydroxamate assay to detect the production of acetyl phosphate (1, 2, 6, 8). A standard 200-μl reaction mixture contained 0.5 mM thiamine pyrophosphate (TPP), 1 mM DTT, 5 mM magnesium chloride, and 50 mM morpholineethanesulfonic acid (at pH 6.0 for L. plantarum Xfp assays and pH 5.5 for C. neoformans Xfp2 assays) with varied concentrations of the substrates F6P and Pi, in the form of sodium phosphate (pH 6.0 for L. plantarum Xfp and pH 5.5 for C. neoformans Xfp2). Reactions were initiated by the addition of 38 to 46 μg enzyme and allowed to proceed for 30 min at 37°C for L. plantarum Xfp and 40°C for C. neoformans Xfp2. After 30 min, 100 μl of 2 M hydroxylamine hydrochloride (pH 7.0) was added, and reaction mixtures were allowed to incubate at room temperature for 10 min to fully convert all acetyl phosphate to acetyl hydroxamate. Reactions were terminated by the addition of 600 μl of a 50:50 mixture of 2.5% ferric chloride in 2N hydrochloric acid and 10% trichloroacetic acid to generate the ferric-hydroxamate complex. The color change due to product formation was measured by a change in absorbance at 540 nm. All data sets correspond to reactions performed in triplicate.
L. plantarum Xfp kinetic analysis.
To determine the L. plantarum Xfp apparent kinetic parameters Km app and kcat app, one substrate was varied while the other substrate was held constant at a saturating concentration (60 mM for F6P and 8 mM for Pi). Since the commercial availability of X5P has been discontinued, all kinetic parameters were determined using F6P. Data were plotted using KaleidaGraph software (Synergy), and kinetic parameters were found by applying the Hill equation (equation 5) (9, 10) to the data set, where V0 is initial velocity, [S] is substrate concentration, V is maximum velocity, K0.5 is substrate concentration at half-maximal velocity, and h is the Hill constant.
| (5) |
Determination of inhibitor IC50 values (individually and in combination).
The half-maximal inhibitory concentration (IC50) was determined for each L. plantarum Xfp inhibitor and an additional C. neoformans Xfp2 inhibitor not previously described by measuring the decrease in enzyme activity at K0.5 substrate concentrations (11 mM F6P and 1 mM Pi for L. plantarum Xfp and 16 mM F6P and 13 mM Pi for C. neoformans Xfp2) in the presence of increasing inhibitor concentration. The IC50 was found by fitting the data with a log (inhibitor) versus response curve in Graphpad Prism 5 software. In order to determine if inhibitors share the same or overlapping allosteric binding sites, the effect on IC50 of one inhibitor in the presence of another inhibitor was measured.
RESULTS
Effect of pH on L. plantarum Xfp activity.
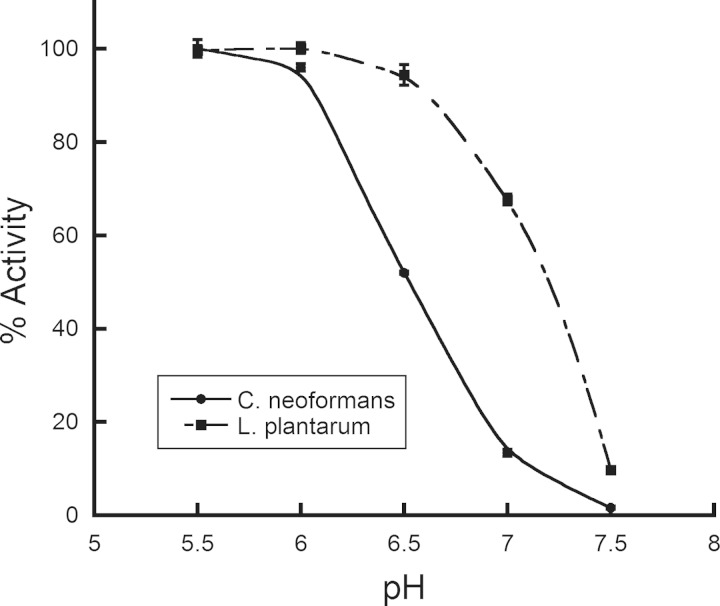
Recombinant L. plantarum Xfp was produced and purified using nickel affinity chromatography and kinetically characterized. C. neoformans Xfp2 activity is significantly reduced with increasing pH, and maximal activity occurs between pH 4.5 and 6.0 (6). As with C. neoformans Xfp2, L. plantarum Xfp activity decreases with increasing pH. However, the decrease in activity occurs at pH 6.5 versus at pH 6.0 for C. neoformans Xfp2 (Fig. 1), suggesting that L. plantarum Xfp is slightly more tolerant to elevated pH than C. neoformans Xfp2. Maximum L. plantarum Xfp activity occurs around pH 6.0, which was used in all kinetic assays.
FIG 1.
Effect of pH on L. plantarum Xfp and C. neoformans Xfp2 activity. Enzyme reactions were performed using F6P and Pi K0.5 concentrations for L. plantarum Xfp and C. neoformans Xfp2. Activity begins to decrease at pH values above 6.0 for C. neoformans Xfp2 and above 6.5 for L. plantarum Xfp.
Kinetic characterization of L. plantarum Xfp.
We have recently shown that C. neoformans Xfp2 displays substrate cooperativity and is subject to allosteric regulation (6), neither of which has been reported for any of the bacterial Xfp enzymes (1, 4, 5, 11), including the L. plantarum Xfp (2). To establish whether substrate cooperativity exists for L. plantarum Xfp, apparent kinetic parameters (Table 1) were determined in the acetyl phosphate-forming direction for the substrates F6P and Pi by fitting experimental data to the Hill equation (equation 5), in which a Hill constant (h) greater than 1.0 represents positive cooperativity and a Hill constant less than 1.0 represents negative cooperativity (12, 13). L. plantarum Xfp displays negative cooperativity for Pi, as indicated by a Hill constant of 0.68 ± 0.02 (Table 1), similar to the Hill constant of 0.59 ± 0.03 for the C. neoformans enzyme (6). The K0.5 value of 1.0 ± 0.1 mM for Pi (K0.5 is a kinetic parameter for enzymes that display substrate cooperativity and is similar to Km in defining the substrate concentration required to obtain half-maximum activity) is similar to the Km previously determined for the L. plantarum enzyme (2).
TABLE 1.
Apparent kinetic parameters for L. plantarum Xfp and C. neoformans Xfp2
| Enzyme and substrate | K0.5 (mM) | kcat app (s−1) | kcat app/K0.5 (s−1 mM−1) | H |
|---|---|---|---|---|
| L. plantarum Xfp | ||||
| F6P | 11.0 ± 1.4 | 1.05 ± 0.05 | 0.10 ± 0.01 | 0.99 ± 0.04 |
| Pi | 1.0 ± 0.1 | 1.12 ± 0.03 | 1.11 ± 0.09 | 0.68 ± 0.02 |
| C. neoformans Xfp2a | ||||
| F6P | 15.9 ± 1.3 | 3.47 ± 0.10 | 0.22 ± 0.01 | 1.41 ± 0.11 |
| Pi | 13.3 ± 1.5 | 4.22 ± 0.13 | 0.32 ± 0.03 | 0.59 ± 0.03 |
Previously reported kinetic parameters (6).
Unlike that for C. neoformans Xfp2, the L. plantarum Xfp Hill constant of approximately 1.0 does not indicate the existence of substrate cooperativity in regard to F6P binding. F6P progress curves for L. plantarum Xfp were found to fit both the Michaelis-Menten and Hill equations equally well, with R values of around 0.99. Using the Michaelis-Menten equation, Km app for F6P was determined to be 10.8 ± 0.8 mM, while a K0.5 for F6P of 11.0 ± 1.4 mM was calculated using the Hill equation for the same data set (Table 1). Both F6P K0.5 and Km app are on the same order of magnitude as the Km app for F6P previously determined by Yevenes and Frey (2).
L. plantarum Xfp is inhibited by PEP and OAA but unaffected by AMP and ATP.
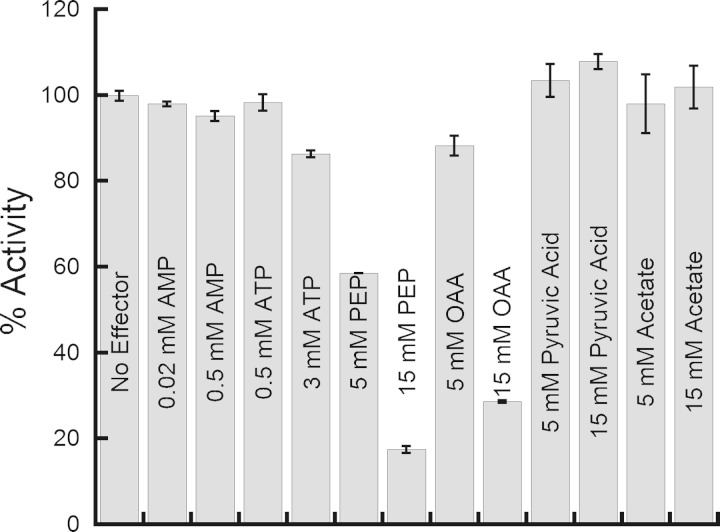
The same ligands examined as possible allosteric effectors of C. neoformans Xfp2 (6) were tested to determine their effect on L. plantarum Xfp activity. As with C. neoformans Xfp2, L. plantarum Xfp is inhibited by phosphoenolpyruvate (PEP) and oxaloacetic acid (OAA) (Fig. 2) and slightly inhibited by citrate (data not shown). L. plantarum Xfp activity was unaffected by the presence of AMP or ATP, the primary allosteric activator and inhibitor, respectively, of C. neoformans Xfp2. Additional ligands tested, such as acetate and pyruvic acid, that had no effect on C. neoformans Xfp2 activity also had no effect on L. plantarum Xfp activity.
FIG 2.
Effect of various ligands on L. plantarum Xfp activity. Reactions were performed using L. plantarum Xfp F6P and Pi K0.5 substrate concentrations. Two concentrations of each coenzyme or metabolic intermediate were tested. Reactions were performed in triplicate. Activity is reported as the percentage of activity with no ligand present.
Glyoxylate inhibits both L. plantarum Xfp and C. neoformans Xfp2 activity.
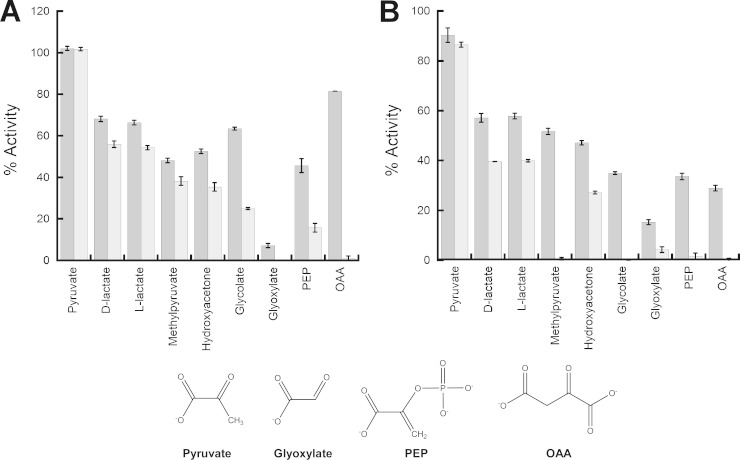
Since PEP and OAA serve as common allosteric effectors for bacterial L. plantarum Xfp and eukaryotic C. neoformans Xfp2, various PEP analogs were tested to determine the specificity of this allosteric site for PEP and the primary chemical moiety that contributes to the allosteric inhibitory effect. Nonphosphorylated PEP analogs previously have been utilized to determine the chemical moiety that contributes to allostery in muscle pyruvate kinase (14), and each of these PEP analogs were used to test L. plantarum Xfp inhibition (Fig. 3A). Interestingly, glyoxylate was found to inhibit Xfp in addition to PEP and OAA. L. plantarum Xfp was almost fully inhibited by 8 mM glyoxylate, with only 7.2% ± 1.0% activity remaining, but was only partially inhibited by 8 mM PEP (45.7% ± 3.4% activity remaining). Pyruvate showed no inhibition, while the PEP analogs d-lactate, l-lactate, methyl pyruvate, hydroxyacetone, and glycolate displayed intermediate levels of inhibition. These same PEP analogs were used to test their inhibitory effect against C. neoformans Xfp2 as well, and the results were similar to those found for the L. plantarum Xfp. Glyoxylate also inhibits C. neoformans Xfp2 activity, with 8 mM reducing activity to 15.3% ± 1.0%. Similar to L. plantarum Xfp, pyruvate had no effect on C. neoformans Xfp2 activity, while all other PEP analogs show intermediate inhibition between that of pyruvate and glyoxylate (Fig. 3B).
FIG 3.
Effect of nonphosphorylated PEP analogs on L. plantarum Xfp and C. neoformans Xfp2 activities. Reactions were performed using L. plantarum Xfp (A) and C. neoformans Xfp2 (B) F6P and Pi K0.5 substrate concentrations. Concentrations of 8 mM (dark gray) and 16 mM (light gray) for each nonphosphorylated PEP analog were tested in triplicate. Activity is reported as a percentage of activity with no ligand present.
Determination of allosteric effector IC50s.
The half-maximal inhibitory concentration (IC50) was determined for each L. plantarum Xfp inhibitor (Table 2) using K0.5 substrate concentrations. The IC50 for glyoxylate was lower than those for PEP and OAA by approximately 3-fold and 5-fold, respectively. In order to determine if any of these inhibitors share the same site, the IC50 of one inhibitor was determined in the presence of a second inhibitor held constant at its IC50. If two inhibitors share the same site or if their sites overlap, then approximately half the sites should be occupied by the inhibitor held constant, thereby lowering the amount of the varied inhibitor required to reduce activity by an additional 50%.
TABLE 2.
Half-maximal inhibitory concentrations (IC50s)
| Enzyme | Inhibitor |
IC50 (mM) | |
|---|---|---|---|
| Varied | Constant | ||
| L. plantarum Xfp | Glyoxylate | 1.93 ± 0.05 | |
| PEP | 6.70 ± 0.12 | ||
| OAA (10.5 mM) | 1.14 ± 0.12 | ||
| Glyoxylate (2 mM) | 6.04 ± 0.11 | ||
| OAA | 10.5 ± 0.07 | ||
| Glyoxylate (2 mM) | 7.78 ± 0.20 | ||
| C. neoformans Xfp2 | Glyoxylate | 4.93 ± 0.04 | |
| PEP (8 mM) | 3.13 ± 0.18 | ||
| PEP | 5.31 ± 0.13 | ||
| Glyoxylate (5 mM) | 4.01 ± 0.07 | ||
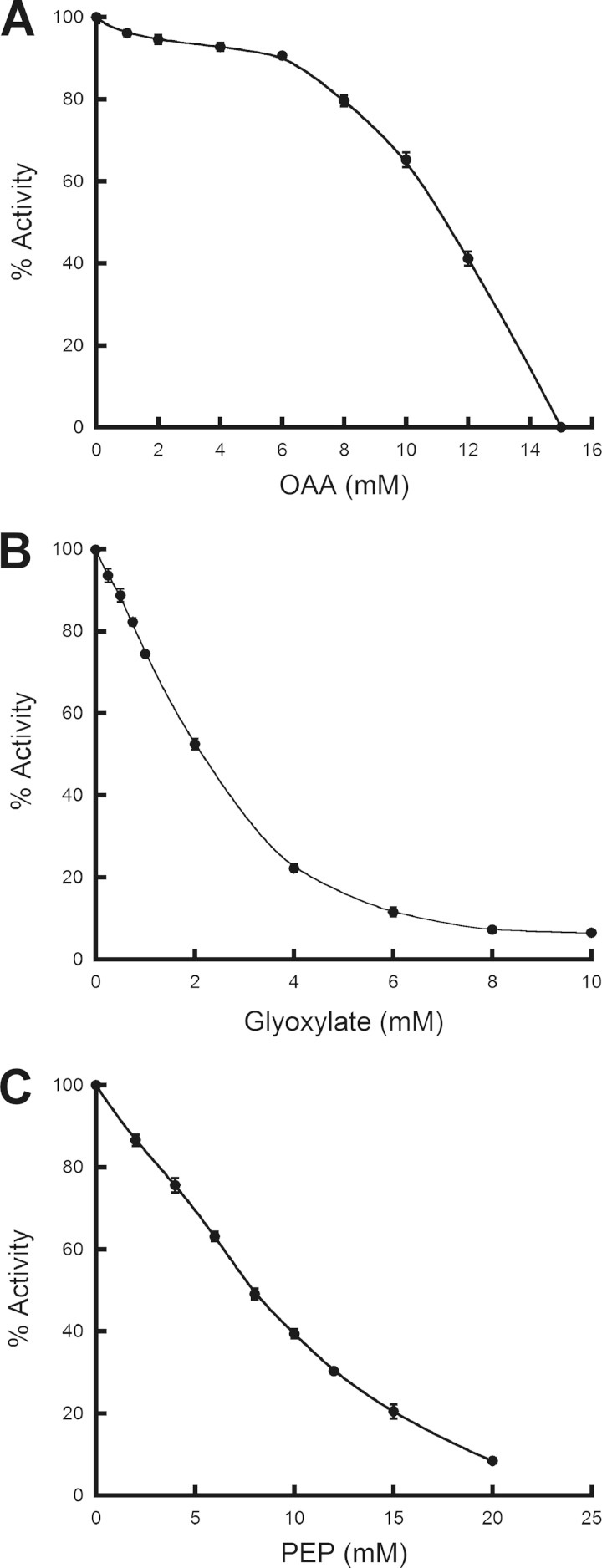
In the presence of 10.5 mM OAA, the PEP IC50 decreased by more than half of the original value, demonstrating that the L. plantarum PEP and OAA binding sites are the same or overlapping, as indicated previously for C. neoformans Xfp2 (6). The reason the IC50 of PEP is reduced by more than half in the presence of OAA most likely is due to the unique inhibitory effect of OAA (Fig. 4A) not seen in glyoxylate (Fig. 4B) or PEP (Fig. 4C) inhibition profiles. High concentrations of OAA were required to initiate L. plantarum Xfp inhibition, followed by a sharp decrease in activity in the presence of additional OAA (Fig. 4A). This inhibitory concentration threshold suggests that PEP and OAA interact with the binding site differently or that the PEP and OAA binding sites are overlapping instead of identical. The IC50 of PEP does not change significantly in the presence of 2 mM glyoxylate, while the IC50 of OAA decreases by approximately 26%. Therefore, it appears that the PEP/OAA site is separate from the glyoxylate site, since neither PEP nor OAA IC50 display close to a 50% decrease in the presence of 2 mM glyoxylate.
FIG 4.
Inhibition of L. plantarum Xfp by OAA, PEP, and glyoxylate. Reactions were performed in triplicate using L. plantarum Xfp F6P and Pi K0.5 substrate concentrations. Activity was measured in the presence of increasing concentrations of OAA (A), glyoxylate (B), or PEP (C) and are reported as a percentage of maximal activity with no ligand present.
Since glyoxylate was not previously recognized as an Xfp inhibitor (6), the IC50 of glyoxylate was determined for C. neoformans Xfp2 (Table 2). The C. neoformans Xfp2 PEP IC50 of 5.31 ± 0.13 mM, slightly lower than the 8 mM IC50 previously reported, (6) decreased to 4.01 ± 0.07 mM in the presence of 5 mM glyoxylate. Since the IC50 of PEP only decreases by about 24% in the presence of 5 mM glyoxylate, it is likely that the binding of PEP and glyoxylate occur at separate sites on C. neoformans Xfp2 as well.
Allosteric inhibitors influence F6P binding.
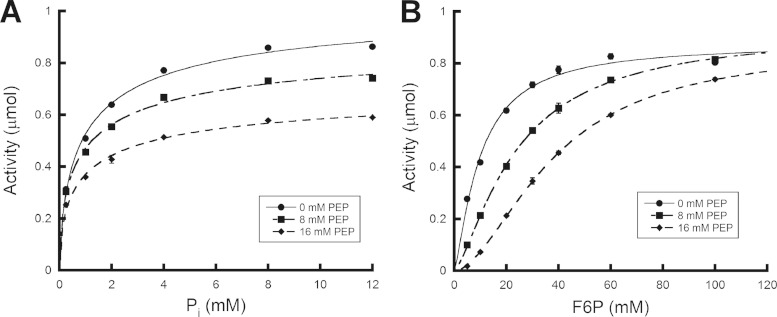
Progress curves of substrate concentration versus activity were generated for L. plantarum Xfp substrates F6P and Pi in the presence of increasing PEP concentrations (Fig. 5). The presence of PEP had the same effect on L. plantarum Xfp kinetic parameters as it did for C. neoformans Xfp2 (6). In regard to Pi, the presence of PEP had little effect on K0.5, which remained between 1.0 ± 0.03 mM and 1.3 ± 0.30 mM at 0 mM PEP and 16 mM PEP, respectively, or Hill constant, which ranged between 0.66 ± 0.01 and 0.49 ± 0.02. However, a gradual reduction in Vmax from 1.06 ± 0.01 μmol of product formed per 30-min reaction at 0 mM PEP to 0.80 ± 0.04 μmol at 16 mM PEP was observed. The K0.5 of F6P increased from 9.8 ± 0.2 mM in the absence of inhibitor to 37.8 ± 0.9 mM in the presence of 16 mM F6P, and the Hill constant also increased from 1.31 ± 0.01 to 1.79 ± 0.01. The presence of PEP had little effect on F6P maximal activity, with Vmax ranging between 0.93 ± 0.02 and 0.87 ± 0.01 μmol of product formed per 30-min reaction. Since PEP and OAA bind at the same or overlapping sites on L. plantarum Xfp, PEP was utilized to represent the effect of both PEP and OAA inhibition on substrate binding. The influence that PEP inhibition has on the F6P K0.5 and Hill constant suggests that, similar to C. neoformans Xfp2 (6), the binding of PEP and OAA directly influences the binding of F6P to the L. plantarum Xfp active site. Glyoxylate binding did not significantly influence F6P K0.5 but had a greater influence on Vmax (data not shown).
FIG 5.
Effect of PEP on L. plantarum Xfp substrate progress curves. Progress curves were generated in the presence of 0, 8, and 16 mM PEP for the substrates Pi (a) and F6P (b). Reactions were performed in triplicate, and activities were reported as μmol of product formed.
DISCUSSION
Bacteria with nonstandard (i.e., not “E. coli-like”) central metabolic pathways play major roles in the human microbiome. These include large numbers of Lactobacillus and Bifidobacterium species, which metabolize sugars by glycolytic pathways that are quite different from the conventional Emden-Meyerhoff-Parnas pathway. One of those pathways, the pentose phosphoketolase pathway, requires Xfp, an enzyme that has been neglected until quite recently.
We recently determined that a fungal Xfp, C. neoformans Xfp2, displayed both substrate cooperativity (positive cooperativity for F6P and negative cooperativity for Pi) and allosteric regulation in the form of inhibition through the binding of ATP, PEP, and OAA and activation by the binding of AMP (6). Here, we report the discovery of substrate cooperativity and allosteric regulation for bacterial L. plantarum Xfp. This report describes the first indication that substrate cooperativity and allosteric regulation also exists among at least some bacterial Xfp enzymes. Kinetic parameters for L. plantarum Xfp originally were determined by Yevenes and Frey by fitting substrate progress curves with the Michaelis-Menten equation to determine the apparent Km for F6P and Pi, which were found to be 24 ± 4 mM and 2.9 ± 0.5 mM, respectively (2). We produced the recombinant L. plantarum Xfp and demonstrated that this enzyme displays negative cooperativity in regard to Pi binding with a Hill constant less than one but little cooperativity in regard to F6P binding with a Hill constant roughly equal to one.
In addition to substrate cooperativity, L. plantarum Xfp, like C. neoformans Xfp2, is allosterically regulated. It is inhibited by PEP and OAA, but unlike C. neoformans Xfp2, the presence of ATP or AMP had little to no effect on activity. Initially, L. plantarum Xfp kinetic parameters did not suggest the presence of substrate cooperativity for F6P binding alone; however, the presence of the inhibitor PEP induces positive cooperativity, as demonstrated by the sigmoidal progress curves and increase in F6P Hill constant shown in Fig. 5. Unlike PEP, glyoxylate inhibits enzyme activity without greatly influencing F6P binding, since both F6P Km and the Hill constant were not greatly altered. In regard to C. neoformans Xfp2 and L. plantarum Xfp regulation by excess PEP and OAA, the presence of these intermediates indicate the energy needs of the cell have been met. The cell can switch from glycolysis, the breakdown of glucose, to gluconeogenesis to synthesize and ultimately store glucose until it is needed; therefore, Xfp may be inhibited by these intermediates in order to limit additional energy production through the Xfp/Ack pathway (6).
In addition to Xfp, L. plantarum can produce acetyl phosphate from pyruvate using pyruvate oxidase (Pox) and from acetyl-CoA using phosphotransacetylase (Pta). It has been shown that at least some Acks in heterofermentative bacteria are allosterically regulated (15). Perhaps Xfp regulation by the presence of ATP and AMP is less necessary in L. plantarum, which has additional sources for acetyl phosphate production and may also possess an allosterically regulated Ack, although this has not been experimentally proven. Xfp regulation by ATP and AMP may have evolved in C. neoformans, where there is no evidence of an allosterically regulated Ack (C. Ingram-Smith, A. Guggisberg, S. Henry, J. Welch, K. Laws, A. Mattison, A. Bizhanova, and K. Smith, unpublished data) or the presence of additional acetyl phosphate producing enzymes, such as Pox and Pta. Therefore, the control of ATP production by Ack in C. neoformans may rest solely on the production of acetyl phosphate by Xfp.
Interestingly, acetyl phosphate has been shown in several bacteria (e.g., E. coli, Salmonella enterica, Xenorhabdus nematophilus, Borrelia burgdorferi, and Campylobacter) to donate its phosphoryl group to certain response regulators, with physiologically important consequences (for a review, see reference 16). Two groups have recently shown that acetyl phosphate is the acetyl donor for most of the protein acetylation that occurs in E. coli (17, 18). Acetyl phosphate-dependent phosphorylation or acetylation has not yet been reported in bacteria that possess Xfp, but it is reasonable to imagine that these posttranslational modifications would occur, especially considering acetyl phosphate is a key central metabolite.
Nonphosphorylated PEP analogs were tested to determine the primary chemical moiety that contributes to PEP allosteric inhibition for both L. plantarum Xfp and C. neoformans Xfp2. Interestingly, pyruvate displayed no inhibition, while glyoxylate, which differs from pyruvate by the absence of a single methyl group, inhibits L. plantarum Xfp more than PEP or OAA at the same concentration. Our results suggest that PEP and OAA bind to the same or overlapping sites in both L. plantarum Xfp and C. neoformans Xfp2, but glyoxylate binds at a distinct site.
Since glyoxylate is an allosteric inhibitor presumably with its own allosteric site on both L. plantarum Xfp and C. neoformans Xfp2, there is likely a metabolic connection between the presence of excess glyoxylate and the inhibition of Xfp, consequently limiting the production of acetyl phosphate from X5P and F6P. The glyoxylate cycle functions as a bypass of the decarboxylation steps of the tricarboxylic acid cycle, allowing for the use of simple two-carbon compounds, such as acetate and ethanol, to generate malate from the combined action of the enzymes isocitrate lyase (Icl) and malate synthase (Mls) (19–21). The glyoxylate cycle is utilized when glucose is limiting (19), but the production of excess glyoxylate indicates that other 2-carbon compounds, such as acetate, are prevalent. Since we hypothesize that C. neoformans Xfp2 partners with Ack to generate acetate and ATP, the presence of excess glyoxylate indicates that acetate is in abundance, so glyoxylate inhibits Xfp, thereby inhibiting the production of acetyl phosphate and consequently the production of acetate by Ack.
We have failed to identify genes encoding the enzymes Icl and Mls within the L. plantarum genome, suggesting it lacks a glyoxylate cycle. However, L. plantarum cell extract is capable of metabolizing glyoxylate (22). An ORF within the L. plantarum genome designated 2-hydroxyacid dehydrogenase (accession number YP_004888759) has 61% identity to a gene designated a glyoxylate reductase (equation 6) in Lactobacillus otakiensis, and it also has been shown in Rhizobium etli that an enzyme previously labeled as 2-hydroxyacid dehydrogenase displays glyoxylate reductase activity (23).
| (6) |
In plants, glyoxylate reductase is believed to function as a way of removing excess reducing equivalents like NADPH (24), and glyoxylate reductase in L. plantarum could serve a similar purpose. A possible connection between L. plantarum Xfp inhibition by excess glyoxylate is that the presence of glyoxylate indicates the presence of excess NADPH. The pentose phosphate pathway (PPP) serves as a major source of the NADPH produced in the cell. Thus, inhibiting Xfp, which utilizes PPP end products, may hinder the PPP from producing additional end products, thereby also reducing the amount of NADPH produced upstream. The regulation of Xfp by glyoxylate in L. plantarum may serve as a means of balancing the production and utilization of NADPH by the PPP and glyoxylate reductase, respectively, to aid in cellular redox balance.
Concluding remarks.
Substrate cooperativity and allosteric regulation no longer can be considered a purely eukaryotic Xfp phenomenon with the discovery of its existence in at least some bacterial Xfps, specifically L. plantarum Xfp. However, there are differences between the degree of substrate cooperativity and the allosteric effectors that inhibit or activate eukaryotic Xfp and bacterial Xfp from C. neoformans and L. plantarum, respectively. Both C. neoformans Xfp2 and L. plantarum Xfp share PEP, OAA, and glyoxylate as allosteric inhibitors, but C. neoformans Xfp2 also is inhibited by ATP and activated by AMP, while an activator of L. plantarum Xfp has yet to be discovered. Additionally, regulation by glyoxylate appears to result from different phenomena in C. neoformans, which primarily produces glyoxylate through the glyoxylate cycle, versus L. plantarum, which appears to lack a glyoxylate cycle. Allosteric regulation of both the bacterial and eukaryotic Xfps suggests that tight control over the pathway is important for responding to the different environmental stresses that ultimately influence cellular metabolism.
ACKNOWLEDGMENTS
We thank Cheryl Ingram-Smith for her critical reading of the manuscript.
This work was supported by awards from the National Institutes of Health (GM084417-01A1), National Science Foundation (award 0920274), and South Carolina Experiment Station Project SC-1700340.
Footnotes
This article represents Technical Contribution 6266 of the Clemson University Experiment Station.
REFERENCES
- 1.Meile L, Rohr LM, Geissmann TA, Herensperger M, Teuber M. 2001. Characterization of the D-xylulose 5-phosphate/D-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J Bacteriol 183:2929–2936. doi: 10.1128/JB.183.9.2929-2936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yevenes A, Frey PA. 2008. Cloning, expression, purification, cofactor requirements, and steady state kinetics of phosphoketolase-2 from Lactobacillus plantarum. Bioorg Chem 36:121–127. doi: 10.1016/j.bioorg.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Ingram-Smith C, Martin SR, Smith KS. 2006. Acetate kinase: not just a bacterial enzyme. Trends Microbiol 14:249–253. doi: 10.1016/j.tim.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki R, Katayama T, Kim BJ, Wakagi T, Shoun H, Ashida H, Yamamoto K, Fushinobu S. 2010. Crystal structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism. J Biol Chem 285:34279–34287. doi: 10.1074/jbc.M110.156281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrareanu G, Balasu MC, Vacaru AM, Munteanu CV, Ionescu AE, Matei I, Szedlacsek SE. 2014. Phosphoketolases from Lactococcus lactis, Leuconostoc mesenteroides, and Pseudomonas aeruginosa: dissimilar sequences, similar substrates but distinct enzymatic characteristics. Appl Microbiol Biotechnol 98:7855–7867. doi: 10.1007/s00253-014-5723-6. [DOI] [PubMed] [Google Scholar]
- 6.Glenn K, Ingram-Smith C, Smith KS. 2014. Biochemical and kinetic characterization of xylulose 5-phosphate/fructose 6-phosphate phosphoketolase 2 (Xfp2) from Cryptococcus neoformans. Eukaryot Cell 13:657–663. doi: 10.1128/EC.00055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Lipmann F, Tuttle LC. 1945. A specific micromethod for determination of acyl phosphates. J Biol Chem 159:21–28. [Google Scholar]
- 9.Hill AV. 1910. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol 40:iv–vii. [Google Scholar]
- 10.Motulsky H. 2004. Fitting Models to biological data using linear and nonlinear regression: a practical guide to curve fitting. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 11.Takahashi K, Tagami U, Shimba N, Kashiwagi T, Ishikawa K, Suzuki E. 2010. Crystal structure of Bifidobacterium longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium. FEBS Lett 584:3855–3861. doi: 10.1016/j.febslet.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 12.Traut T. 2008. Allosteric regulatory enzymes. Springer, New York, NY. [Google Scholar]
- 13.Copeland RA. 2000. Enzymes: a practical introduction to structure, mechanism, and data analysis, 2nd ed. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 14.Urness JM, Clapp KM, Timmons JC, Bai X, Chandrasoma N, Buszek KR, Fenton AW. 2013. Distinguishing the chemical moiety of phosphoenolpyruvate that contributes to allostery in muscle pyruvate kinase. Biochemistry 52:1–3. doi: 10.1021/bi301628k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puri P, Goel A, Bochynska A, Poolman B. 2014. Regulation of acetate kinase isozymes and its importance for mixed-acid fermentation in Lactococcus lactis. J Bacteriol 196:1386–1393. doi: 10.1128/JB.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. 2013. Acetyl phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Rude TH, Toffaletti DL, Cox GM, Perfect JR. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun 70:5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2:e00103–11. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 22.Hasan N, Nassif N, Durr IF. 1972. The metabolism of glyoxylate by a cell-free extract of Lactobacillus plantarum. Int J Biochem 3:607–612. doi: 10.1016/0020-711X(72)90019-5. [DOI] [Google Scholar]
- 23.Fauvart M, Braeken K, Daniels R, Vos K, Ndayizeye M, Noben JP, Robben J, Vanderleyden J, Michiels J. 2007. Identification of a novel glyoxylate reductase supports phylogeny-based enzymatic substrate specificity prediction. Biochim Biophys Acta 1774:1092–1098. doi: 10.1016/j.bbapap.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Tolbert NE, Yamazaki RK, Oeser A. 1970. Localization and properties of hydroxypyruvate and glyoxylate reductases in spinach leaf particles. J Biol Chem 245:5129–5136. [PubMed] [Google Scholar]